Molecular and morphological analysis confirms two additional species of python (Antaresia) on Cape York Peninsula, Queensland
D. J. D. Natusch A B * , L. McIntyre A , B. J. Muller A and D. Esquerré C
A B * , L. McIntyre A , B. J. Muller A and D. Esquerré C
A
B
C
Abstract
Antaresia is a genus of small (<1.4 m) python species inhabiting mainland Australia, with one species (A. papuensis) recently described from New Guinea and several islands in the Torres Strait. Currently, only Antaresia maculosa peninsularis is formally known to occur in Cape York Peninsula in northern Queensland on the Australian mainland. We used molecular and morphological data to examine whether populations of Antaresia in northern Cape York comprised several distinct taxa (namely: A. childreni, A. maculosa peninsularis, and A. papuensis). A phylogenetic analysis of the mitochondrial cytochrome b locus recovered samples of Antaresia from far northern Cape York as part of the same clade as A. papuensis from New Guinea and islands in the Torres Strait. Further to the south, we found that A. m. peninsularis and A. childreni occur in sympatry together. With the possible exception of colouration and pattern, we found no aspects of scalation that allow for easy and consistent visual distinction between the three taxa on Cape York. We discuss our finding in the context of biogeography of the region.
Keywords: Antaresia childreni, Antaresia maculosa, Antaresia papuensis, Cape York, New Guinea, phylogenetics, snake, Torres Strait.
Introduction
Pythons within the genus Antaresia are among the world’s smallest species of python (<1.4 m snout–vent length: Cogger 2014). Until recently, the group was thought to be endemic to Australia before being discovered in neighbouring New Guinea (both in Indonesia and Papua New Guinea: O’Shea et al. 2004; Natusch and Lyons 2011). A recent integrative study using genomic and morphological data revealed that what was thought to be two species (Antaresia childreni and A. stimsoni) is actually one widespread species across much of the Australian continent – A. childreni (Esquerré et al. 2021). On the east coast of Australia, the spotted python (A. maculosa) was found to comprise two divergent lineages, which were classified as two subspecies: A. maculosa maculosa, occurring along the eastern seaboard from south of Brisbane to approximately Cairns in the north, and A. maculosa peninsularis, occurring from Cairns in the south to Cape York in the north (Esquerré et al. 2021). Finally, based on molecular data, specimens of Antaresia from New Guinea and the Torres Strait were found to be consistently divergent from the Australian mainland populations and were thus described as a new species: A. papuensis (Esquerré et al. 2021).
Despite the comprehensive analysis by Esquerré et al. (2021), the status of Antaresia near the northern tip of Cape York Peninsula remained unresolved. Visual examination of specimens from northern Cape York suggested considerable variability in colour and pattern among different specimens, with morphs sometimes resembling A. childreni, A. maculosa peninsularis, or A. papuensis (D. Natusch pers. obs., 2001–2025). This variability begs the question as to whether all three taxa might occur in northern Cape York. Such a finding would significantly increase the known range of A. childreni into northern Queensland and be the first record of A. papuensis from the Australian mainland. In the present study, we used molecular and morphological data to clarify that question.
Methodology
Specimen sampling and location
Tissue samples or purified DNA used for our molecular genetic analyses were obtained from the Australian Biological Tissue Collection (ABTC) at the South Australian Museum or from specimens serendipitously located at night while undertaking other fieldwork in northern Cape York. For our molecular analysis, we sequenced 13 samples from Cape York, one from Torres Strait and one from New Guinea (Supplementary Table S1). For our morphological analyses we examined a total of 58 Cape York, Torres Strait, and New Guinea specimens from two sources. First, we examined pythons in the following museum collections: Australian Museum, Sydney (AMS); Australian National Wildlife Collection, Canberra (ANWC); Museum Victoria, Melbourne (MV); Queensland Museum, Brisbane (QM); South Australian Museum (SAMA), Western Australian Museum (WAM); and the Papua New Guinea National Museum (PNGM). Secondly, we examined pythons captured in Cape York as part of other studies on snakes in the region (e.g. Natusch et al. 2018).
Molecular analysis
DNA was extracted from the tissue samples using a salt extraction protocol (Miller et al. 1988). To amplify and sequence the mitochondrial locus cytochrome b, we designed the new primers PythonCytbF (5′-AACTACAAAAACATGCCCCA-3′) and PythonCytbR (5′-TTGGTTTACAAGAACAATGCTTT-3′) using Geneious Prime 2022 (Biomatters, Auckland, New Zealand, 2021). For the PCR we used MyTaq polymerase and buffer mastermix DNA (Bioline, UK), following the manufacturer’s protocol. We ran a denaturation cycle at 94°C for 5 min, then cycled 35 times at 94°C for 30 s, 52°C for 30 s, 72°C for 120 s, and then a final extension step at 72°C for 5 min. Sequencing was performed through Azenta Life Sciences. We checked and edited the sequences using Geneious Prime 2022 and aligned using MAFFT v.7 (Katoh and Standley 2013).
To infer the phylogenetic relationships of the samples from the tip of Cape York with other Antaresia we aligned the new sequences with those provided by Esquerré et al. (2021) and inferred a phylogenetic tree using a Maximum Likelihood approach with the program IQTree v. 2.0 (Minh et al. 2020). We used Morelia bredli as a representative of the most closely related genus to Antaresia as an outgroup to root the tree (Esquerré et al. 2020, 2022) We partitioned the dataset by codon position and used Model Finder (Kalyaanamoorthy et al. 2017) to estimate the best partition and nucleotide substitution model, and computed branch support with 1000 ultrafast bootstrap replicates (Hoang et al. 2018). The best model was a fully codon-partitioned model with substitution models HKY + F + I + G4 for first codon, HKY + F + I for second codon and TIM2 + F + G4 for third codon.
Morphometric analysis
To test whether the Antaresia taxa in our study region can be distinguished morphologically, we divided specimens into four groups based on geographic location, colouration, pattern, and the early results of our molecular analyses: (1) A. maculosa peninsularis from Cairns north to the Heathlands Ranger Base (approximately 11.8°S); (2) A. papuensis from Indonesia, Papua New Guinea, and the Torres Strait; (3) unassigned specimens from northern Cape York resembling A. childreni; and (4) unassigned specimens from northern Cape York Peninsula resembling A. papuensis (Table 1). We examined the following meristic characters considered taxonomically important in previous studies of Antaresia (Esquerré et al. 2021): number of ventral scales; dorsal midbody rows; subcaudal scales; prefrontal scales; postocular scales; loreal scales; supralabial and infralabial scales; and the number of thermoreceptive pits in the infralabial scales. Finally, we qualitatively examined the external pattern of the different specimens of Antaresia in line with Esquerré et al. (2021). To test for significant differences in scalation among specimens from our assigned populations, we used analysis of variance (ANOVA) with population as the factor and scale measures as the dependent variable where measures conformed to a normal distribution. For those measures that did not approximate a normal distribution and for which transformation was not possible, we used non-parametric Kruskal–Wallis tests. All analyses were conducted in JMP 16 Pro (SAS Institute, Cary, NC).
| Antaresia maculosa peninsularis | Antaresia papuensis from NG and Torres Straight | Antaresia papuensis from mainland Australia | Antaresia childreni from northern Cape York | ||
|---|---|---|---|---|---|
| N | 37 | 5 | 11 | 3 | |
| Ventral scales | 272.6 ± 1.9 | 270.6 ± 5.0 | 276.9 ± 3.2 | 253 ± 0.8 | |
| 250–296 | 253–284 | 266– 295 | 252–254 | ||
| Subcaudal scales | 44.1 ± 0.4 | 42.8 ± 1.5 | 44.0 ± 0.8 | 45 ± 1.1 | |
| 40–48 | 40–48 | 42– 48 | 43–47 | ||
| Midbody scale rows | 40.5 ± 0.3 | N/A | 43.1 ± 0.9 | 39.6 ± 0.4 | |
| 38–43 | N/A | 40–48 | 39–40 | ||
| Prefrontal scales | 4 ± 0 | 3 ± 0.4 | 4 ± 0 | 4 ± 0 | |
| 4–4 | 2–4 | 4–4 | 4–4 | ||
| Postocular scales | 3.1 ± 0.05 | 2.8 ± 4.2 | 2.9 ± 0.2 | 2.7 ± 0.3 | |
| 3–4 | 2–3 | 2–4 | 2–3 | ||
| Loreal scales | 3.8 ± 0.1 | 4 ± 0.4 | 3.3 ± 0.2 | 3 ± 0 | |
| 3–7 | 3–5 | 3–4 | 3–3 | ||
| Infralabial scales | 13.3 ± 0.1 | 12 ± 0.6 | 14.1 ± 0.2 | 13.3 ± 0.3 | |
| 12–15 | 10–14 | 13–15 | 13–14 | ||
| Supralabial scales | 10.6 ± 0.1 | 10.6 ± 0.4 | 10.3 ± 0.2 | 11 ± 0 | |
| 10–12 | 10–12 | 10–11 | 11–11 | ||
| Heat pits in infralabials | 4 ± 0 | 4 ± 0 | 4 ± 0 | 4 ± 0 | |
| 4–4 | 4–4 | 4–4 | 4–4 |
The values are mean ± s.e. and ranges.
NG, New Guinea; SVL, snout-vent length.
Results
Molecular analysis
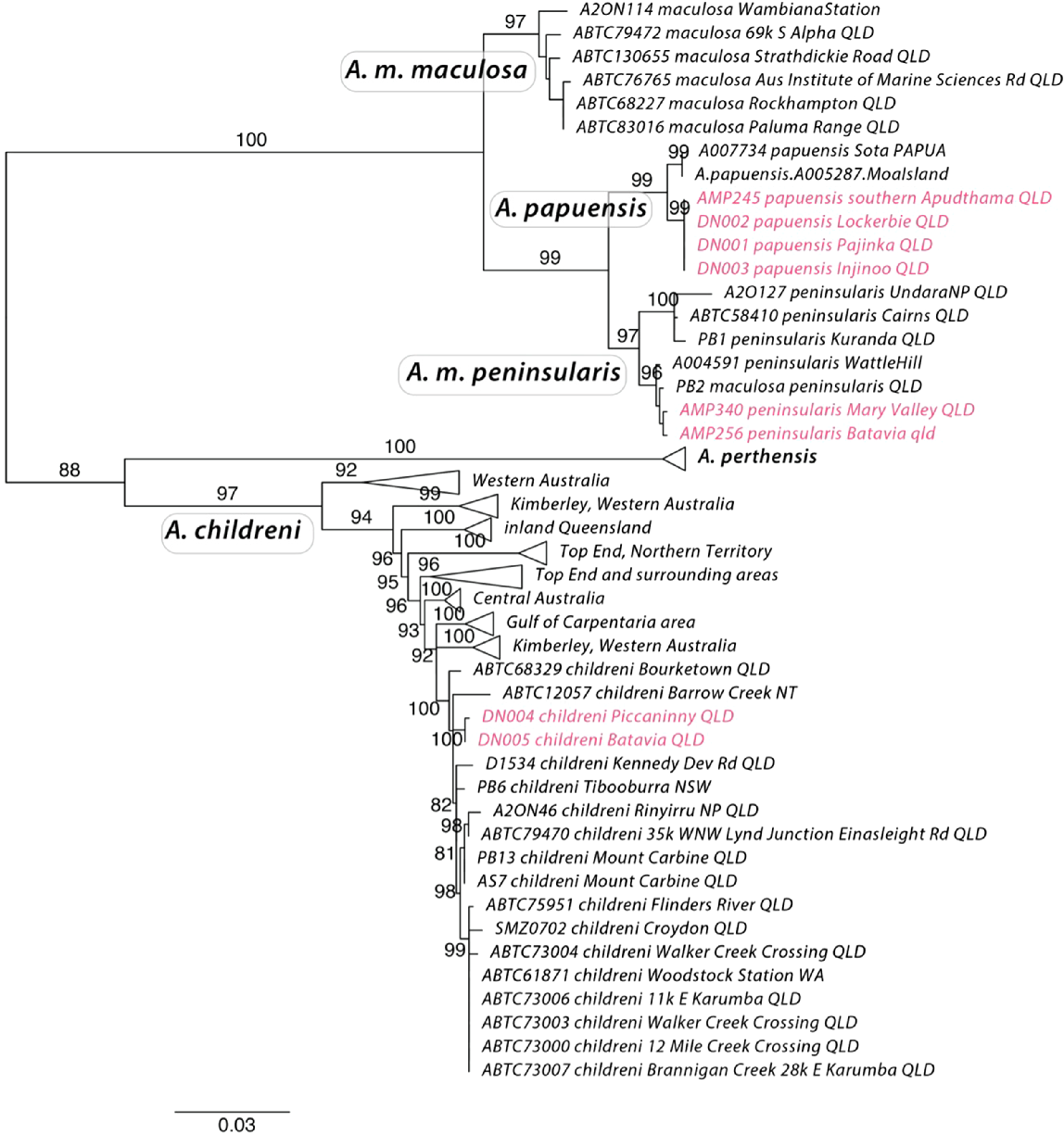
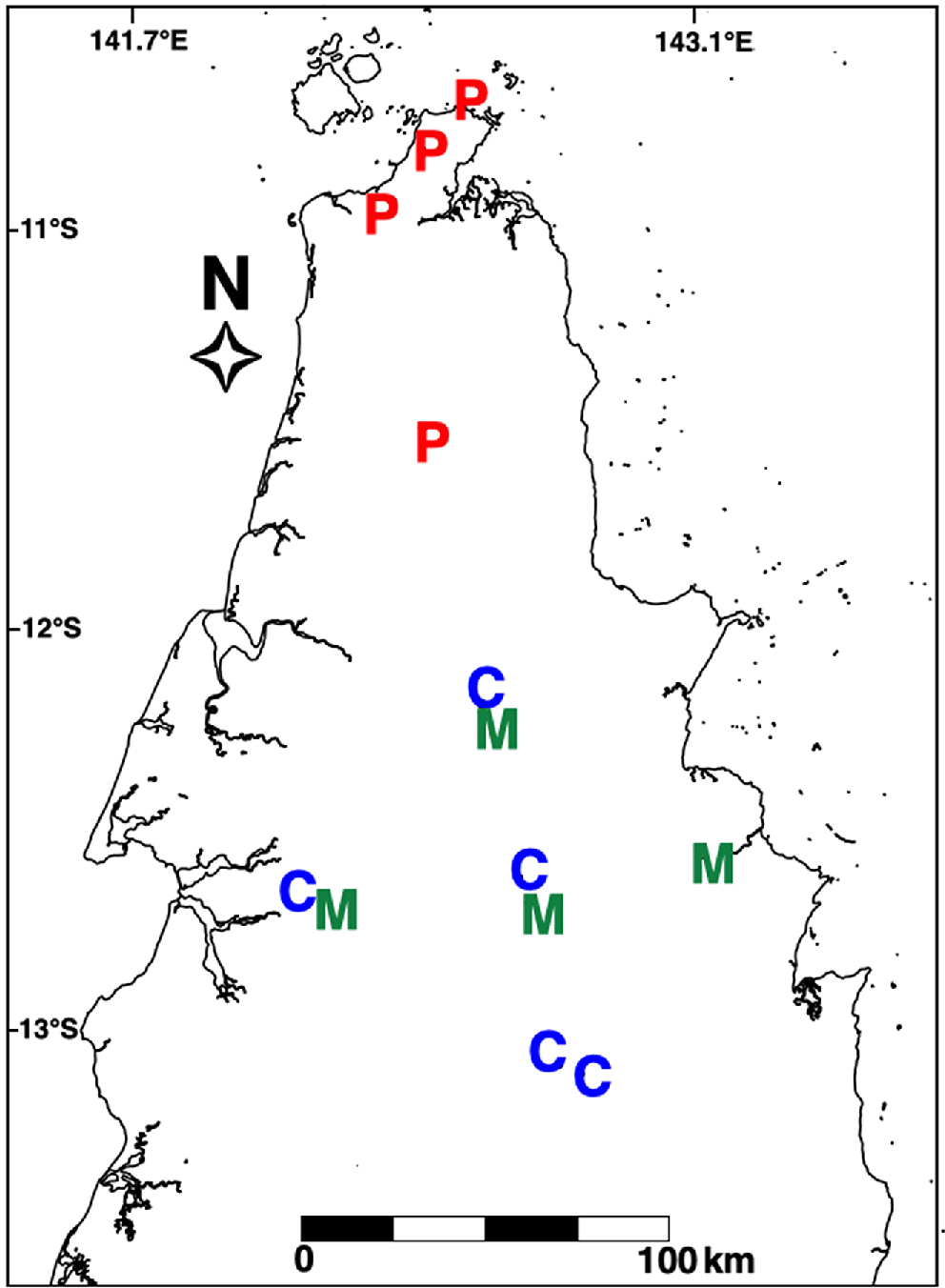
The phylogenetic tree estimated using cytochrome b recovers well supported clades that include what is currently described as Antaresia papuensis, A. maculosa maculosa, A. m. peninsularis and A. childreni, all with high branch support (bootstraps > 97%). Interestingly, it infers A. m. peninsularis as more closely related to A. papuensis than to A. m. maculosa. The samples from the extreme northern tip of Cape York are inferred to be most closely related to A. papuensis, rather than to A. maculosa pensinsularis or A. childreni (Figs 1 and 2). The phylogeny also confirms the presence of A. childreni on Cape York (Fig. 2). The southernmost confirmed record of A. papuensis on Cape York was located at 11.4973°S, 142.4316°E (Fig. 2). The presence of A. childreni was confirmed as far north as 12°S and occurring in sympatry with A. maculosa peninsularis (Fig. 2).
Morphology
We performed a morphological analysis to test whether there are consistent phenotypic differences between the main clades inferred by our molecular analysis. Our analysis of Cape York Antaresia meristic characters revealed that the Cape York Antaresia childreni had significantly fewer ventral scales than either A. maculosa peninsularis or A. papuensis (ANOVA: F3,47 = 2.9, P = 0.046) (Table 1). Antaresia papuensis from the tip of Cape York possessed a greater number of midbody rows than specimens of A. maculosa peninsularis or A. childreni (ANOVA: F2,26 = 6.87, P = 0.004) (Table 1). Finally, Antaresia papuensis from New Guinea and the Torres Strait had significantly fewer prefrontal scales than A. childreni, A. m. peninsularis, or conspecifics from mainland Australia (Kruskal–Wallis test: χ2 = 19.9, d.f. = 3, P = 0.0002) (Table 1). However, it should be noted that the sample sizes for these different groups were small, and that with the exception of prefrontal scales there was overlap in the ranges of these measures – suggesting they may not be useful for diagnostic purposes (Table 1). There was no significant difference among any of the other measures taken of specimens from the different groupings (all P > 0.05). Based on qualitative assessment, the patterning of A. papuensis from New Guinea, the Torres Strait, and the tip of Cape York consisted of background colouration ranging from light grey to dark brown with small and irregular, ragged-edged, dark brown blotches along the dorsal surface (Fig. 3a). By contrast, A. m. peninsularis exhibits a background colouration ranging from greyish brown to a light sandy (or ‘blonde’) colour. The spots or blotches on the dorsal surface are dark, ranging from chocolate brown to a reddish dark brown. The shape and distribution of the spots is variable, but overall are larger and less ragged than in A. papuensis (Fig. 3b). Antaresia childreni on Cape York possessed the most unique patterning, consisting of dark tan markings that form into distinct bands along the dorsal surface, rather than blotches (Fig. 3c).
Antaresia species from northern Cape York Peninsula. The specimen in (a) exhibits typical patterning of Antaresia papuensis from Cape York. Specimens in (b) (Antaresia maculosa peninsularis) and (c) (A. childreni) were located 3.1 km from one another and display typical colouration and pattern of these taxa in northern Cape York.

Discussion
Our molecular genetic analysis supports the presence of three Antaresia species in northern Cape York. Essentially, northern Cape York is a contact point for western (A. childreni), eastern (A. m. peninsularis), and northern (A. papuensis) lineages of Antaresia, further exemplifying its significance as an area of faunal interchange (Nix and Kalma 1972; Kikkawa et al. 1981). Our findings also confirm Cape York as the Australian hotspot for python diversity, with eight species now recorded from the region (Antaresia childreni, A. maculosa peninsularis, A. papuensis, Aspidites melanocephalus, Liasis fuscus, Morelia spilota, M. viridis, and Simalia amethistina).
Our analysis based on cytochrome b indicates relatively shallow divergence between A. papuensis and A. m. peninsularis, and provides evidence for these taxa being more closely related with each other than to A. m. maculosa. The much more thorough analysis by Esquerré et al. (2021), using thousands of genome-wide single nucleotide polymorphisms, infers A. m. maculosa and A. m. peninsularis as sister taxa and A. papuensis as the sister lineage to that clade. The mitochondrial samples of A. papuensis from the tip of Cape York form a well-supported clade with specimens of known A. papuensis for which the nuclear analyses were conducted. Moreover, population structure analysis, isolation by distance analysis, and species delimitation analysis all indicate that A. papuensis is on an independent evolutionary trajectory from A. m. peninsularis (Esquerré et al. 2021). Nevertheless, future studies should focus on gathering a denser and more comprehensive sampling of the Cape York, Torres Strait, and New Guinean populations. Genomic data will enhance our understanding of the evolutionary history and boundaries between the lineages present there, as inferences from single loci are more prone to be biased by incomplete lineage sorting and introgression (Maddison 1997).
The morphological divergence we report among the Antaresia in our study is minor. We thus consider that, at this time, there are few scale characters that can be used to morphologically distinguish A. papuensis, A. maculosa peninsularis, and A. childreni on Cape York. All species grow to similar body sizes; and although some specimens of A. papuensis possessed only two postocular scales, this was the exception rather than the rule. Moreover, the only specimens of A. papuensis to possess fewer than four prefrontal scales were two specimens from Moa Island in the Torres Strait (in which prefrontals were fused). It is possible that this may relate to lower levels of genetic diversity within this small insular population, rather than a diagnostic trait for this taxon, which is known to occur in other snake species (Brown et al. 2017). All specimens of A. papuensis from mainland Australia examined during this study possessed four prefrontal scales. Greater sample sizes of A. childreni from Cape York would be useful for further testing morphological divergence between the taxa. In the meantime, however, the three species of Antaresia can be easily distinguished by their colouration and pattern (Fig. 3). All of the specimens assigned by us to different lineages a priori, based on visual examination, were confirmed upon subsequent molecular analyses – suggesting that colouration and pattern are the most useful diagnostic characters for these taxa.
It is difficult to determine whether geographic or reproductive barriers separate the three taxa at the tip of Cape York. Specimens of A. childreni and A. m. peninsularis were located 3.1 km apart and likely occur in sympatry. Similarly, the southernmost record of A. papuensis was located approximately 55 km north of the other taxa, with no obvious barriers to gene flow. Although greater sampling in potential contact zones is needed, it appears that all specimens north of 11.4973°S, 142.4316°E (Fig. 2) are A. papuensis. Antaresia m. peninsularis appears to occur on both the east and west coasts of northern Cape York, whereas specimens of A. childreni are yet to be confirmed from the far eastern coast of the Peninsula (Fig. 2). Further south in Cape York, specimens of A. childreni have been recorded as far east as Rinyurru (Lakefield) National Park and Mt Carbine, suggesting that the Great Dividing Range may have played a role in the divergence of A. childreni and A. maculosa in that area of Australia. Regardless of the historical mechanisms restricting gene flow in Antaresia in Cape York, the lack of contemporary geographic barriers helps strengthen earlier conclusions by Esquerré et al. (2021) that suggest reproductive isolation of these three taxa.
Data availability
Genetic data have been uploaded to Genbank. Morphological data are available from the authors upon request.
Declaration of funding
This work was undertaken with support from People For Wildlife. Molecular work was funded by the University of Wollongong.
Acknowledgements
We thank the Traditional Owners of Cape York and the Apudthama Land Trust for allowing us to collect data from snakes on their country. We thank Sebastian Hoeffer for donating some additional samples for the molecular analysis. This research was carried out under Queensland Department of Environment and Science permits (WISP12944313 and P-SPP-100705267) and University of Sydney and Queensland Department of Primary Industries animal ethics committee guidelines (approval numbers: L04/3-2013/3/5969 and CA 2024/07/1871). The research was funded and facilitated by People For Wildlife and the University of Wollongong. Finally, we thank two anonymous reviewers for comments that significantly improved the quality of this manuscript.
References
Brown GP, Madsen T, Dubey S, Shine R (2017) The causes and ecological correlates of head scale asymmetry and fragmentation in a tropical snake. Scientific Reports 7, 11363.
| Crossref | Google Scholar |
Esquerré D, Donnellan SC, Brennan IG, Lemmon AR, Moriarty Lemmon E, Zaher H, Grazziotin FG, Keogh JS (2020) Phylogenomics, biogeography, and morphometrics reveal rapid phenotypic evolution in pythons after crossing Wallace’s line. Systematic Biology 69, 1039-1051.
| Crossref | Google Scholar | PubMed |
Esquerré D, Donnellan S, Pavón-Vázquez CJ, Fenker J, Keogh JS (2021) Phylogeography, historical demography and systematics of the world’s smallest pythons (Pythonidae, Antaresia). Molecular Phylogenetics and Evolution 161, 107181.
| Crossref | Google Scholar |
Esquerré D, Brennan IG, Donnellan S, Keogh JS (2022) Evolutionary models demonstrate rapid and adaptive diversification of Australo-Papuan pythons. Biology Letters 18, 20220360.
| Crossref | Google Scholar |
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35, 518-522.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Maddison WP (1997) Gene trees in species trees. Systematic Biology 46, 523-536.
| Crossref | Google Scholar |
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 16, 1215.
| Crossref | Google Scholar |
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear F (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530-1534.
| Crossref | Google Scholar | PubMed |
Natusch DJD, Lyons JA (2011) The harvest of Antaresia maculosa (Pythonidae) from West Papua, Indonesia. Herpetological Review 42, 509-511.
| Google Scholar |
Natusch DJD, Lyons JA, Dubey S, Shine R (2018) Ticks on snakes: the ecological correlates of ectoparasite infection in free-ranging snakes in tropical Australia. Austral Ecology 43, 534-546.
| Crossref | Google Scholar |
O’Shea M, Sprackland RG, Bigilale IH (2004) First record for the genus Antaresia (Squamata: Pythonidae) from Papua New Guinea. Herpetological Review 35, 225-227.
| Google Scholar |