Investigating the Bivalve Tree of Life – an exemplar-based approach combining molecular and novel morphological characters
Rüdiger Bieler A N , Paula M. Mikkelsen B , Timothy M. Collins C , Emily A. Glover D , Vanessa L. González E , Daniel L. Graf G , Elizabeth M. Harper H , John Healy A I , Gisele Y. Kawauchi E , Prashant P. Sharma F , Sid Staubach A , Ellen E. Strong J , John D. Taylor D , Ilya Tëmkin J K , John D. Zardus L , Stephanie Clark A , Alejandra Guzmán E M , Erin McIntyre E , Paul Sharp C and Gonzalo Giribet EA Invertebrates, Field Museum of Natural History, 1400 South Lake Shore Drive, Chicago, IL 60605, USA.
B Paleontological Research Institution, 1259 Trumansburg Road, and Department of Ecology & Evolutionary Biology, Cornell University, Ithaca, NY 14850, USA.
C Department of Biological Sciences, AHC 1 Bldg, Rm 319C, Florida International University, Miami, FL 33199, USA.
D Department of Life Sciences, The Natural History Museum, London SW7 5BD, UK.
E Museum of Comparative Zoology & Department of Organismic and Evolutionary Biology, Harvard University, 26 Oxford Street, Cambridge, MA 02138, USA.
F American Museum of Natural History, Division of Invertebrate Zoology, 200 Central Park West, New York City, NY 10024, USA.
G University of Wisconsin-Stevens Point, Biology Department, 800 Reserve Street, Stevens Point, WI 54481, USA.
H Department of Earth Sciences, University of Cambridge, Downing Street, Cambridge, CB2 3EQ, UK.
I Queensland Museum, PO Box 3300, South Brisbane, Qld 4101, Australia.
J Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, PO Box 37012, MRC 163, Washington, DC 20013, USA.
K Biology Department, Northern Virginia Community College, 4001 Wakefield Chapel Road, Annandale, VA 22003, USA.
L Department of Biology, The Citadel, 171 Moultrie Street, Charleston, SC 29409, USA.
M Stanford University, 300 Pasteur Drive, Stanford, CA 94305, USA.
N Corresponding author. Email: rbieler@fieldmuseum.org
Invertebrate Systematics 28(1) 32-115 https://doi.org/10.1071/IS13010
Submitted: 15 March 2013 Accepted: 17 November 2013 Published: 20 March 2014
Journal Compilation © CSIRO Publishing 2014 Open Access CC BY-NC-ND
Abstract
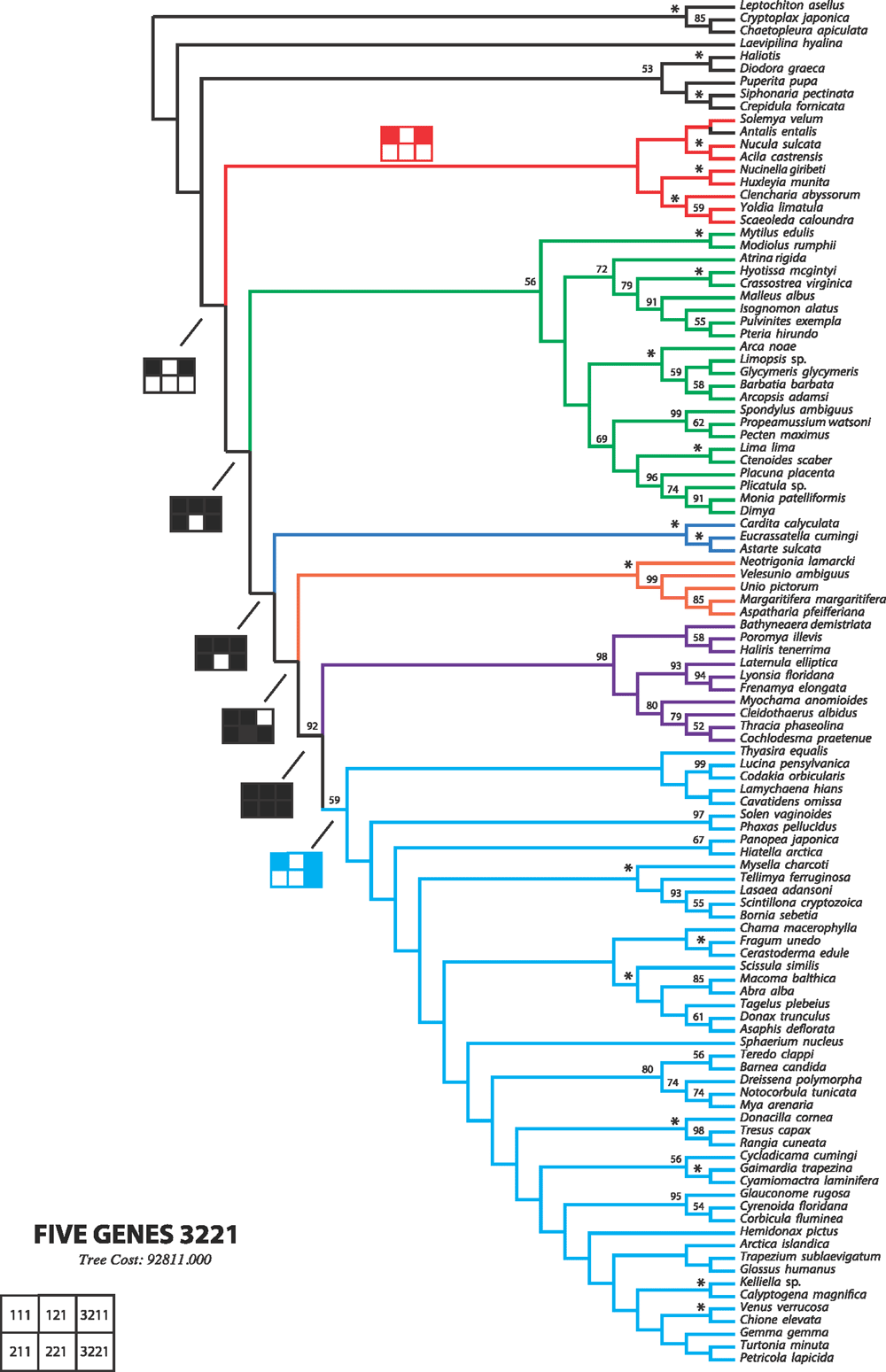
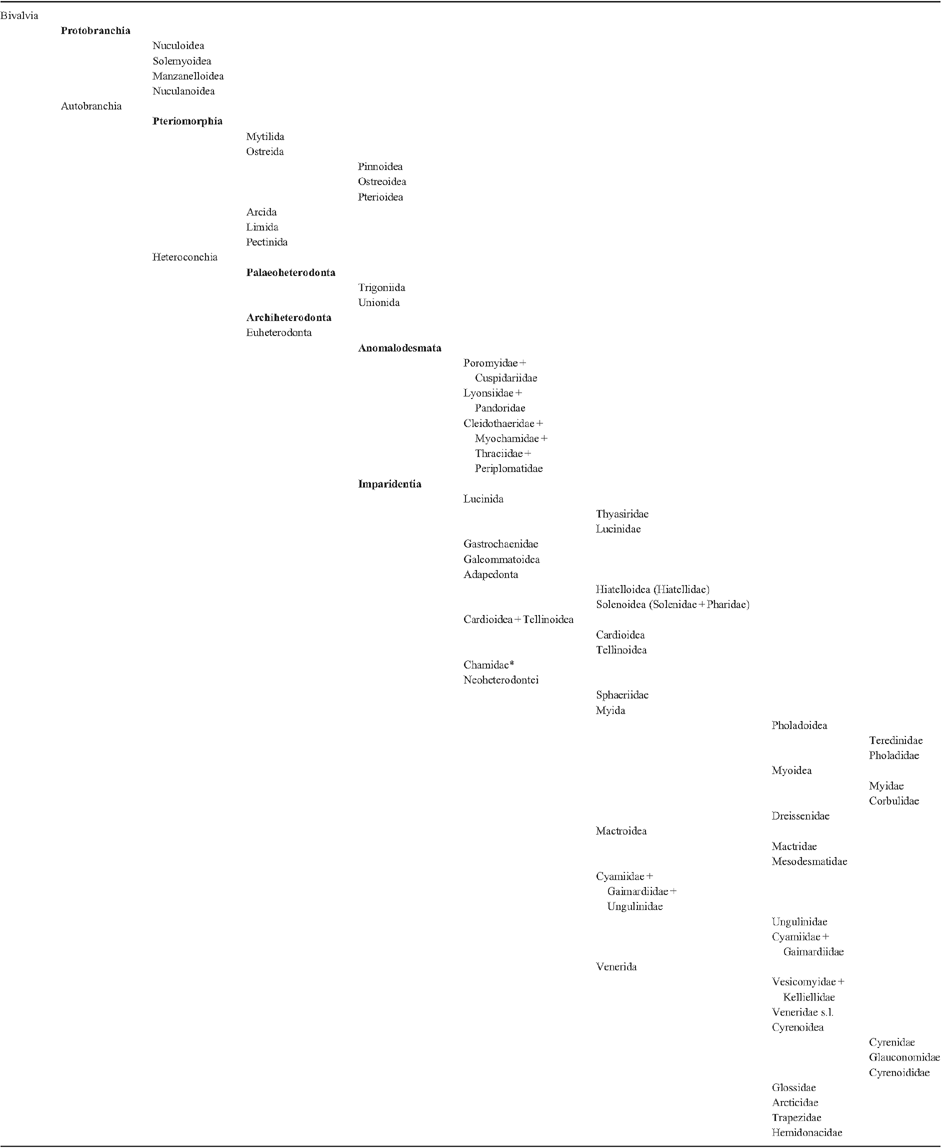
To re-evaluate the relationships of the major bivalve lineages, we amassed detailed morpho-anatomical, ultrastructural and molecular sequence data for a targeted selection of exemplar bivalves spanning the phylogenetic diversity of the class. We included molecular data for 103 bivalve species (up to five markers) and also analysed a subset of taxa with four additional nuclear protein-encoding genes. Novel as well as historically employed morphological characters were explored, and we systematically disassembled widely used descriptors such as gill and stomach ‘types’. Phylogenetic analyses, conducted using parsimony direct optimisation and probabilistic methods on static alignments (maximum likelihood and Bayesian inference) of the molecular data, both alone and in combination with morphological characters, offer a robust test of bivalve relationships. A calibrated phylogeny also provided insights into the tempo of bivalve evolution. Finally, an analysis of the informativeness of morphological characters showed that sperm ultrastructure characters are among the best morphological features to diagnose bivalve clades, followed by characters of the shell, including its microstructure. Our study found support for monophyly of most broadly recognised higher bivalve taxa, although support was not uniform for Protobranchia. However, monophyly of the bivalves with protobranchiate gills was the best-supported hypothesis with incremental morphological and/or molecular sequence data. Autobranchia, Pteriomorphia, Heteroconchia, Palaeoheterodonta, Archiheterodonta, Euheterodonta, Anomalodesmata and Imparidentia new clade ( = Euheterodonta excluding Anomalodesmata) were recovered across analyses, irrespective of data treatment or analytical framework. Another clade supported by our analyses but not formally recognised in the literature includes Palaeoheterodonta and Archiheterodonta, which emerged under multiple analytical conditions. The origin and diversification of each of these major clades is Cambrian or Ordovician, except for Archiheterodonta, which diverged from Palaeoheterodonta during the Cambrian, but diversified during the Mesozoic. Although the radiation of some lineages was shifted towards the Palaeozoic (Pteriomorphia, Anomalodesmata), or presented a gap between origin and diversification (Archiheterodonta, Unionida), Imparidentia showed steady diversification through the Palaeozoic and Mesozoic. Finally, a classification system with six major monophyletic lineages is proposed to comprise modern Bivalvia: Protobranchia, Pteriomorphia, Palaeoheterodonta, Archiheterodonta, Anomalodesmata and Imparidentia.
Additional keywords: Bivalvia, evolution, gills, labial palps, Mollusca, phylogeny, shell microstructure, sperm ultrastructure, stomach.
Introduction
Bivalves constitute a commercially and ecologically important group of molluscs related to gastropods and scaphopods (Kocot et al. 2011; Smith et al. 2011) and are the second most species-rich molluscan class after Gastropoda. Their membership in the phylum Mollusca is undisputed and bivalve monophyly, although challenged in early molecular analyses, has found support in more recent studies using large datasets. Bivalve species have become the focus of numerous lines of interdisciplinary research, including the recent publication of the draft genomes of two pteriomorphian species (Takeuchi et al. 2012; Zhang et al. 2012) and new transcriptomic resources (Clark et al. 2010; Kocot et al. 2011; Milan et al. 2011; Smith et al. 2011; Coppe et al. 2012). Translational medical studies on bivalves include aging research (Ungvari et al. 2011) and the discovery of new antibiotics produced by the bacterial gill symbionts of shipworms (Elshahawi et al. 2013). Bivalves have also been used as models for understanding diversification in the deep sea (Etter et al. 2005, 2011; Rex et al. 2005; Sharma et al. 2013), and their rich fossil record has emerged as one of the most powerful tools for explaining global ecological and biogeographical patterns (Jablonski et al. 2006; Mittelbach et al. 2007; Krug et al. 2009; Roy et al. 2009; summarised by Bieler et al. 2013). Nevertheless, composition and interrelationships of its constituent clades have remained a matter of heated debate.
Hypotheses about the higher-level internal structure of Bivalvia have, of course, changed over time, driven by corroborating and conflicting signals from character systems derived from shell morphology (especially hinge teeth and muscle scars that are also recognisable in the rich fossil record), and organisational patterns of the gills or stomachs. Numerous classifications have been proposed (summarised and compared, for example, by Newell 1965; Beesley et al. 1998; Amler et al. 2000; Schneider 2001). Among the commonly recognised subgroups are Protobranchia (‘primitive’ bivalves with plesiomorphic ctenidia with a solely respiratory function) and Autobranchia (formerly Autolamellibranchiata or Autolamellibranchia; bivalves with hypertrophied gills used for filter-feeding in addition to respiration). The latter initially excluded members with highly modified septibranch gills (e.g. Nevesskaja et al. 1971). The gill-based concept of Protobranchia was largely equivalent to Palaeotaxodonta in hinge-based classifications and the latter either included (e.g. Pojeta 1987) or excluded (Cope 1996) the Solemyidae (which was variously placed as Cryptodonta or Lipodonta). Consensus developed to break down the Autobranchia into Pteriomorphia (with or without the Mytilida – the latter also classified separately as Isofilibranchia), Palaeoheterodonta (including Trigoniida and Unionida among extant taxa), Heterodonta (a large group of bivalves including Venerida and Myida), as well as the Anomalodesmata (comprising an array of strange and highly specialised marine bivalves with mostly prismato-nacreous shells and modified eulamellibranchiate or septibranch ctenidia that include Pholadomyida and the previously mentioned Septibranchia) (e.g. Newell 1965; Amler 1999). The advent of cladistic methodology and molecular techniques allowed for reinvestigation of hypothesised taxa and their interrelationships. Among the new results was the recognition of Anomalodesmata as a clade nested within Heterodonta, rendering the latter subclass paraphyletic (e.g. Giribet and Wheeler 2002; Harper et al. 2006). Anomalodesmata constitutes the most-basal clade of Euheterodonta – an unranked clade introduced by Giribet and Distel (2003) for the heterodonts excluding Carditoidea and Crassatelloidea. Other previously unrecognised clades within Heterodonta were also supported, such as Neoheterodontei – an unranked group erected by Taylor et al. (2007b) that includes Sphaerioidea, Myida, Gaimardioidea, Mactroidea, Ungulinoidea, Cyrenoidea ( = Corbiculoidea), Chamoidea and Veneroidea. Archiheterodonta was introduced by Giribet (2008); the name was used by Taylor et al. (2007b) citing ‘Giribet (in press)’ for the members of the superfamilies Carditoidea and Crassatelloidea (the order Carditida sensu Bieler et al. 2010), which form a well supported clade, not nested within the more traditional heterodont group, both in molecular as well as morphological analyses, and are united most notably by the presence of intracellular haemoglobin and sperm ultrastructure.
Two recent family-level classifications have attempted to provide synopses of the bivalve system, arranging the more than 1000 family-group names for extant and extinct Bivalvia into a classification informed by shell-morphology, anatomy and other knowledge gathered from the published literature. The two classifications differ substantially, resulting from the fact that one (Bieler et al. 2010) avoided making decisions in unresolved cases of conflicting or missing information (resorting to alphabetical or ‘classical’ arrangement of lower ranked taxa in such cases), whereas the other (Carter et al. 2011) opted to assign all taxa into a finely dissected system of eighteen Linnaean ranks.
Previous attempts to analyse the phylogeny of the entire class employed numerical (and later cladistic) approaches based on morphological (Purchon 1978, 1987b; Salvini-Plawen and Steiner 1996; Cope 1997, 2000; Waller 1998; Carter et al. 2000; Giribet and Wheeler 2002) or molecular (Steiner and Müller 1996; Adamkewicz et al. 1997; Campbell et al. 1998; Hoeh et al. 1998; Giribet and Carranza 1999; Steiner 1999; Campbell 2000; Steiner and Hammer 2000; Giribet and Wheeler 2002; Giribet and Distel 2003; Giribet et al. 2006; Wilson et al. 2010; Plazzi and Passamonti 2010; Plazzi et al. 2011, 2013; Sharma et al. 2012) characters. Only Giribet and Wheeler (2002) analysed morphological and molecular characters simultaneously, and used a combination of shell morphological data based on specimen observations and literature-derived data for anatomy, the latter often not based on the same material (or same species-level taxon) as the specimens employed for the molecular component of the study.
In this work, we employ an exemplar approach (sensu Prendini 2001) by basing shell-morphological, gross-anatomical, ultrastructural and molecular data, whenever possible, on the specimens from the same population (usually from the same collecting event and sometimes even the same specimen) (see species habitus and/or shells in Figs 1–5). Each of these exemplar taxa was investigated for a series of morphological and anatomical character suites. These include ‘classic’ morphological features of bivalve systematics such as stomach morphology and other features of the alimentary tract (investigating and disassembling ‘stomach types’ as used by, for example, Purchon 1985, 1987a), and the morphology and interrelationship of gills and labial palps (a rich past field of study that led to a system of widely employed ‘gill types’; Stasek 1963). To this was added a detailed investigation of shell microstructure (building upon and greatly expanding earlier work by, for example, Bøggild 1930; Taylor et al. 1969, 1973; Carter 1990b), establishing a new system for homologising the different shell layers. We also present an intensive effort to obtain and analyse sperm ultrastructure data for the majority of species included here (building on earlier efforts by, for example, Dan and Wada 1955; Franzén 1955, 1983; Hodgson and Bernard 1986; Healy 1995, 1996; Healy et al. 2000, 2008a).
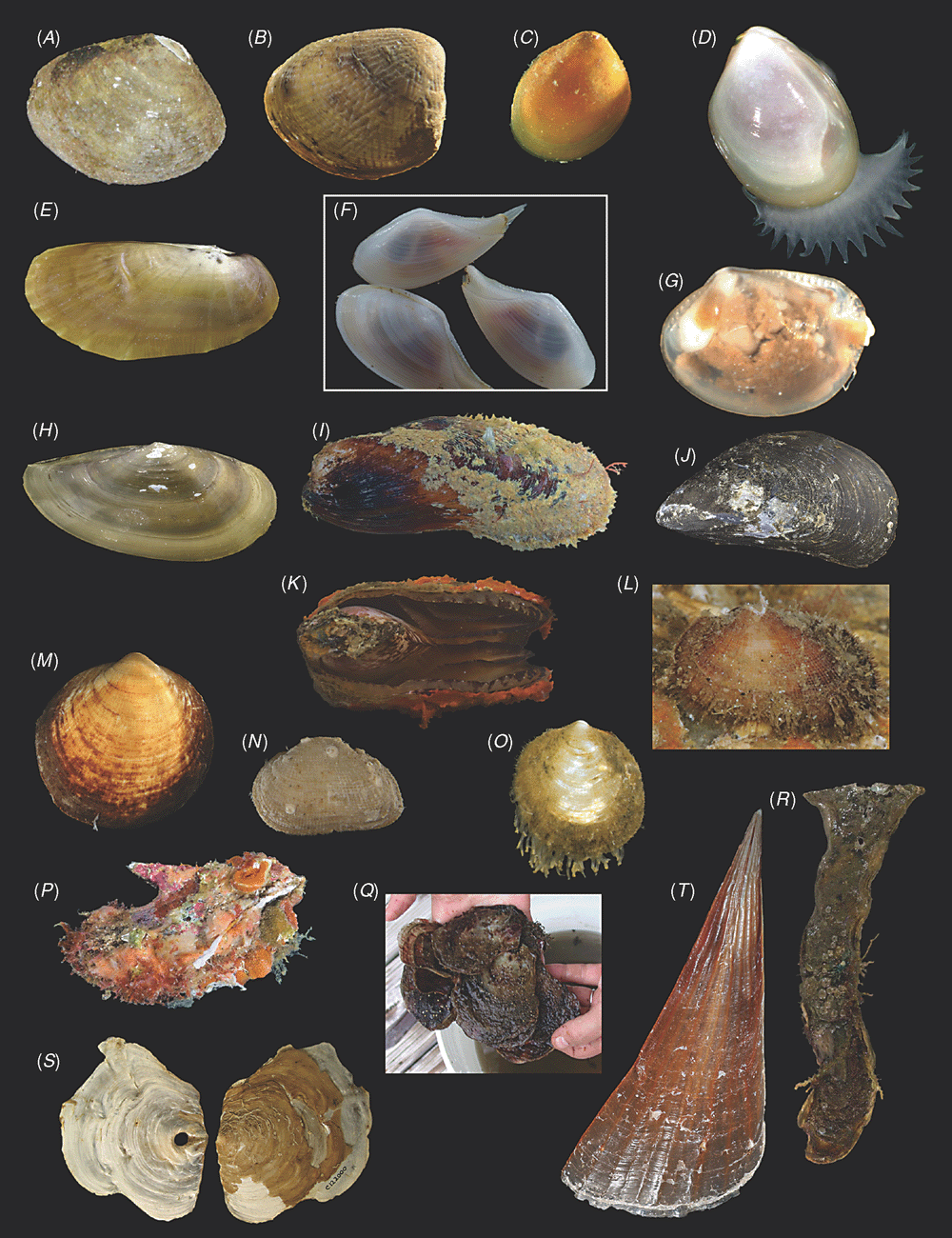
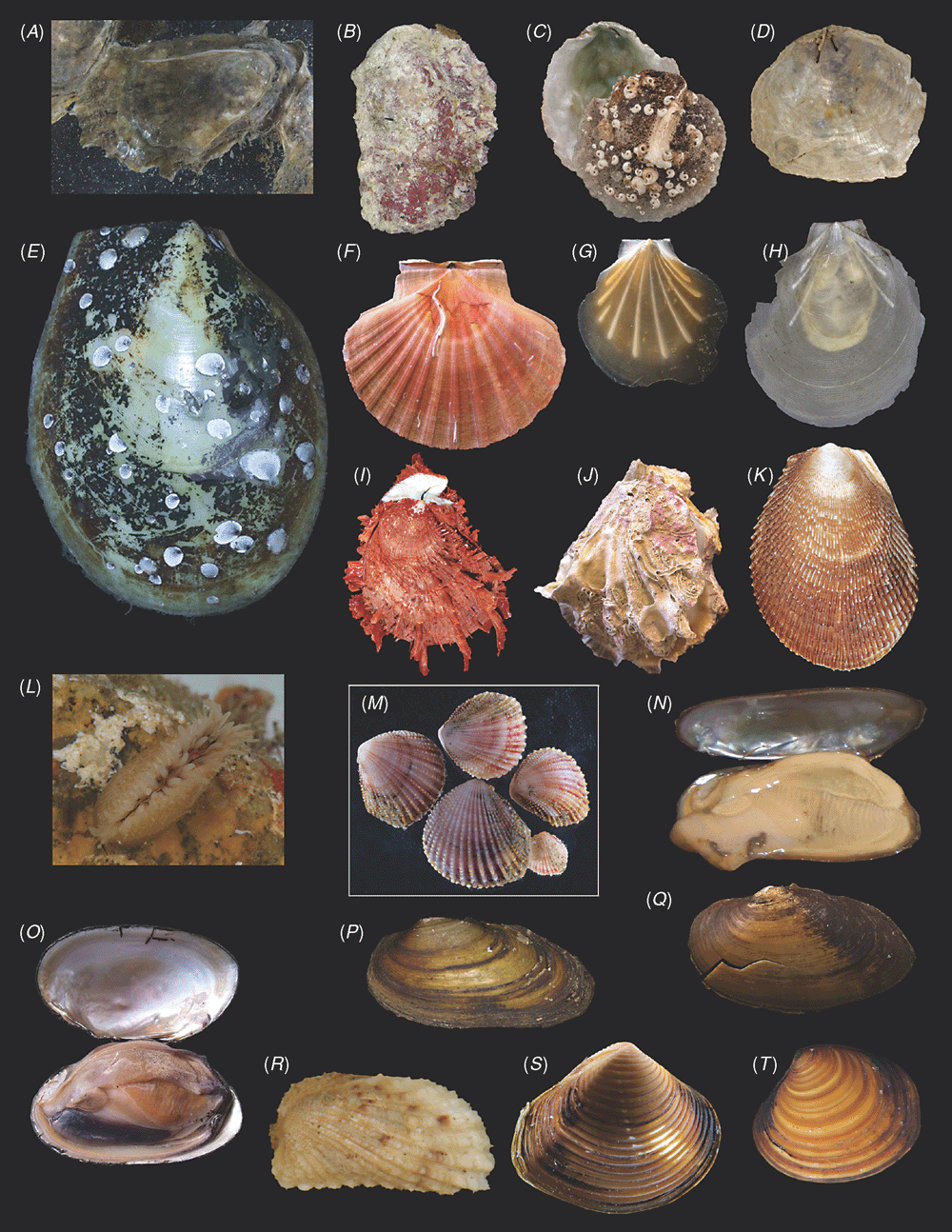
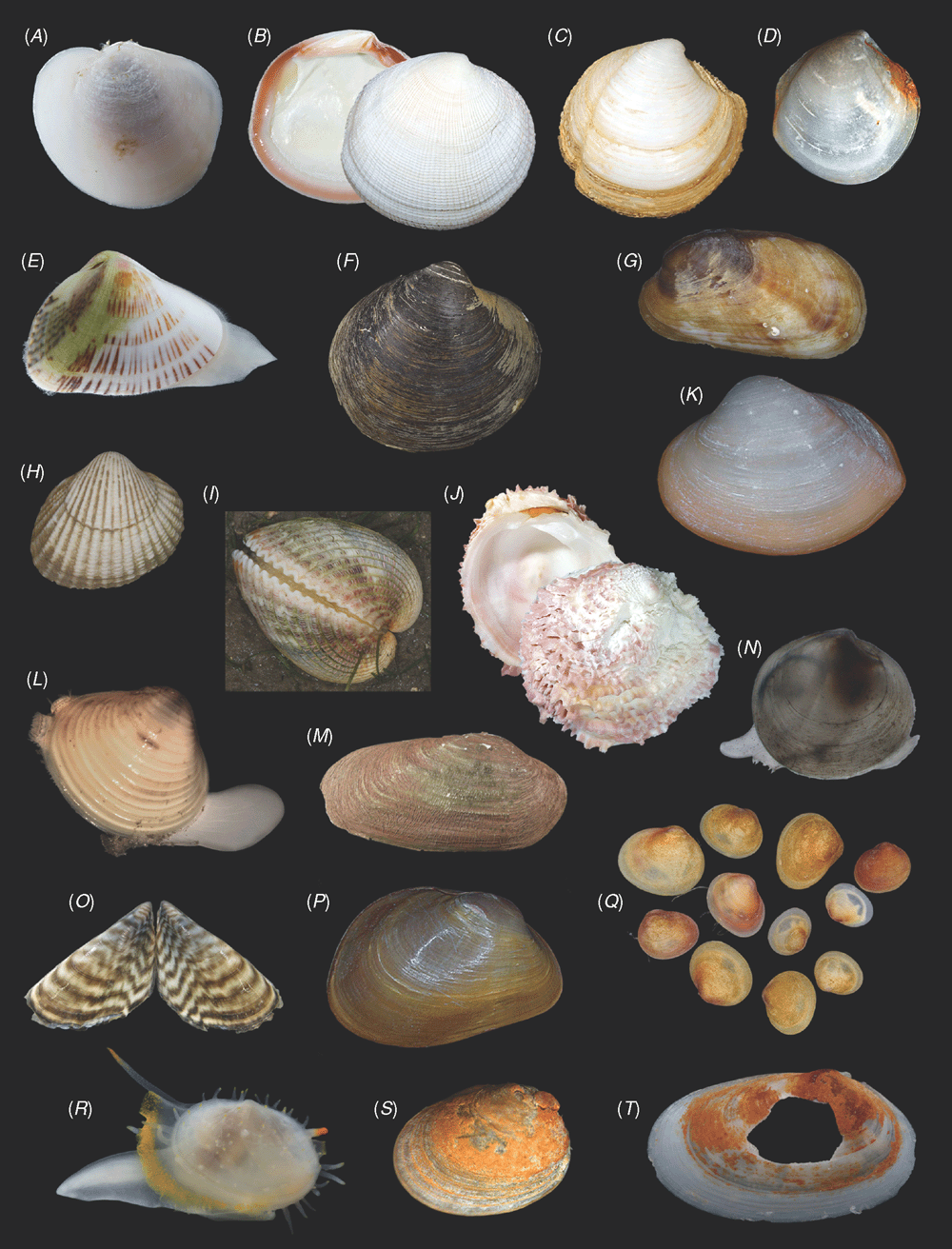
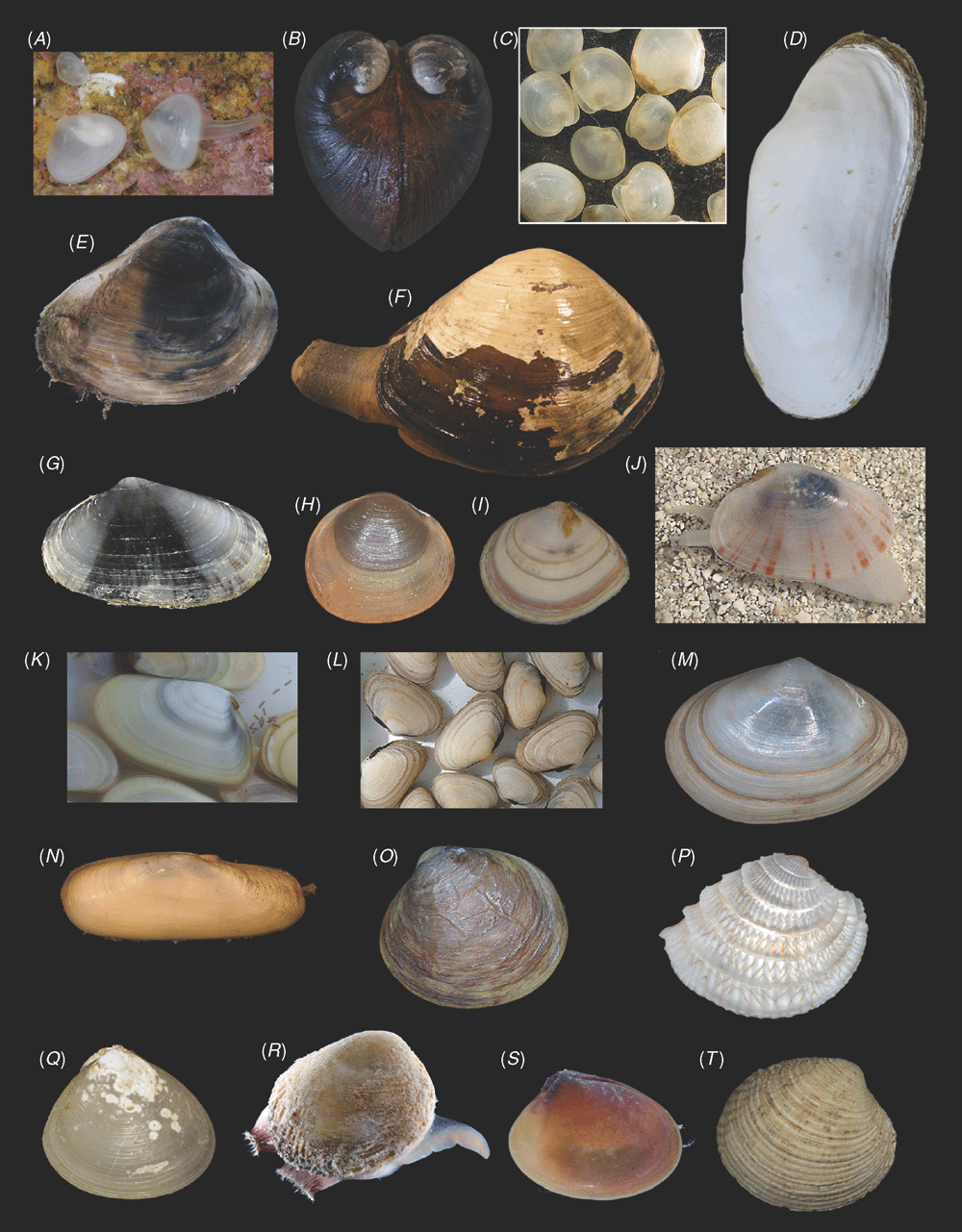
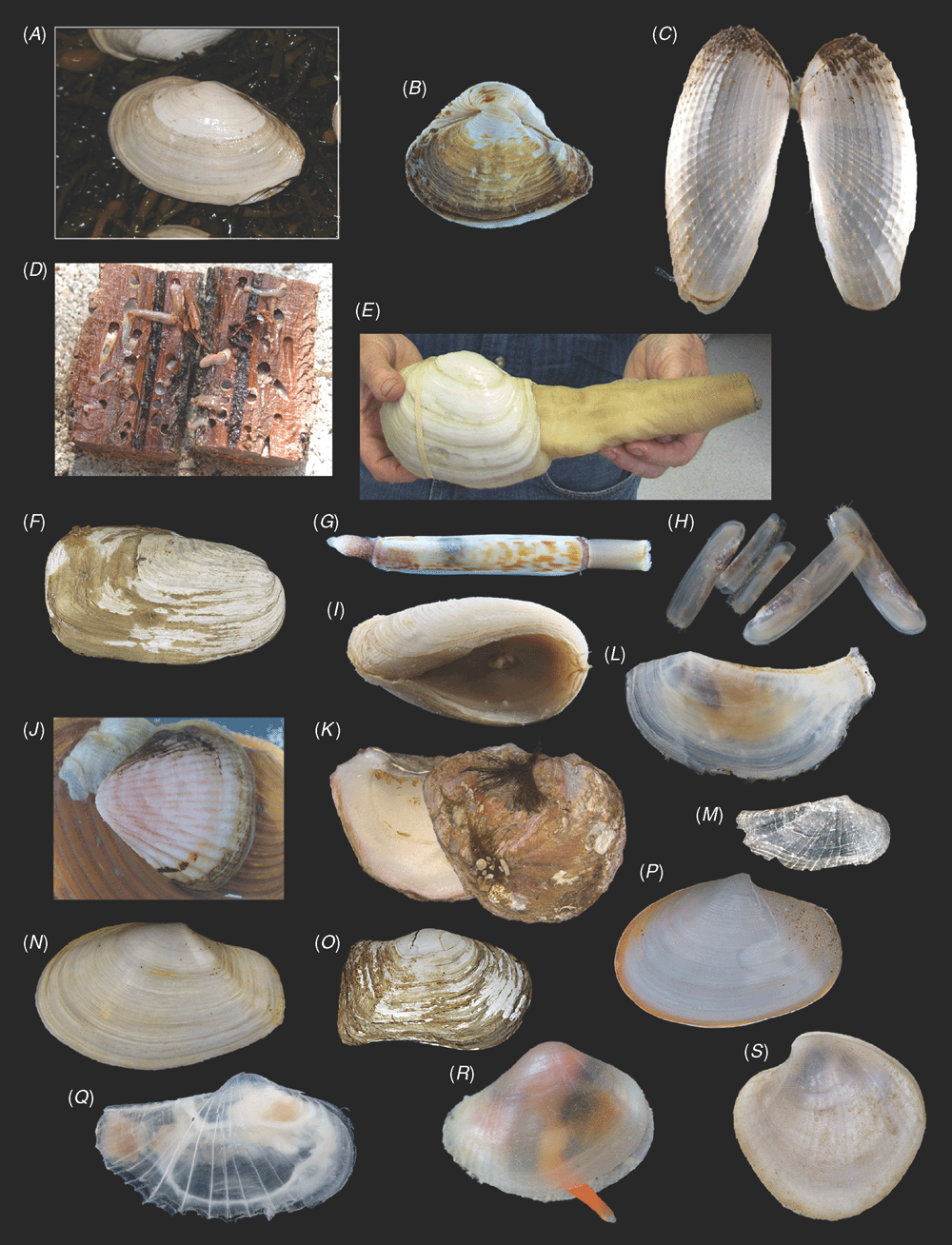
Sampling across the bivalve system, this study includes novel morphological and molecular data for 8 protobranchs (Fig. 1A–H), 27 pteriomorphians (Figs 1I–T, 2A–L), 5 paleoheterodonts (Fig. 2M–Q), 3 archiheterodonts (Fig. 2R–T) and 60 euheterodonts (including 11 anomalodesmatans) (Figs 3–5). The data were analysed using an array of modern methods for estimating the phylogeny and diversification times of the major bivalve lineages. Our results are largely consistent with many of the previous schemes of bivalve phylogenetics, but also provide new insights and strengthen support for some nodes that were unsupported in earlier molecular studies. Finally, we propose a refined classification system for bivalves entirely based on well supported phylogenetic results.
Materials and methods
Specimens
Taxonomic authorities and dates, higher classification, localities and accession numbers for all taxa used in this analysis are found in Table 1. This analysis is part of a larger study; some species in the figures were not employed in this particular analysis, but are representative of the illustrated characters and character states.

|
Ingroup choices
Exemplar species were chosen to maximise coverage of previously recognised or assumed branches of the Bivalvia, including single species of most families except for the larger and morphologically diverse groups (e.g. Lucinidae, Veneridae, Tellinidae), where multiple representatives were included. Because of the demands on the quality of tissue preservation and stage of the life cycle (e.g. for sperm ultrastructural work), focus was placed whenever possible on recollectable species and primarily obtained from a limited number of regions in Florida (USA), Spain, the UK, and Queensland (Australia), in sites accessible to the authors through multiple collecting seasons. Choice of taxa was also coordinated with past and ongoing taxon-driven work in sub-branches of the bivalve tree (e.g. work on lucinids (Taylor and Glover 2006; Taylor et al. 2011), unionoids (Graf and Cummings 2006; Whelan et al. 2011) and anomalodesmatans (Dreyer et al. 2003; Harper et al. 2006)) as well as with parallel studies on other aspects of bivalve biology and anatomy (e.g. Simone et al. in press). In a few cases there was a mismatch between the specimens used for morphology and molecular analysis, and in those cases we refer to the highest common taxon in the matrices combining both sets of characters. Such is the case for: Panopea, where morphology was obtained from P. globosa specimens obtained from a fish market in Hong Kong while the molecular data come from P. japonica from a fish market in Fukuoka, Japan; Pinnidae, where the morphology is from BivAToL (Bivalve Assembling the Tree of Life Project)-15 for Pinna carnea and molecules are from Atrina rigida (BivAToL-170) and Propeamussiidae, which consists of morphological data from Parvamussium jeffreysii (BivAToL-307) and molecular data for Propeamussium watsoni (BivAToL-179). A few additional cases like this result from combining morphology from BivAToL specimens and molecules from GenBank (see Table 1).
Outgroups
Outgroup taxa were selected based on prior work for the Assembling the Protostome Tree of Life project from the US National Science Foundation (Principal Investigator: G. Giribet). Most of these sequences were published by Giribet et al. (2006) or by Wilson et al. (2010) (see Table 1).
New fieldwork and utilisation of museum specimens
Multiple collecting fieldtrips, fully or in part for the express purpose of obtaining fresh material for the BivAToL project, were undertaken in: Fort Pierce, FL, USA (Smithsonian Marine Station, 2009); Florida Keys, USA (2007–2012); along a deep-water transect from Gay Head, MA, USA to Bermuda (2008); England and Wales, UK (2009); Western Scotland, UK (2008); Catalonia and Andalusia, Spain (2008, 2011); Moreton Bay, Queensland, Australia (Moreton Bay Research Station, 2008); Hong Kong (Swire Institute of the Marine Sciences, 2011); Singapore (2010); New South Wales, Australia (2010); Zambia (2007, 2008); and Illinois, USA (2009). In addition, specimens from the Philippines, Mozambique, Antarctica and a few additional locations were obtained (see Table 1 for details). Specimens for some taxa were obtained from the Marine Biological Laboratory, Woods Hole (MA, USA) or commercial fish markets. To include targeted taxa that were especially difficult to obtain (e.g. deep-sea protobranchs) or that came from previous campaigns (e.g. Wilson et al. 2009), preserved material contributed from colleagues and existing museum collections was sometimes used that was not appropriately preserved or in sufficient quantity to be studied for all character suites. For details, see Table 1.
Collecting and initial preservation
Whenever possible, multiple (~21) specimens from the same collecting site were obtained and preserved in a variety of preservation fluids (RNAlater or 96% ethanol for molecular work; 3.5% buffered glutaraldehyde for transmission (TEM) and scanning (SEM) electron microscopy; Bouin’s fixative solution for gross anatomical dissections; 70% ethanol for shell microstructural observations and vouchering purposes). Specimens targeted for anatomical study were either cold-relaxed (by temporarily chilling warm-water species) or anaesthetised in isotonic MgCl2 solution, when available. In some cases with extremely rare material that we could not collect ourselves, or where appropriate chemicals could not be obtained in certain field situations, high-percentage ethanol (95–100%) and formalin-preservatives were used for molecular and morphological samples, respectively.
Specimen management and documentation
All newly obtained material for this project (BivAToL) has been organised in a ‘Specimen Central’ collection at the Field Museum of Natural History (FMNH), with collection, preservation and subsequent study records maintained in a KE EMu (www.kesoftware.com/) database. These data are available via workgroup-accessible interactive tools at http://bivatol.org. Each specimen lot of a BivAToL exemplar species was assigned a unique BivAToL registration number that remained with the material regardless of changes in species-level identification or transfer to other vouchering institutions. Material was dispersed to the various collaborating laboratories from Specimen Central according to their specimen and/or preservation needs. Whenever possible, selected specimens were photographed alive before fixation, featuring characters of the animal, such as siphons and extended foot shape, colouration of mantle and mantle features, etc. (see Figs 1–5).
Morphological research
Gross morphology
Specimens for anatomical research were fixed in Bouin’s solution, washed twice in 70% ethanol and subsequently stored in 70% ethanol. The shell was removed and the mantle of one side carefully dissected. Gill and palp observations were accomplished with a Leica MZ8 dissecting microscope with drawing tube. Dissections for alimentary system anatomy were completed using Wild M8 and Leica MZ 12.5 dissecting microscopes with camera lucida. For gross morphology of the stomach and intestinal coiling patterns, the alimentary system was completely excised from the surrounding viscera. For internal anatomy, the stomach, oesophagus and style sac were sliced longitudinally along the anteroposterior axis, yielding more or less symmetrical right and left halves. Visualisation of internal structures was enhanced through the use of aqueous toluidine blue.
Microstructure
Initial observations of shells were made with a stereomicroscope. These enabled the study of gross microstructural detail, such as the character of the periostracum, the presence of organic sheets within the shell, and also the selection of regions of the shell for further study with SEM. Samples were prepared for SEM in several ways: (1) surface views; (2) fractures through the shell; and (3) polished and etched sections. Some of the shells requiring polishing and etching were first set in epoxy resin blocks before cutting them along the desired line, polishing with carborundum grit and then etching in 1% HCl for ~20 s. A few samples were partially cleared of organic material by immersion in domestic bleach (NaClO), followed by washing in distilled water. All samples for SEM were cleaned ultrasonically before mounting on SEM stubs and sputter coated with gold or gold/palladium. A variety of scanning electron microscopes were used (JEOL 820, Philips XL30, Zeiss Ultraplus).
Electron microscopy
For gill and palp studies, entire soft bodies or, in the case of larger specimens, their isolated gills and labial palps, were fixed in cold 3.5% glutaraldehyde in Sørensen buffer plus 10% sucrose (pH 7.3) and cold-stored for a minimum of 24 h. For SEM investigations, the fixed material was dehydrated through a graded ethanol series (25/50/70/99/99/99%), followed by chemical critical point drying using three flushes of hexamethyldisilazane (HMDS, Ted Pella, Inc.), gold sputter coating (Desk IV, Danton Vacuum, LLC), and examination using a Leo EVO 60 scanning electron microscope.
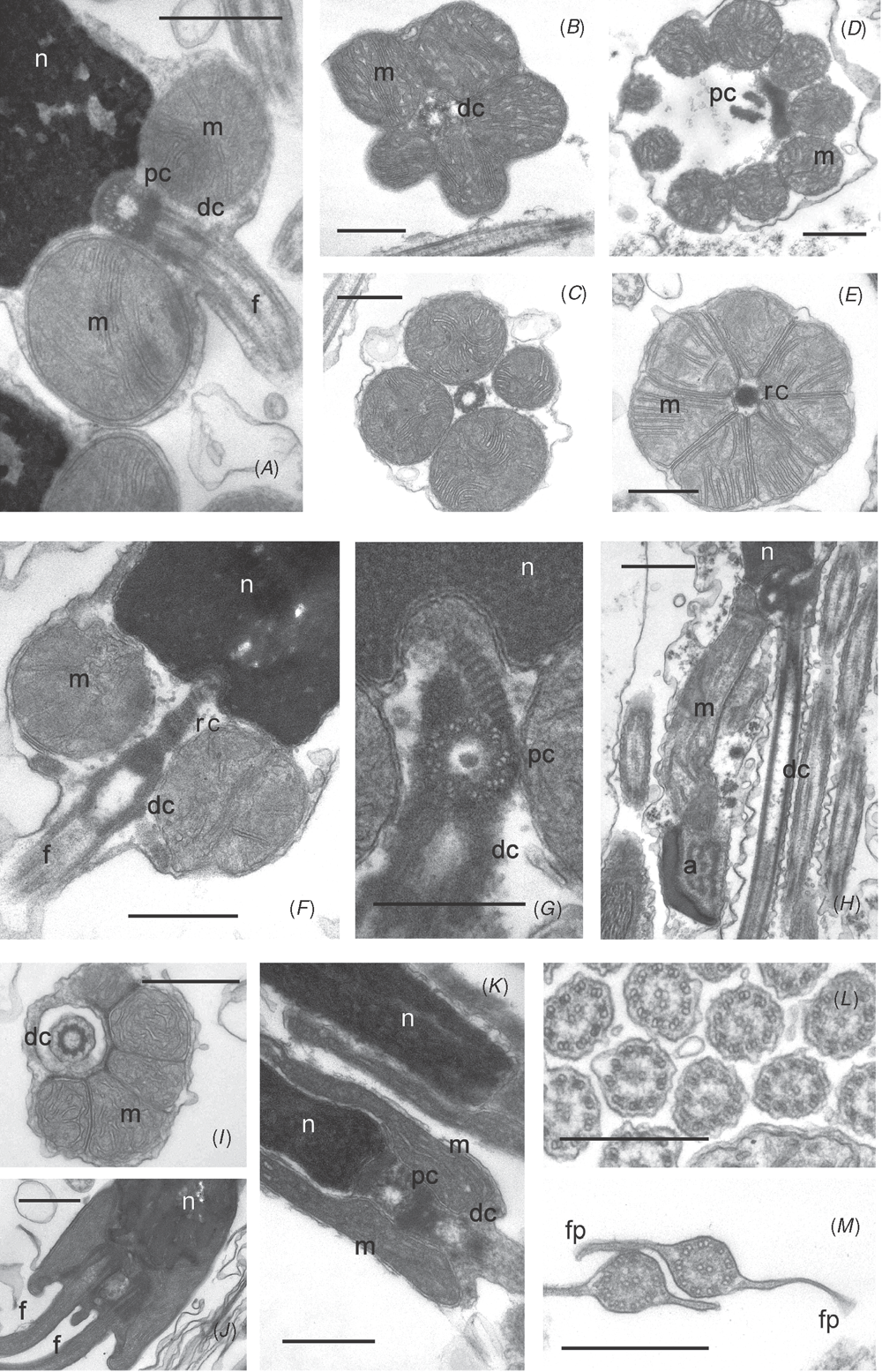
For sperm ultrastructure, small blocks of testicular tissue were fixed in 3% glutaraldehyde in 0.1M phosphate buffer with 10% sucrose. Samples were then processed using a BioWave microwave oven containing a ColdSpot (Pelco, Ted Pella Inc.), following the manufacturer’s instructions. Tissues were post-fixed in 1% osmium tetroxide in 0.1M phosphate buffer with 10% sucrose. After fixation, the specimens were dehydrated through a graded series of ethanol and infiltrated with Epon before overnight polymerisation in a conventional oven at 60°C. Thin sections (70–80 nm) were obtained using an Ultracut ultramicrotome (Leica EM UC6), stained with lead citrate and uranyl acetate, and photographed following observation on a Jeol 1011 transmission electron microscope equipped with a digital camera, operating at 80 kV.
For study of the gross morphology of spermatozoa, suspensions of fixed spermatozoa that had settled to the bottom of the container were washed and resuspended in 0.1M phosphate buffer with 10% sucrose and then allowed to settle on glass coverslips coated with poly-l-lysine. The coverslips with attached spermatozoa were then dehydrated through a graded ethanol series and either critical-point dried (Autosamdri-815, Tousimis) or allowed to dry overnight in HMDS. The coverslips were then mounted on stubs and sputter coated with gold and viewed with a Jeol NeoScope (JCM 5000).
Molecular research
Genomic DNA was extracted from specimens preserved in 96% ethanol (EtOH), RNAlater or frozen at −80°C, using the DNeasy Tissue Kit from QIAGEN. DNA was extracted from a fragment of the foot or mantle (for large specimens) or the entire body (for small specimens). Purified genomic DNA was used as a template for PCR amplification. Molecular markers consisted of two nuclear ribosomal genes (complete 18S rRNA and a ~2.2 kb fragment of 28S rRNA) and fragments of one nuclear protein-encoding gene (histone H3), one mitochondrial ribosomal gene (16S rRNA) and one mitochondrial protein-encoding gene (cytochrome c oxidase subunit I; COI). Complete 18S rRNA was amplified according to Giribet and Wheeler (2002) and Giribet and Distel (2003). COI, 16S rRNA and histone H3 fragments were amplified using standard primers (Xiong and Kocher 1991; Folmer et al. 1994; Edgecombe et al. 2002). 28S rRNA was amplified in three fragments, using the primers 28Srd1a-28Srd5b, 28Srd1a-28Srd4b, or 28Ssip1–28Srd5b for the first fragment, 28Sa-28Sb or 28Sa-28Srd5b for the second fragment, and 28Srd4.8a-28Srd7b1 for the third fragment (see Giribet and Shear 2010). PCRs were performed in 25 µL volume according to standard protocols with annealing temperatures between 34 and 54°C for coding genes and between 40 and 59°C for ribosomal genes. Primer sequences are indicated in Table 2.

|
The resulting amplified samples were purified using an Eppendorf vacuum and Millipore Multiscreen® PCRµ96 Cleanup Filter Plates following manufacturer’s instructions. After performing a sequencing reaction, the BigDye-labelled PCR products were cleaned with Performa DTR V3 96-Well Short Plates (Edge BioSystems) and directly sequenced using an automated ABI Prism® 3730 Genetic Analyzer in the Harvard Bauer Center for Genomic Research. Chromatograms were read and sequences assembled using Sequencher™ v. 4 to 5.0.1 (Gene Codes Corporation, Ann Arbor, MI, USA). All new sequences have been deposited in GenBank under accession codes KC429087–KC429518 (see Table 1). Sequence files for each gene were prepared with MacGDE (Linton 2005).
Data matrix
A MorphoBank project (http://www.morphobank.org) served as a collaborative platform for morphological character/state management and documentation, as well as matrix building. The matrix, associated data and images are provided as supplementary material to this paper on the MorphoBank website (Project 790).
Analyses
Phylogenetic analyses – morphology
The morphological dataset, a matrix with more than 20 000 scored cells, was analysed under parsimony in POY v. 4.1.2 (Varón et al. 2010) with 100 random addition sequences and TBR branch swapping. The morphological data matrix was further analysed in combination with molecular sequence data under parsimony in POY and under Bayesian inference in MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2005), where the morphological data partition was assigned a discrete equal-rates model (Lewis 2001). Details on combined analyses are provided below.
Phylogenetic analyses – dynamic homology under parsimony
Parsimony analysis under direct optimisation (Wheeler 1996) used POY v. 4 (Varón et al. 2010) on 4–6 processors on a Quad-Core Intel Xeon 3 GHz Mac Pro. Timed searches (multiple Wagner trees followed by SPR + TBR + ratchet and tree fusing) of 2–6 h each were run for the combined analyses of all molecules under six analytical parameter sets (see below). Two additional rounds of sensitivity analysis tree fusing (SATF) (Giribet 2007), taking all input trees from the previous round of analyses, were conducted for the combined analysis of molecules under the multiple parameter sets evaluated. These were also 6-h timed searches, and the resulting tree lengths were plotted to check for heuristic stability. Once a parameter set stabilised and the optimal result was found multiple times, we stopped that inquiry, but continued with additional rounds of searches for those parameter sets that continued improving or that found the optimal solution only once.
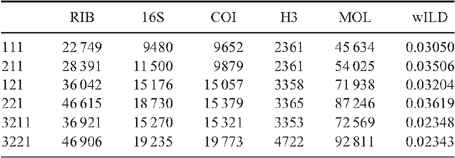
Because a broad parameter space has already been explored in detail in earlier studies, we restricted the dynamic homology analyses to six parameter sets, namely 111, 121, 211, 221, 3221 and 3211. Parameter set 3221 (indel opening cost = 3; indel extension cost = 1; transversions = transitions = 2) has been favoured in many analyses and has been justified philosophically as the best way of analysing data under direct optimisation (De Laet 2010). In addition, we explored a parameter set, namely 3211, in which transversions and transitions receive different costs (indel opening cost = 3; indel extension cost = 1; transversion cost = 2; transition cost = 1), extending the idea of mixed-parameter sets of Sharma et al. (2011). In an effort to limit the difference between indel costs and transformation costs (Spagna and Álvarez-Padilla 2008), four additional parameter sets 111, 121, 211 and 221, often optimal in other analyses, were explored. To calculate the Wheeler incongruence length difference metric (WILD) (Wheeler 1995; Sharma et al. 2011), each individual partition, or the combination of the two nuclear rRNA partitions, was run with a similar search strategy as described above with a 2-h timed search. The resulting WILD values are presented in Table 3.
|
A jackknife resampling analysis (Farris et al. 1996) with 1000 replicates and a probability of deletion of each character of 0.36 was applied to assess nodal support. Because resampling techniques can be meaningless under dynamic homology, different strategies can be applied (see Giribet et al. 2012). Dynamic characters can be converted to a static set, but this tends to inflate support values, because it is based on the implied alignment that favours the topology (Giribet et al. 2012). Instead, we resampled characters that were static a priori (morphology and the pre-aligned protein-coding gene histone H3), as well as fragments of the dynamic characters by using both the number of fragments (20 fragments for 18S rRNA, 5 fragments for each of the three 28S rRNA files and 8 fragments for COI) as well as the command auto_sequence_partition, which evaluates each predetermined fragment. When a long region appears to have no indels, then the fragment is broken inside that region (Giribet et al. 2012).
Phylogenetic analyses – probabilistic approaches
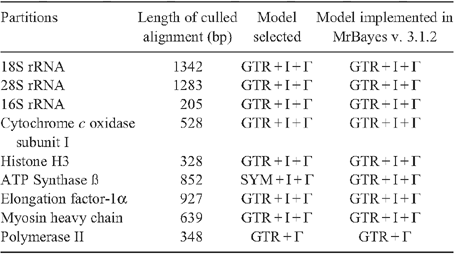
Model-based analyses were conducted on static alignments, which were inferred as follows. Sequences of ribosomal genes were aligned using MUSCLE v. 3.6 (Edgar 2004) using default settings, and subsequently treated with GBlocks v. 0.91b (Castresana 2000) to cull positions of ambiguous homology. Sequences of the protein-encoding genes COI and histone H3 were aligned using MUSCLE v. 3.6 with default settings as well, but alignments were confirmed using protein sequence translations before treatment with GBlocks. The size of data matrices for each gene subsequent to treatment with GBlocks is provided in Table 4.
|
Maximum likelihood (ML) analysis was conducted using RAxML v. 7.2.7 (Stamatakis 2006) on 24 CPUs of a computer cluster (Odyssey) at Harvard University. A unique GTR model of sequence evolution with correction for rate heterogeneity (GTR + Γ) was specified for each data partition. Two hundred and fifty independent searches were conducted. Nodal support was estimated via the rapid bootstrap algorithm (1000 replicates) using the GTR-CAT model (Stamatakis et al. 2008).
Bayesian Inference (BI) was conducted using MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2005) on 24 CPUs of the computing cluster at Harvard University. A unique GTR model of sequence evolution with correction for rate heterogeneity and a proportion of invariant sites (GTR + Γ + I) was specified for each partition, as recommended by jModeltest (Guindon and Gascuel 2003; Posada 2008). Four runs, each with four chains (default distribution of hot and cold chains), were conducted for 20 million generations. The list of models selected by jModeltest for all data partitions is provided in Table 4. Runs were examined in Tracer v.1.5 (Rambaut and Drummond 2009) to check for stationarity.
Nine-gene analyses
In addition to the new data generated and analysed here, we conducted a nine-gene analysis by combining the five-gene dataset with a four-gene dataset, the latter consisting of nuclear protein-encoding genes ATP synthase β, elongation factor-1α, myosin heavy chain type II and RNA polymerase II recently published by Sharma et al. (2012).
The nine-gene dataset was analysed for the subset of 42 taxa represented in the Sharma et al. (2012) study, and also for all 108 terminals (wherein the protein-encoding genes were missing for a subset of species). The datasets were analysed as described above with respect to ML and BI approaches.
Finally, a total evidence analysis of all data (nine genes plus morphology) was conducted under BI using MrBayes v. 3.1.2. In the total evidence analysis, the morphological data partition was assigned a discrete equal-rates model (Lewis 2001). Four runs, each with four chains (default distribution of hot and cold chains) were conducted for 20 million generations.
Estimation of divergence times and lineage diversification
Ages of clades were inferred using the nine-gene dataset in BEAST v.1.7.4 (Drummond and Rambaut 2007). Substitution models assigned to each partition are indicated in Table 4. The branch length distribution was examined and fitted with an exponential distribution, and outlier taxa with excessively long terminal branches were removed (Cavatidens omissa, Chama macerophylla, Myochama anomioides and Poromya illevis). An uncorrelated lognormal clock model was inferred for each partition, and a birth–death speciation process was assumed for the tree prior. Other priors were sequentially optimised in a series of iterative test runs (data not shown). Two Markov chains were run for 108 generations, sampling every 104 generations. Convergence diagnostics were assessed using Tracer v.1.5 (Rambaut and Drummond 2009).
Ten calibration points were used to constrain divergence times based on fossil taxa. We opted to constrain nodes using normal or uniform distributions spanning the earliest fossil appearance of clades, rather than lognormal or truncated lognormal distributions. This is due to the predilection of BEAST v.1.7.4 to infer Precambrian diversification for all subclasses of Bivalvia upon use of lognormal distribution priors, which is inconsistent with the bivalve fossil record. The constraints that we employed were as follows. (1) The root age of Bivalvia was constrained using a uniform distribution prior between 520.5 and 530 Ma, based on the age of the earliest crown group bivalve (Fordilla troyensis Barrande, 1881 from the Tommotian of Siberia; Pojeta et al. 1973; Pojeta and Runnegar 1974; Parkhaev 2008) and the age of the oldest known shelled molluscs (univalved molluscs from the Tommotian; Caron et al 2007a, 2007b). (2) Anomalodesmata was constrained using a normal distribution prior spanning 478.6–488.3, based on Ucumaris conradoi Sánchez & Vaccari, 2003 (from the Tremadocian). (3) Arcida was constrained using a normal distribution prior spanning 471.8–488.6 Ma, based on Glyptarca serrata Cope, 1996 (Arenigian; Cope 1997). (4) Cardiidae was constrained using a normal distribution prior spanning 204–228 Ma, based on Tulongocardium nequam Healey, 1908 (Norian; Schneider 1995). (5) Mactroidea was constrained using a normal distribution prior spanning 112–125 Ma, based on Nelltia elliptica (Whitfield, 1891) (Aptian; Saul 1973). (6) Nucinellidae was constrained using a normal distribution prior spanning 197–201.6 Ma, based on Nucinella liasina (Bistram, 1903) (Hettangian; Conti 1954). (7) Ostreoidea was constrained using a normal distribution prior spanning 237–245 Ma, based on the Muschelkalk of Germany (Anisian; Hautmann and Hagdorn 2013). (8) Palaeoheterodonta was constrained using a normal distribution prior spanning 471–478 Ma, based on the early Ordovician genus Noradonta Pojeta & Gilbert-Tomlinson, 1977 (Arenigian; Babin 1982; Cope 2000). (9) Pterida was constrained using a normal distribution prior spanning 476–488.4 Ma, based on the Ordovician genus Pterinea Goldfuss, 1826 (Bassler 1915), taking a conservative approach to the placement of this fossil. (10) Tellinoidea was constrained using a normal distribution prior spanning 197–201.6 Ma, based on Tancredia securiformis (Dunker, 1846) (Hettangian).
To observe the tempo of diversification through time, we generated log-lineage through time plots (LTT) using the R package ape v. 3.0–11 (Paradis et al. 2004).
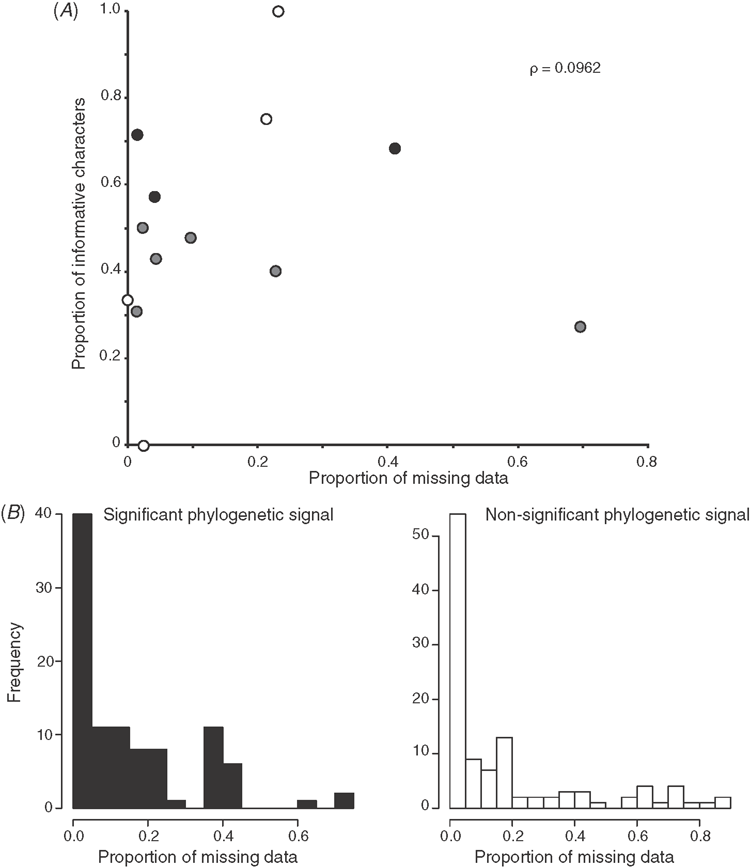
Analysis of phylogenetic signal
We assessed phylogenetic signal inherent to each of the 221 morphological characters using Mesquite v. 2.75 (Maddison and Maddison 2011). Characters were optimised using equal weights parsimony on the total evidence tree (nine genes plus morphology). Null distributions were generated by 500 replicates of randomly reshuffled trips. Characters with parsimony steps on the total evidence tree that differed significantly from the null distribution (below 5th percentile) were scored as having more phylogenetic structure than expected by chance. Tests for correlation between amount of missing data and degree of phylogenetic signal were conducted using the Spearman rank correlation coefficient.
Use of name endings at the ordinal rank
The ordinal endings -ida and -oida have both been extensively employed in bivalve literature. We follow the suggested standardisation to -ida as used in the bivalve classifications by Scarlato and Starobogatov (1979), Bieler et al. (2010) and Carter et al. (2011), among others. No change in inferred rank is implied by this adjustment. These names, as well as those with informal endings, are used as labels of their clades. We are aware that the formal endings imply certain ranks in the Linnaean hierarchy, but we have not attempted to re-rank all hypothesised clades resulting from this study as we consider such a step as premature until a denser family-level sampling is presented.
Results
Morphological characters and states
One-hundred of the 210 characters (48%) used in this analysis have never been used before in a phylogenetic analysis. Others have been previously employed but are coded here in modified form. The final matrix consists of 22 680 cells, with 16.6% of missing data, most of which belong to hard-to-obtain character systems like sperm ultrastructure or larval characters.
Shell characters
01 Adult shell shape: (0) univalved with a single aperture; (1) univalved with two apertures; (2) bivalved with adductor muscles to close the valves. This character is included to distinguish gastropod (state 0) and scaphopod (1) outgroups from bivalves (state 2). This character was also coded by Giribet and Wheeler (2002: char. 16). In previous studies, this character was combined with a muscle system character – the presence/absence of adductor muscles. Adductor muscles do not exist in the absence of a bivalve shell (except in a few derived shelled opisthobranchs, not included in this matrix), and bivalved shells do not exist without adductor muscles, so we have retained only this character here.
02 Lateral expansions of the shell (auricles) at each side of the umbo: (0) absent; (1) present (Fig. 2F). This character was also coded by Giribet and Wheeler (2002: char. 19).
03 Byssal gape: (0) absent; (1) present. Most/all juvenile bivalves produce a byssus as an aid to settlement (Yonge 1962). Coding is restricted to bivalves that produce a byssus in the adult. This character was also coded by Giribet and Wheeler (2002: char. 20).
04 Anterior adductor muscle (or scar): (0) present; (1) reduced in size with respect to the posterior adductor; (2) absent (monomyarian condition). This character was also coded by Giribet and Wheeler (2002: char. 21).
05 Posterior adductor muscle (or scar): (0) present; (1) reduced in size with respect to the anterior adductor; (2) absent (monomyarian). This character was also coded by Giribet and Wheeler (2002: char. 22), although only character states 0 and 1 were used.
06 Position of posterior pedal retractor scar relative to posterior adductor scar: (0) anterodorsal; (1) inset on the anterior, concave face of a crescentic posterior adductor scar. This character was also coded by Giribet and Wheeler (2002: char. 23).
07 Pallial line: (0) absent; (1) present. We consider a pallial line to be present whether it is continuous or formed by multiple scars. Another character concerning the presence of a discontinuous pallial line could be informative in another context. This character was also coded by Giribet and Wheeler (2002: char. 24).
08 Pallial sinus: (0) absent; (1) present. This character (state 1) is often correlated with the presence of siphons, but some have siphons and lack a sinus (e.g. Cyrenidae). Coding is restricted to taxa with a pallial line. This character was also coded by Giribet and Wheeler (2002: char. 25).
09 Umbo: (0) orthogyrous; (1) prosogyrous; (2) opisthogyrous. This character was also coded by Giribet and Wheeler (2002: char. 26).
10 Purple pigment in the internal shell layer: (0) absent; (1) present. This pigment cannot be extracted through the use of acids or organic solvents (Morton et al. 1998) and is present in members of Cyrenidae (=Corbiculidae) and Veneridae. This character was also coded by Giribet and Wheeler (2002: char. 28).
11 External ligament: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 30).
12 Ligament position: (0) amphidetic; (1) opisthodetic. This character was also coded by Giribet and Wheeler (2002: char. 31).
13 Ligament type: (0) simple; (1) duplivincular; (2) alivincular; (3) transverse; (4) parivincular. See Waller (1990) for definitions. This character was also coded by Giribet and Wheeler (2002: char. 32).
14 Resilifer: (0) absent; (1) present; (2) present as a chondrophore. The ligament can sit in a hollow depression in the hinge plate known as the resilifer, located internally just beneath the umbo. A spoon-shaped, projecting resilifer (e.g. in Mactridae) is termed a chondrophore. Coding is restricted to bivalves with an internal ligament. This character was also coded by Giribet and Wheeler (2002: char. 35).
15 Lithodesma: (0) absent; (1) present. A lithodesma is a calcified ossicle found within the ligament of many anomalodesmatan (see Yonge and Morton 1980) and montacutid (e.g. Allen 2000; Marshall 2002; Jespersen et al. 2004) bivalves. This character was also coded by Giribet and Wheeler (2002: char. 36).
16 Pseudonymphae: (0) absent; (1) present. A pseudonymph is a ridge-like support for a dorsal (external) ligament that faces the median plane of the shell, not rotated dorsally as in a true nymph, thus not enhancing the dorsal arching of the ligament. Coding is restricted to those taxa with external ligaments. Although Waller (1990) stated that the structure is limited to the ‘Isofilibranchia’ (here Mytilidae, except absent in Dacrydiinae), Garcia-March et al. (2008) also noted its presence in Recent Pinnidae. This character was also coded by Giribet and Wheeler (2002: char. 37).
17 Operculum: (0) absent; (1) present. An operculum is present in all gastropod larvae, although is lost in the adults of several vetigastropods, many caenogastropods and most euthyneurans (Haszprunar 1988; Ponder and Lindberg 1997). This character was also coded by Giribet and Wheeler (2002: char. 43).
18 Pallets: (0) absent; (1) present. Calcareous pallets that close or protect the burrow when the siphons are retracted are typical of Teredinidae. Coding is restricted to bivalves with siphons (char. 63). This character was also coded by Giribet and Wheeler (2002: char. 59).
19 Umbonal crack or slit: (0) absent; (1) present. A natural crack running from the umbo radially through each valve, often filled with organic material, occurs in members of the families Laternulidae and Periplomatidae. This character was also coded in a phylogenetic study of the anomalodesmatans (Harper et al. 2000: char. 4).
20 Calcareous adventitious tube secreted by the animal: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 183).
21 Cementation to substrate by a calcareous secretion of the mantle margins: (0) absent; (1) present. This type of cementation to the substrate (as opposed to attachment by a calcified byssus, typical of the Anomioidea) is found in various groups including Ostreoidea, certain Pectinidae (Hinnites), Spondylidae, Plicatulidae, some species of Etheriidae, Chamoidea, Hippuritoidea, Myochamidae and Cleidothaeridae (Yonge 1979; Harper 1992; Harper et al. 2000). This character was also coded by Giribet and Wheeler (2002: char. 53).
Hinge characters
Giribet and Wheeler (2002: char. 39) used previously defined hinge types (taxodont, schizodont, heterodont, desmodont, edentate) as a single multistate character, which is here replaced by characters 22–27.
22 Hinge tooth row: (0) many subequal teeth; (1) few dissimilar teeth. This character distinguishes the traditional taxodont and heterodont dentition types, and was used similarly for the larval hinge apparatus by Giribet and Wheeler (2002: char. 38). Edentate species are coded as ‘not applicable’. New character.
23 Hinge tooth row gap at umbo: (0) absent; (1) present. A subumbonal gap in the tooth row is present in many bivalves with taxodont dentition. Coding is restricted to taxa coded as ‘many subequal teeth’ (i.e. taxodont) for hinge tooth row (char. 22: state 0). Edentate species are coded as ‘not applicable’. New character.
24 Secondary teeth: (0) absent; (1) present. Secondary teeth are interlocking hinge teeth presumed not to be homologous with the primary (cardinal and lateral) teeth present throughout Bivalvia. This character was also coded by Giribet and Wheeler (2002: char. 41).
25 Cardinal hinge teeth: (0) absent; (1) present. Cardinal hinge teeth are defined as radiating from a point below the umbo; together with lateral teeth, they comprise the definition of heterodont dentititon. Edentate species are coded as ‘not applicable’. New character.
26 Anterior lateral hinge teeth: (0) absent; (1) present. Lateral hinge teeth are defined as parallel or subparallel teeth (relative to the shell margin) far removed from the umbo; together with cardinal teeth, they comprise the definition of heterodont dentititon. Lateral teeth prevent anteroposterior slippage of the valves when closed. They are present in bivalves as anterior or posterior pairs, or both. Edentate species are coded as ‘not applicable’. New character.
27 Posterior lateral hinge teeth: (0) absent; (1) present. See character 26. New character.
28 Chomata: (0) absent; (1) present. Chomata are small tubercles on short ridges on the hinge of the right valve of members of Ostreidae, Gryphaeidae and Plicatulidae (Harry 1985; Waller 1998). Members of the genus Crassostrea (Ostreidae) do not develop chomata (Slack-Smith 1998a). This character was also coded by Giribet and Wheeler (2002: char. 42).
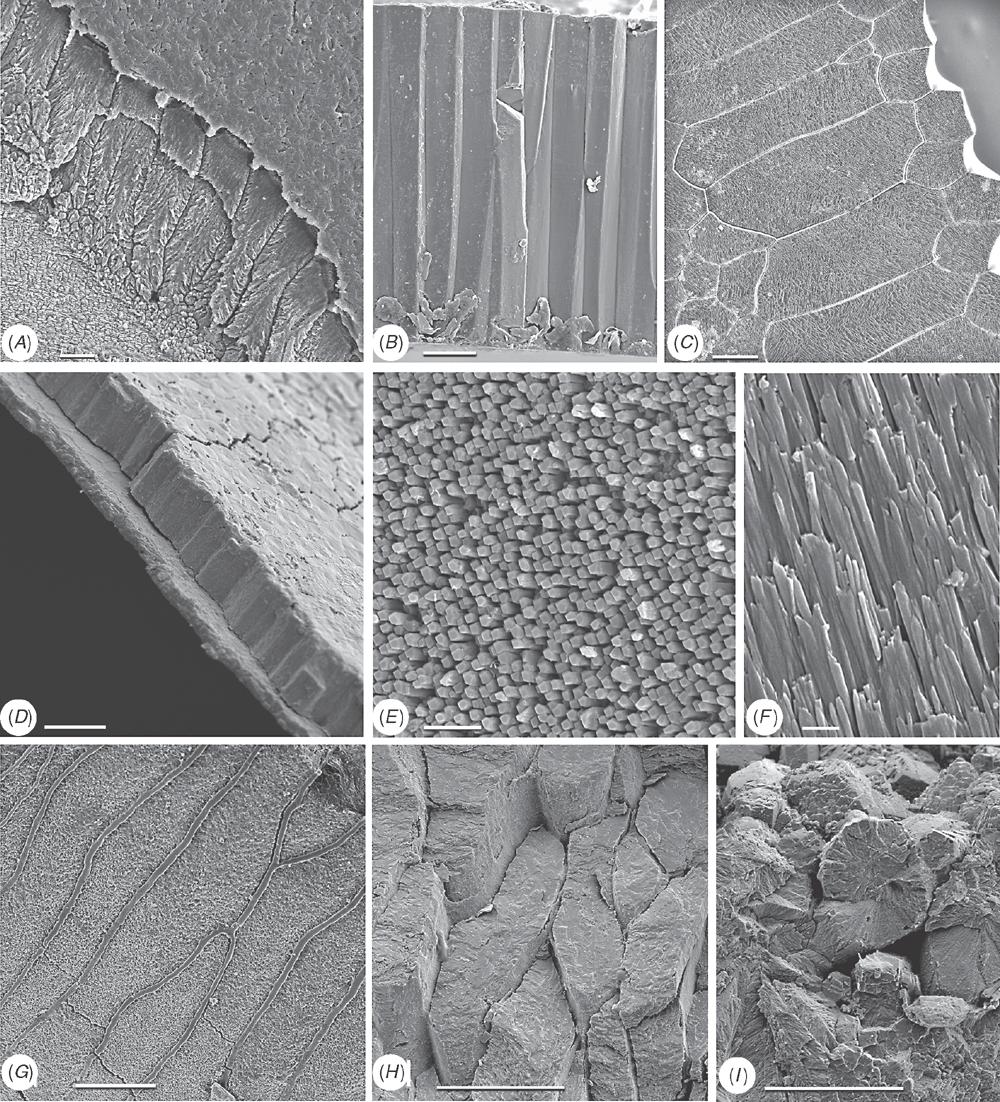
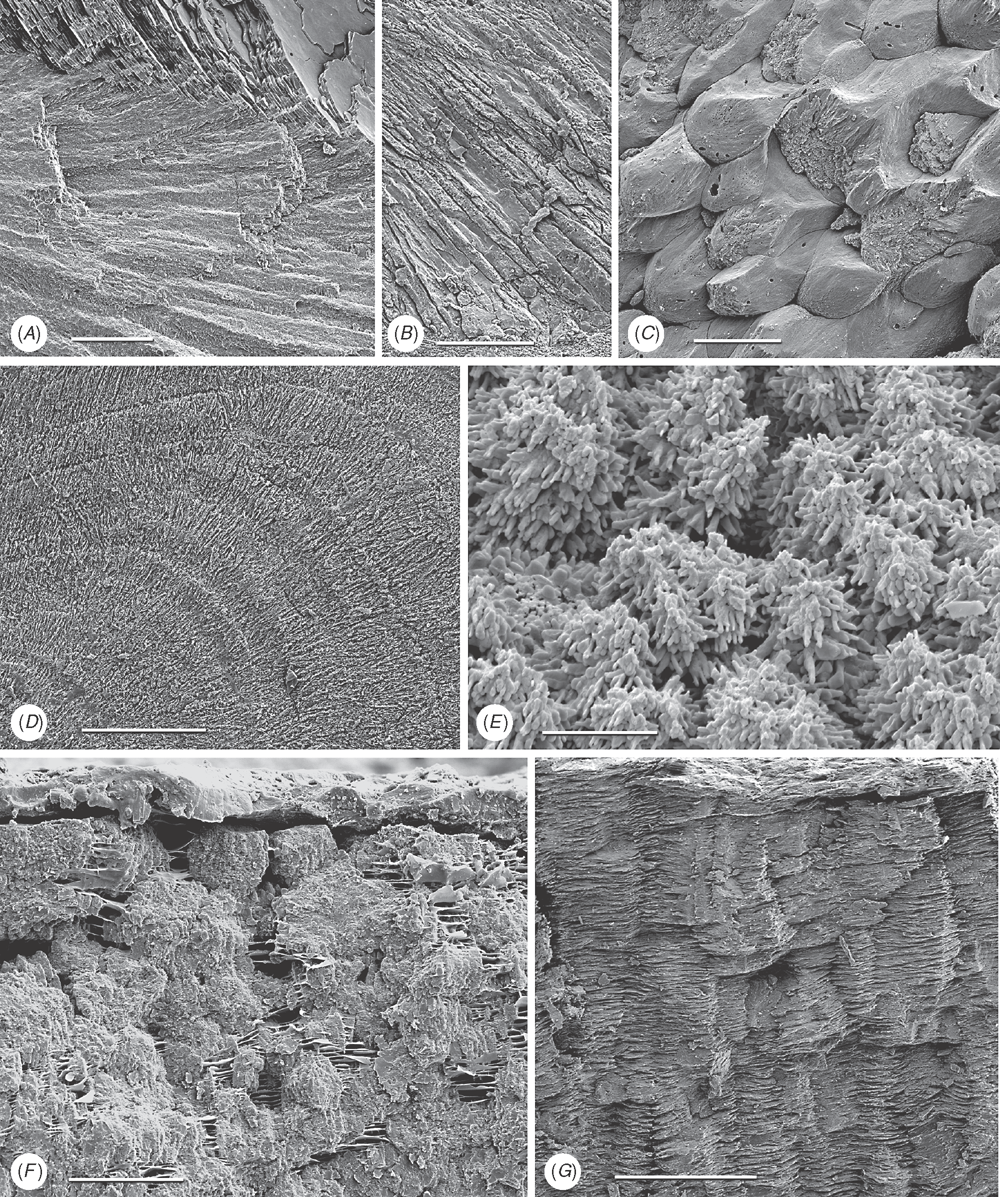
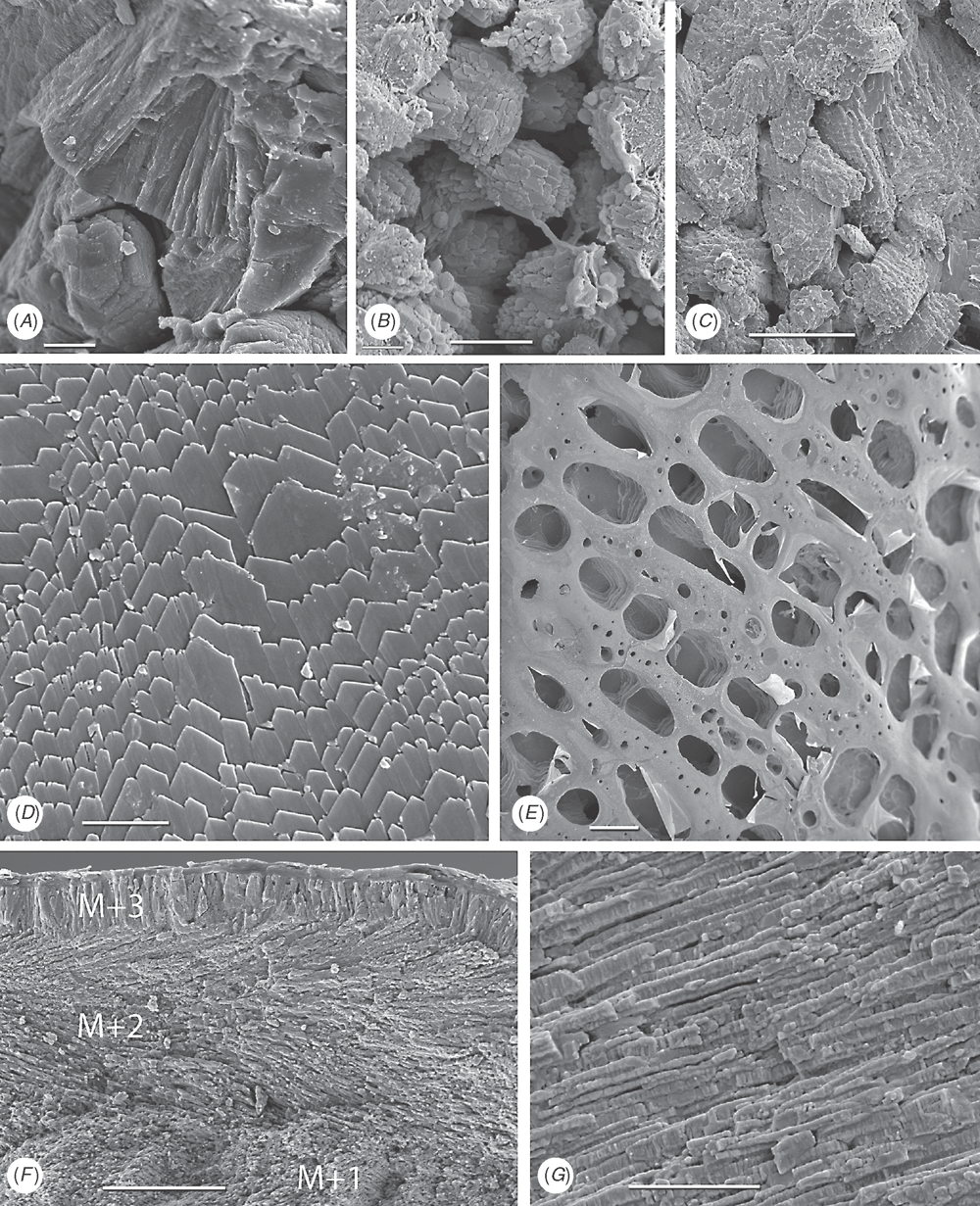
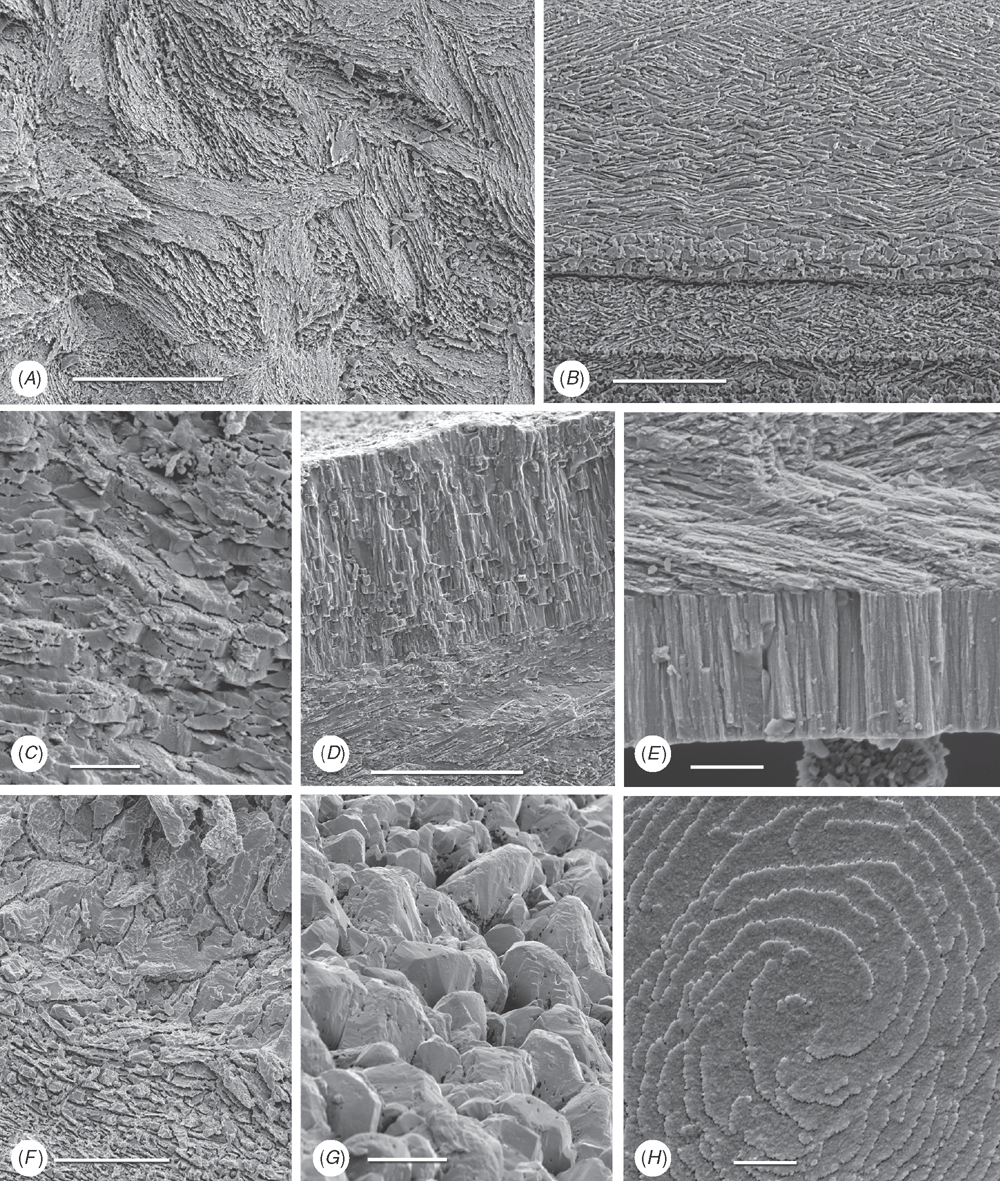
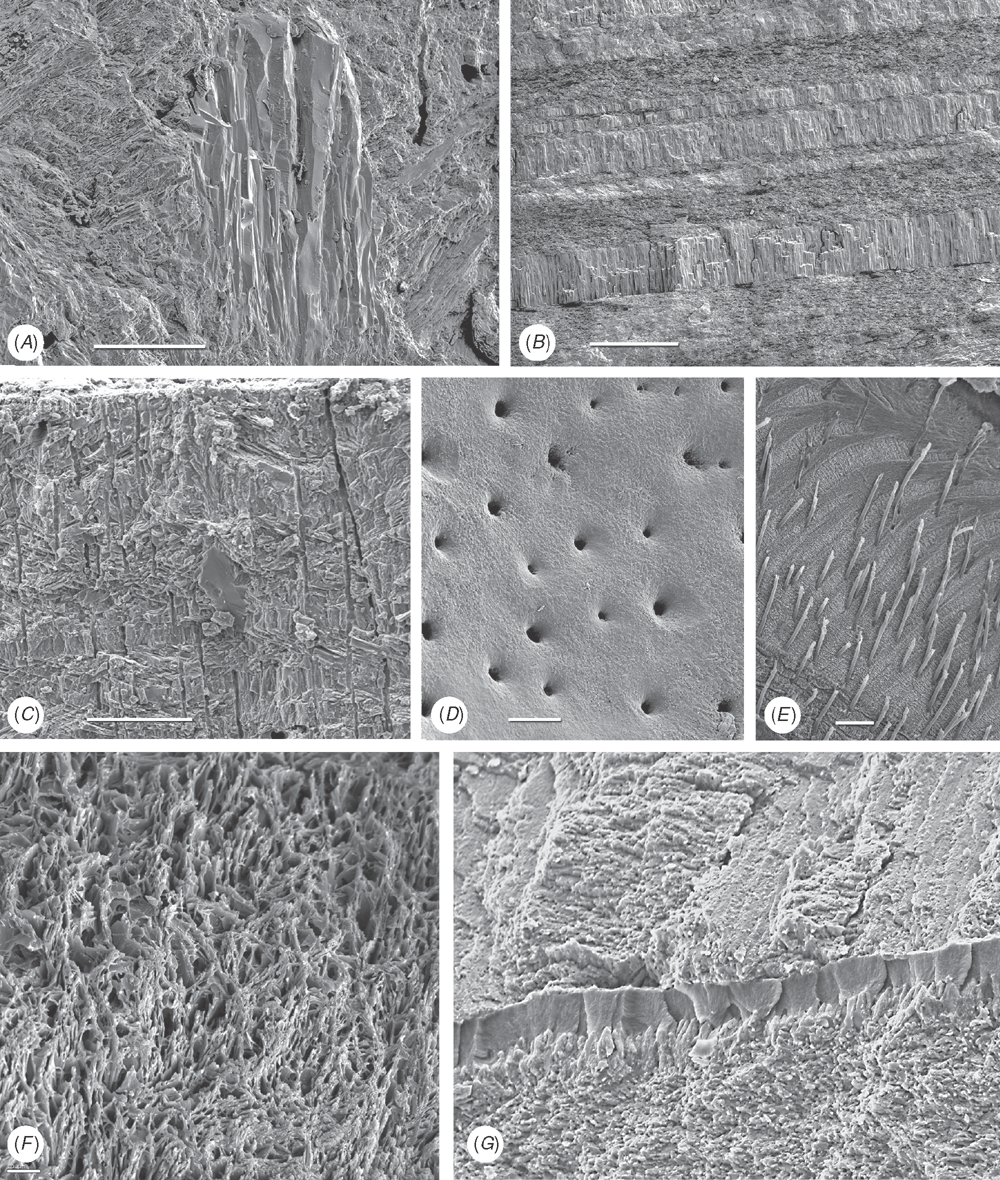
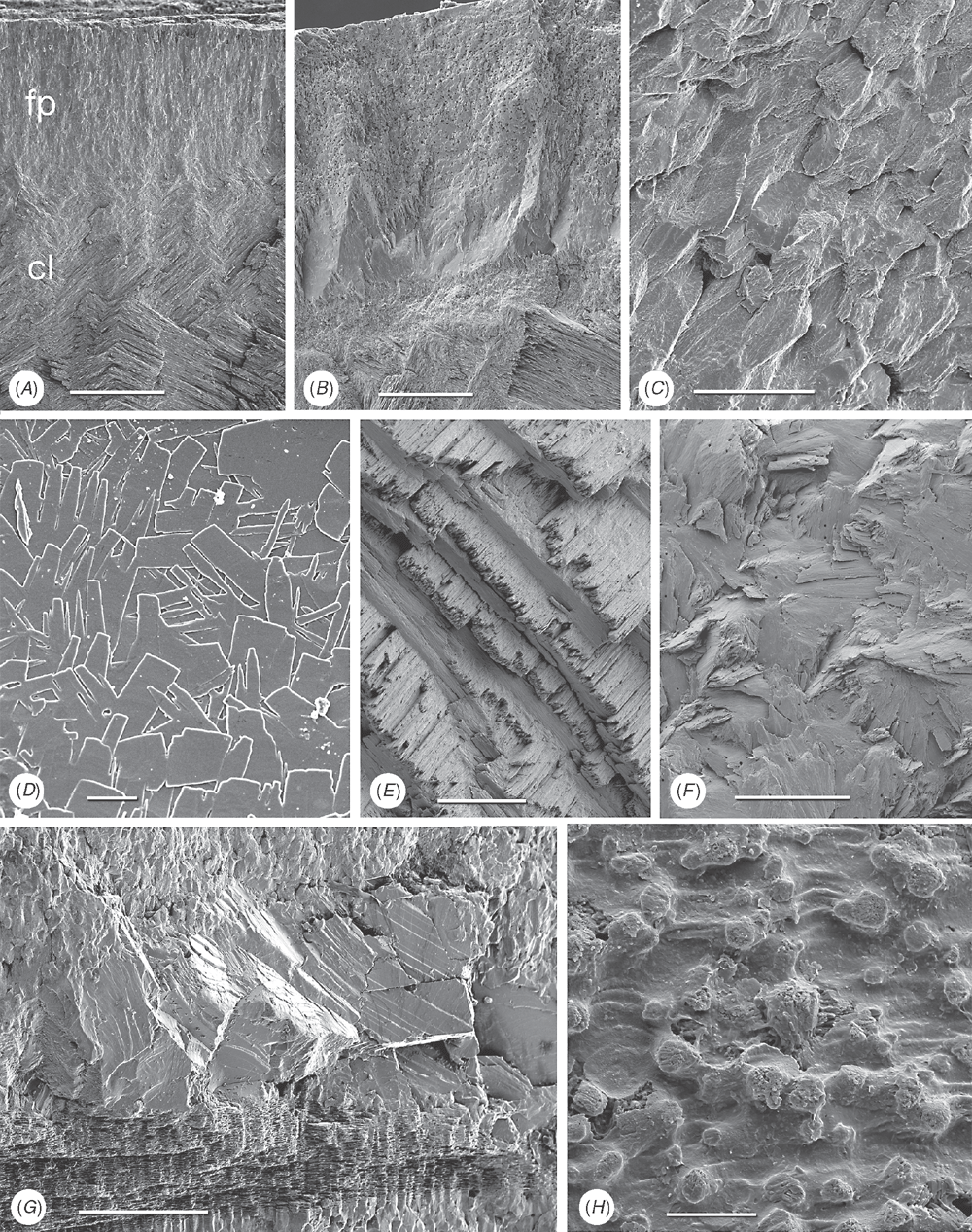
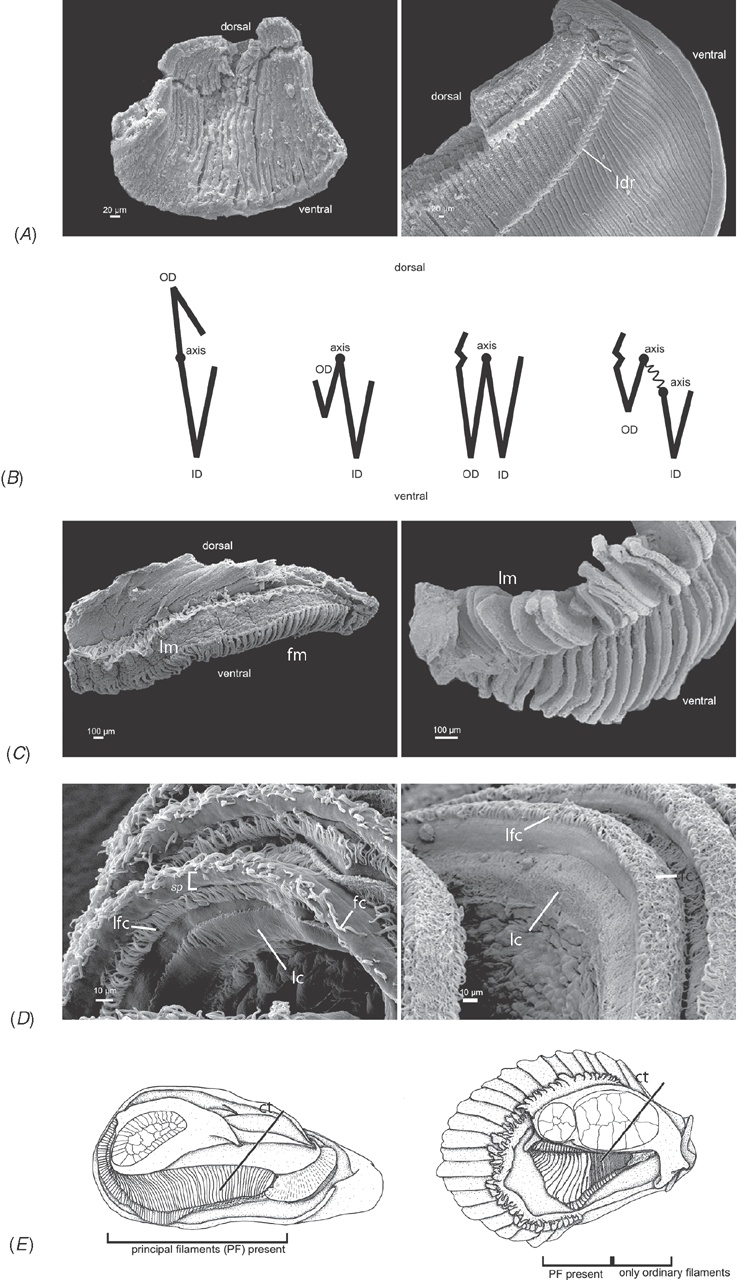
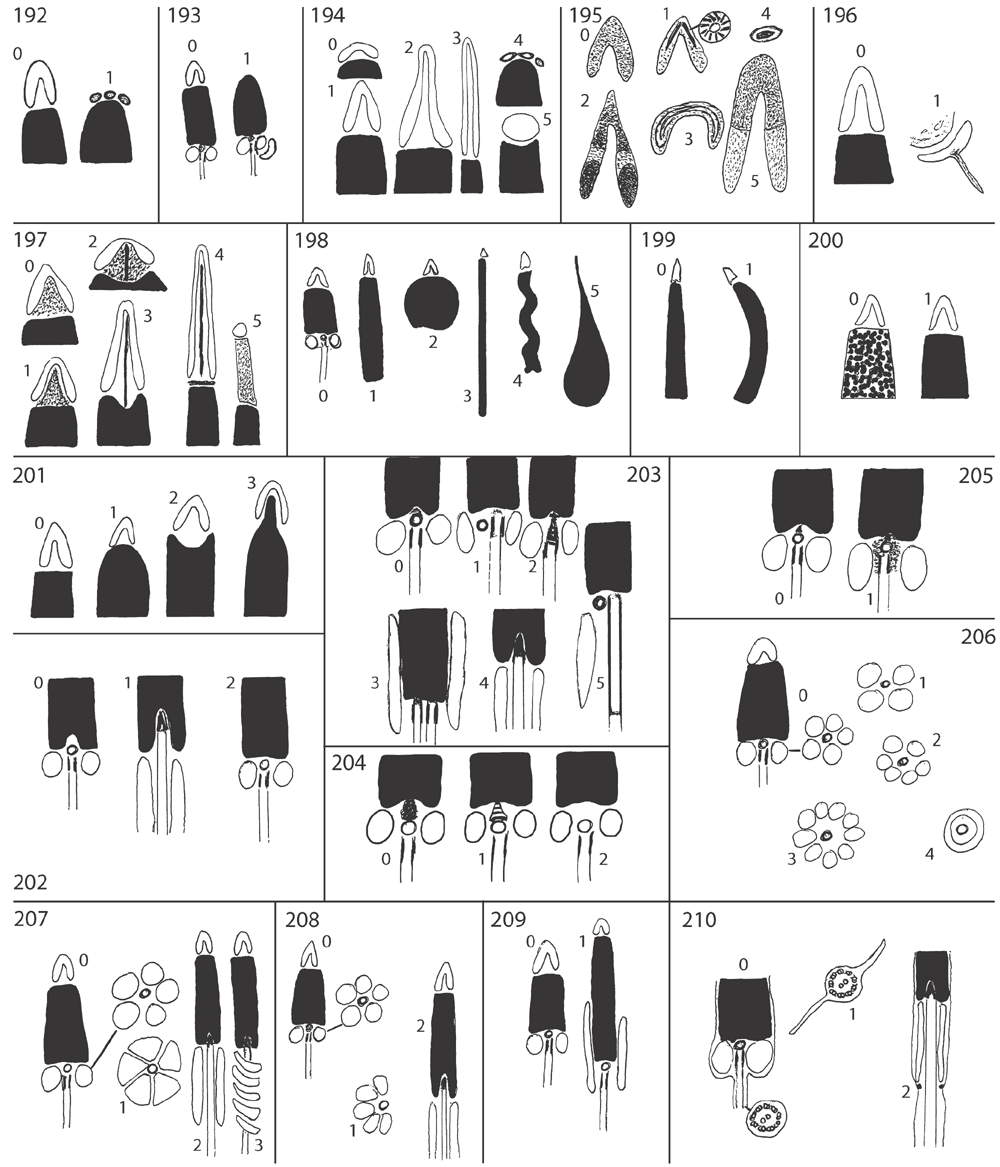
Shell microstructure characters (Figs 6–15)
Bivalve shells are composite structures built of crystals of calcium carbonate (either calcite or aragonite) interspersed in an organic matrix, secreted onto the periostracum, a largely organic outermost shell layer. Variation in the detailed microstructure of bivalves has long been studied (e.g. Bøggild 1930; Taylor et al. 1969, 1973; Carter 1990b). From these studies, it is clear that within the class there are several different microstructural types and that these are, generally, arranged in discrete layers within the shell. Across the Bivalvia, taxa show both different numbers of layers and combinations of microstructural types. These differences are believed to be of phylogenetic and adaptive biomechanical significance.
Although microstructural information has been widely coded as a source of characters in phylogenetic analyses, the standard practice has been to score each individual microstructural type as a present/absent character with no regard for positional information (e.g. Giribet and Wheeler 2002). In this analysis we have attempted to recognise homologous shell layers and used each as a character to be scored according to the microstructure present. The position of the pallial myostracum (present as a thin layer through the shell) was the landmark from which layers were identified, with M+ and M- designations for layers external or internal to the pallial myostracum, respectively. Higher absolute values reflect positions successively farther away from the myostracum. This method was derived from a similar scheme first developed for patelloidan limpets by MacClintock (1967). The greatest variation in microstructure occurs in the outermost layer of the shell, and this is reflected in the large number of states recognised for the M+2 layer.
29 Microstructure asymmetric between the two valves: (0) absent; (1) present. For most taxa, the microstructural arrangements of both left and right valves are the same; however, in some, most notably Pectinoidea, the numbers of microstructural layers and their distributions in the two valves are different. In Pectinoidea, the arrangement shown by the right valve appears to be the less derived (Carter 1990a). New character.
30 Number of shell layers external to the pallial myostracum (trace of pallial line seen in sections through the valve): (0) 2; (1) 1; (2) 3. New character.
31 Number of shell layers inside pallial myostracum: (0) 1; (1) 2. Taxa lacking a pallial myostracum (e.g. oysters) are scored as ‘?’. New character.
32 M+3 layer: (0) foliated prisms (Fig. 6C); (1) irregular spherulitic aragonite prisms (Figs 6I, 15G); (2) fine simple aragonitic prisms (Fig. 8F). New character.
33 M+2 layer: (0) simple aragonite prisms (Neotrigonia-like) (Fig. 6A); (1) simple calcite prisms (Pinna-like) (Fig. 6B); (2) foliated prisms (Fig. 6C); (3) fibrillar calcite prisms (Mytilus-type) (Fig. 6E; (4) denticular composite aragonitic prisms (Nucula-type) (Fig. 7A); (5) fibrillar composite aragonitic prisms (cardiid-type) (Figs 7B, D, 8G); (6) compound composite aragonitic prisms (some venerids, Donax, etc.) (Fig. 7C); (7) elongate prisms (Solemya-type) (Fig. 6G, H); (8) granular aragonitic prisms (Entodesma-type) (Figs 7F, 8B, C); (9) fine fibrillar calcitic prisms (Lima-type) (Fig. 6F); (10) vesicular calcite (e.g. Hyotissa) (Fig. 8E); (11) crossed-lamellar aragonite (Fig. 10C); (12) homogeneous (see Carter 1990a); (13) foliated calcite (Fig. 8D); (14) dendritic aragonitic prisms (e.g. Thracia) (see Checa et al. 2012; Fig. 8E, G) ; (15) blocky aragonite prisms (pandorid type) (Fig. 6D); (16) spherulitic aragonitic prisms (typical anomalodesmatan-type) (Fig. 8A); (17) granular calcite (Neritidae-type) (Fig. 15B); (18) bladed calcite prisms with irregular margins (Fig. 15C); (19) fine aragonite prisms (scaphopod-type) (Fig. 15A). New character.
34 M+1 layer: (0) stacked nacre (aragonite) (Fig. 9A); (1) sheet nacre (aragonite) (Fig. 9B); (2) crossed-lamellar aragonite (Figs 9C, D, 15E); (3) crossed-lamellar grading into homogeneous aragonite (Fig. 9G); (4) crossed-acicular aragonite (Fig. 9E); (5) homogeneous aragonite (heterodont-type) (Figs 9F–H, 10F); (6) homogeneous aragonite (protobranch-type) (see Carter 1990a: figs 16C, D); (7) foliated calcite (Fig. 8D); (8) foliated aragonite (semi-nacre) of monoplacophorans (see Checa et al. 2009; Fig. 15D). New character.
35 M-1 layer: (0) sheet nacre (aragonite) (Fig. 9B); (1) crossed-lamellar aragonite (Fig. 9A); (2) complex crossed-lamellar aragonite (Figs 10A, B, H, 15F); (3) crossed-acicular aragonite (Fig. 9E); (4) homogeneous aragonite (heterodont-type) (Figs 9F, 10C); (5) homogeneous aragonite (protobranch-type) (see Carter 1990a: figs 16C, D); (6) foliated calcite (Fig. 8D); (7) aragonite prisms (Fig. 10D, E, F, G). New character.
36 M-2 layer: (0) sheet nacre (aragonite) (Fig. 9B); (1) complex crossed-lamellar aragonite (Fig. 10A, B, H). New character.
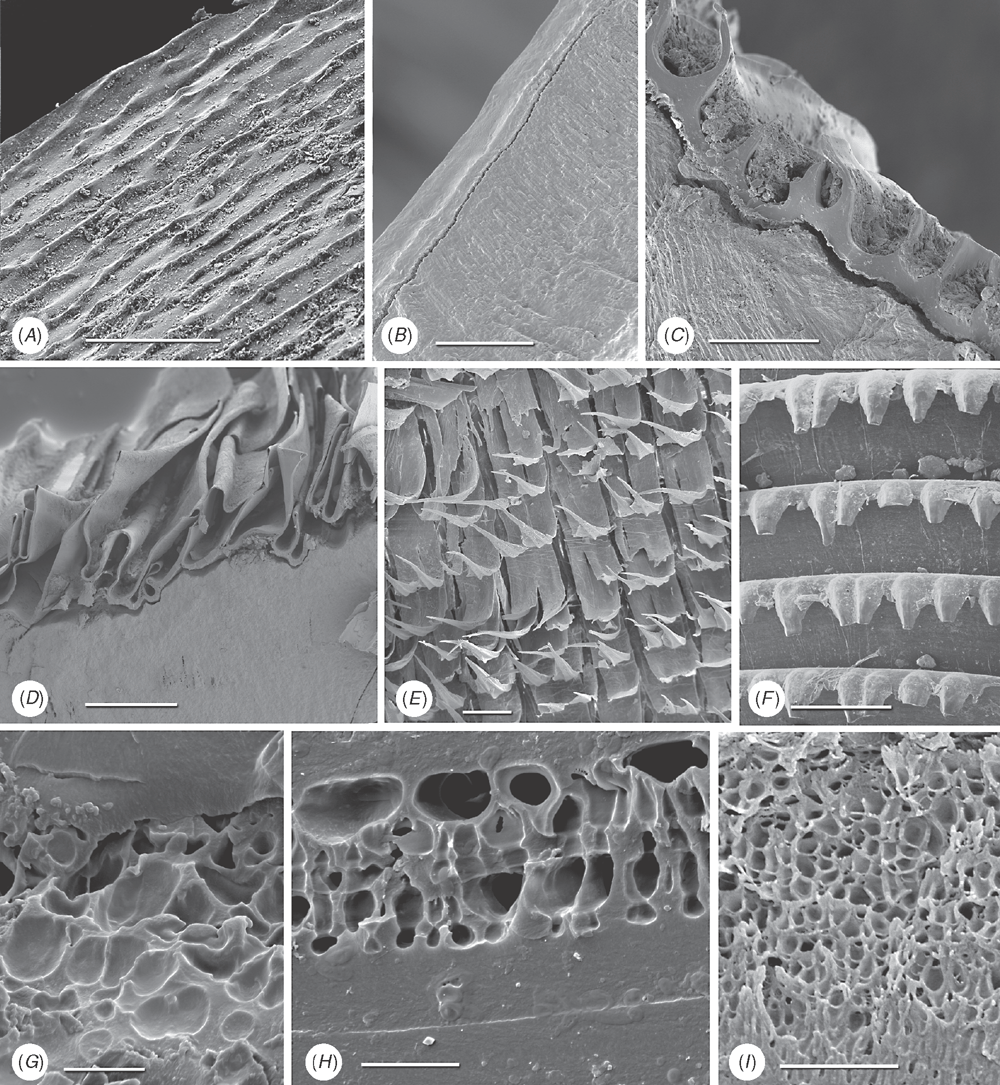
37 Myostracal pillars: (0) absent; (1) present. These are columns of myostracal aragonite that are continuous with the pallial myostracum and that outcrop on the inner shell surface dorsal to the pallial line (e.g. in Chama; see Kennedy et al. 1970) (Fig. 11A). They indicate sites of mantle attachment to the general valve surface. This was coded by Giribet and Wheeler (2002: char. 11).
38 Myostracal prisms in M-1: (0) absent; (1) present. These are sheets of aragonite prisms similar in morphology to those formed under muscle attachment sites (Fig. 12B), variable in thickness and extent. This character was also coded by Carter et al. (2000: char. 97).
39 Tubules: (0) absent; (1) present in M-1 only; (2) present throughout the shell. Tubules are fine cylindrical pores formed by cellular mantle extensions that post-date shell formation (Taylor et al. 1969; Fig. 11C–E). The presence of tubules was also coded as a morphological character in previous studies, for example, Giribet and Wheeler (2002: char. 29), but has been reformulated here to use more precise information on their distribution.
40 Chalky layers: (0) absent; (1) present. These are lenses of loosely bladed calcite crystals, of variable extent that are typical of oysters (Lee et al. 2011; Fig. 11F). This character was coded previously by, for example, Giribet and Wheeler (2002: char. 13).
41 Shell chambers: (0) absent; (1) present. These are vacuolated chambers within the valve, originally fluid-filled, observed in oysters, Etheria, Crassadoma and certain spondylids. New character.
42 Organic layers within shell: (0) absent; (1) present. Multiple distinct organic layers within the calcareous part of the shell are particularly distinctive in Corbulidae (Lewy and Samtleben 1979; Fig. 11G), certain oysters (e.g. Saccostrea; Taylor 1990), Unionidae and Margaritiferidae (Kat 1983). New character.
43 External surface of periostracum: (0) smooth with commarginal growth increments (Fig. 12A); (1) extremely smooth (polished appearance) (Fig. 12B); (2) prominent commarginal lamellae (Fig. 12C, D); (3) projecting shingles, bristles and flaps (Fig. 12E, F). New character.
44 Vacuoles within periostracum: (0) absent; (1) present. Vacuoles are present in the middle layer of the periostracum of Mytilus edulis (see Dunachie 1963) and also Arctica, Glauconome and Corbicula (Fig. 12G–I). New character.
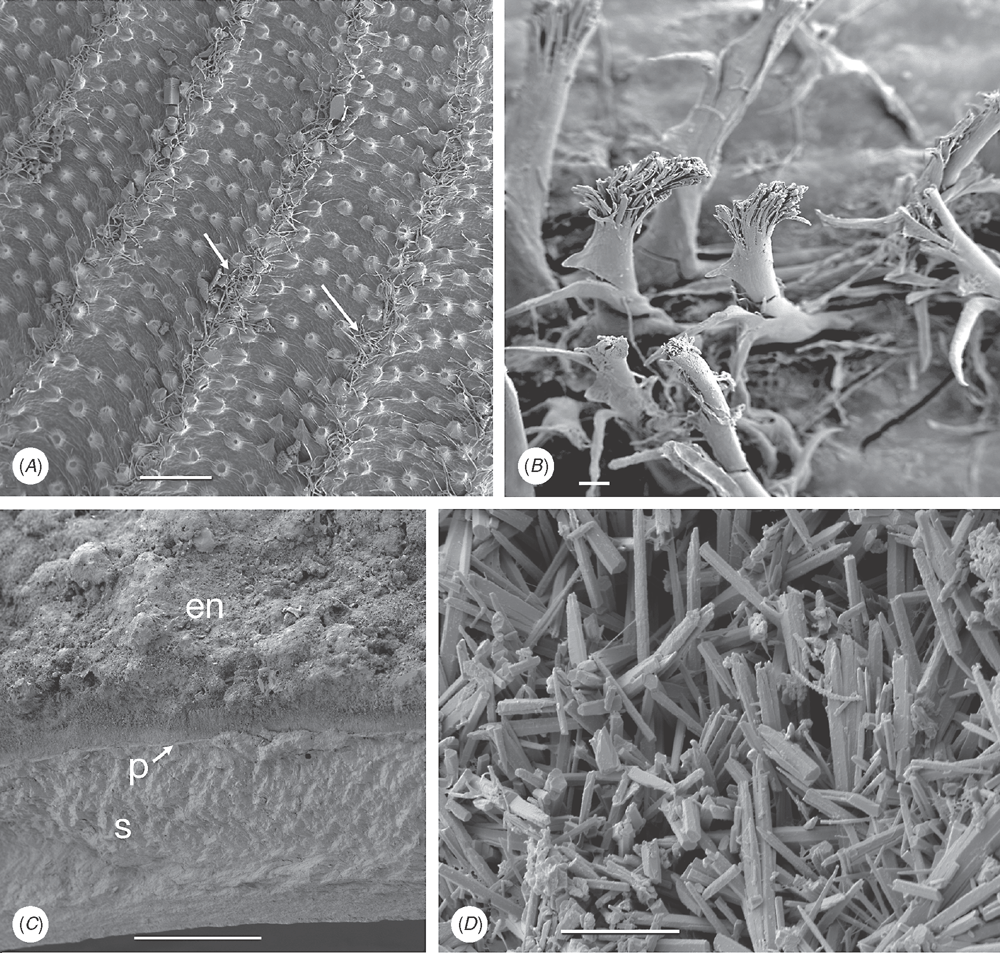
45 Intraperiostracal calcification: (0) absent; (1) small spikes (unionoid-type) (Fig. 13A); (2) large spikes (anomalodesmatan-type) (Fig. 13B); (3) granules (Figs 13C–E, 15H); (4) elongate pins and needles (venerid-type) (Fig. 13F–H); (5) rounded bosses (palaeoheterodont-type) (Fig. 13I). Glover and Taylor (2010) reported ‘needles and pins’ growing through the periostracum in Veneridae and their phylogenetic significance. Spikes growing within the periostracum are characteristic of many anomalodesmatans (Checa and Harper 2010) and unionoids (Zieritz et al. 2011). Intraperiostracal mineralised granules have also been recorded within some Mytilidae and Lucinidae. Although periostracal spikes were observed in several Recent and fossil gastrochaenids by Carter (1978), none was found in our exemplar species, Lamychaena hians, by the latter author or during this study. New character.
46 Arenophilic gland secretions: (0) absent; (1) present over entire shell; (2) present at posterior end only at siphons (e.g. in laternulids). Mantle-secreted adhesive threads arranged on the outer surface of the periostracum are present in most anomalodesmatans (Fig. 14A), usually associated with the adhesion of sediment particles to the shell (see Prezant 1981; Morton 1987). In most taxa (e.g. Lyonsia), they are arranged in radial rows over the entire valve surface, but in laternulids are restricted to the periostracum around the siphons (Sartori et al. 2006). This character was used previously by Harper et al. (2000: char. 14).
47 Applied byssal bristles: (0) absent; (1) present. Bristles on the external surface of the periostracum in certain mytilids (Fig. 14B) are not of mantle origin, instead originate from the byssal gland and are transferred to the periostracum by the foot (Ockelmann 1983). New character.
48 Extraperiostracal encrustations: (0) absent; (1) present. Although inorganic encrustations occur on many bivalves, in some taxa (e.g. Tellimya ferruginosa) they are formed by mantle and siphonal activity (Fig. 14C, D). New character.
49 Periostracal thickness: (0) <1 μm; (1) 1.01–3.16 μm; (2) 3.17–10 μm; (3) 10.1–31.6 μm; (4) 31.7–100 μm; (5) 100.1–316 μm. This continuous character was divided into states using the log scale of Harper (1997). New character.
Developmental and larval characters
50 Torsion: (0) absent; (1) present. This character was coded by Giribet and Wheeler (2002: char. 48).
51 Larval shell hinge (provinculum): (0) simple row of similar teeth; (1) differentiated dentition; (2) edentate. The larval shell hinge (provinculum) forms during the early stages of bivalve shell development. The hinge is generally straight but can have no teeth (edentate) or a few to many similar or differentiated teeth that can be diagnostic for species or families (Rees 1950; Yonge 1978; Le Pennec 1980; Lutz et al. 1982; Lutz 1985). Larval dentition does not necessarily correspond to dentition in the adult shell. A ligament is often associated with the hinge but is a separate structure. This character was coded by Giribet and Wheeler (2002: char. 38).
52 Prodissoconch I length: (0) <149 µm (suggesting planktotrophic development); (1) 150–229 µm (suggesting lecithotrophic development); (2) >229 µm (suggesting direct or brooded development). The larval shell, or prodissoconch, begins growing at the trochophore larval stage as a layer of periostracum secreted from the shell field gland (Eyster and Morse 1984). Prodissoconch I (PI) is complete when periostracum extends fully to cover the embryo and meets along the ventrum at the early veliger stage. This is frequently described as the ‘straight-hinge’ or ‘D-shaped’ larva. PI length is measured in μm as the longest anterior/posterior dimension of PI. Because length is a continuous character, it is divided into three states according to size ranges that, like egg sizes (Ockelmann 1965), have been taken to correspond to developmental mode (Thorson 1950; Jablonski and Lutz 1980, 1983). New character.
53 Prodissoconch II length: (0) prodissoconch II indistinct or absent (suggesting direct or brooded development); (1) <349 µm (suggesting planktotrophic development); (2) >349 µm (suggesting lecithotrophic development). In addition to a PI, most bivalves possess a prodissoconch II (PII) that begins forming after the left and right shell valves of PI have grown to meet at their margins. The PII forms during the veliger stage of development and usually corresponds to a period of feeding and growing in the plankton. Growth of PII (if present) continues into the pediveliger stage, at which stage the larva undergoes metamorphosis and the postlarval shell, or dissoconch, begins forming. The demarcation between PI and PII, and between PII and dissoconch, is usually abrupt and marked by a discontinuity or change in shell sculpture, and thus offers a means for measurement. Larval shell lengths are often reported in the literature but might not indicate the definitive size of the PII shell unless the measurements are taken at metamorphosis or from a postlarval shell also displaying dissoconch growth. Sizes reported from ‘spat’ or ‘settled larvae’ are assumed to represent definitive PII size. Bivalve larvae that do not display a PI–PII boundary are considered to lack the PII stage and are typical of species with protected development, either brooded or in some way encapsulated (Jablonski and Lutz 1980; Gustafson and Reid 1986) (but note discrepancies with protobranch bivalves; Gustafson and Reid 1986). New character.
54 PI/ PII ratio: (0) <0.50; (1) 0.50 or greater. The duration of larval development varies among species and those with a long planktonic period will exhibit a relatively larger growth of PI than those with a short planktonic period. This character is the ratio of PI size divided by PII size. Measurement of shell height (i.e. dorsoventral axis) would provide the most accurate ratio but most sizes reported in the literature are of shell length. Thus, the ratio for this character is calculated from shell length measurements approximately parallel to the hinge line. Coding is restricted to bivalves that show a PII. Lecithotrophic species generally have a ratio of 0.6 or higher and planktotrophic species of 0.4 or lower (Malchus 2004). New character.
55 Embryonic protection: (0) free swimming (unprotected); (1) brooded (in gills or mantle cavity); (2) encapsulated (attached or unattached egg case or capsule outside of parent). During development, larval bivalves can be free-swimming in the plankton, encapsulated in an egg case or capsule outside of the parent (either attached to the benthos or floating in the plankton), or brooded internally by the adult in the mantle cavity (usually the suprabranchial chamber) or chambers of the demibranchs (Zardus and Martel 2002). Brooding or protected development can occur for only a portion of the larval period; so for this character, development is only considered protected if it occurs during the veliger–PII (or equivalent) stage. New character.
56 Larval form: (0) veliger; (1) pericalymma; (2) glochidium; (3) stenocalymma; (4) direct development/no larva. Larval forms among the Bivalvia are more disparate than among the more speciose Gastropoda (Chanley 1968). A veliger larval form with prodissoconch and locomotory velum is typical of most bivalves although it can be modified in brooded forms (e.g. velum reduced or lacking). The pericalymma is typical of protobranch bivalves (Drew 1899; Gustafson and Lutz 1992; Zardus and Morse 1998) and bears resemblance to the scaphopod stenocalymma larva (Buckland-Nicks et al. 2002). The glochidium larval form (hooked or hookless) occurs among many freshwater mussels (Unionida) (Zardus and Martel 2002), and in other species a long-threaded lasidium larva (von Ihering 1891; Bonetto and Ezcurra 1965) is present. Direct development sensu stricto, wherein a larval stage is dispensed with altogether and the embryo develops into a miniature version of the adult, is extremely rare among the Bivalvia, but is common in other molluscs such as cephalopods (Korschelt and Heider 1858). ‘Lasidium’ would be an additional character but is not currently coded in the matrix. This character combines char. 174 and 181 from Giribet and Wheeler (2002), and adds new states.
57 Shape of larval velum: (0) velum absent; (1) oval; (2) bilobed. The velum is the locomotory structure of the veliger larva consisting of a retractable membranous disk with cilia (Cragg 1989) at the rim. When fully extended, the velum can be oval to round in outline or modified into lobes. A velum per se is not present in the larvae of protobranchs (Zardus 2002) but the external, ciliated test of the pericalymma could be a homologous structure. The velum is not otherwise known to be homologous with structures in other larval types. The shape of the velum is diagnostic for some species. New character.
58 Larval eye spot: (0) absent; (1) present. Eyespots are present in several developing bivalve larvae and can be diagnostic for some species (Miyazaki 1962; Chanley and Andrews 1971). Modified from Giribet and Wheeler (2002: char. 180).
59 Larval apical tuft: (0) absent; (1) present. Many bivalve larvae are described as having an apical plate with an apical sense organ (Morse and Zardus 1997). This usually consists of a bundle of cilia (apical tuft), although the cilia can be lacking. The apical tuft often forms at the trochophore stage and persists into the veliger stage. A prominent apical bundle of cilia also occurs in other molluscan larvae (e.g. pericalymma, stenocalymma and gastropod veligers) and is putatively homologous. This character was coded by Giribet and Wheeler (2002: char. 173).
60 Ciliation of larvae: (0) scattered; (1) organised bands. In veliger larvae, bands of cilia occur at the rim of the velum (Cragg 1989), whereas in pericalymma and other larval types that lack a velum, cilia can be organised into bands or scattered across the surface of the larva (Zardus and Martel 2002). New character.
Mantle and sense organ characters
61 Mantle lobes: (0) absent; (1) present. This character was coded by Salvini-Plawen and Steiner (1996) and by Giribet and Wheeler (2002: char. 50).
62 Ventral mantle fusion: (0) absent; (1) present. Ventral mantle fusion is present in scaphopods, mytilids, pteriids and most heterodonts (Salvini-Plawen and Steiner 1996). This character was coded by Giribet and Wheeler (2002: char. 51).
63 Siphons: (0) absent; (1) present. Siphons (posterior tube-like mantle fusions, as opposed to mantle openings or apertures) are present in a wide variety of bivalves, particularly the heterodonts. Some taxa (e.g. Turtonia; Mikkelsen et al. 2006) have only one of the siphons developed and are coded here as ‘siphons present’. Most siphons are equipped with siphonal retractor muscles that leave pallial sinus scars on the inner valves, although in some families (e.g. Pandoridae; Mikkelsen and Bieler 2007), these are less well defined. This character was also coded by Giribet and Wheeler (2002: char. 55).
64 Swimming capacity through valval movement: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 182).
65 Sensory mantle tentacle: (0) absent; (1) present. According to Waller (1998), a single retractile tentacle developed from the middle fold of the mantle in the region of the siphonal embayment is a unique feature of Nuculanoidea. It is apparently absent only in Nuculanidae and in some members of Tindariidae, but is present in all other nuculanoidean taxa (Brooks 1875; Yonge 1939; Allen and Sanders 1982, 1996; Boss 1982; Allen and Hannah 1989). However, it was not seen in our own material of Clencharia, in contrast to the findings of Yonge (1939).
66 Stempell’s organ: (0) absent; (1) present. This tube-shaped organ is situated immediately dorsal to the anterior adductor muscle of some protobranchs (Nucula nucleus, N. delphinodonta and N. sulcata; Stempell 1898; Drew 1901; Haszprunar 1985). Stempell’s organ has also been observed in Acila castrensis (Kurt Schaefer, pers. comm., 2001). It is absent in Malletia inequalis (proxy used for M. abyssorum) and Nuculana pernula (O. Israelson, pers. obs., 1999). This character was also coded by Giribet and Wheeler (2002: char. 136).
Muscles, foot and pedal gland characters
67 Ventral surface (sole) of the foot: (0) present; (1) absent. This character was discussed by Salvini-Plawen and Steiner (1996: 43), Waller (1998: 21), and Giribet and Wheeler (2002: char. 110).
68 Heel of foot: (0) absent or weakly developed as a posteriorly directed, triangular projection of the margin of the sole, but not separated from the sole; (1) distinct and sharply separated from the sole. The members of Nuculidae have a distinct heel sharply separated from the sole (Sanders and Allen 1973). This character was also coded by Giribet and Wheeler (2002: char. 113).
69 Byssus (in adult): (0) absent; (1) present. Giribet and Wheeler (2002: chars 114–115) used two characters to code for the presence of a posterior pedal gland (following Salvini-Plawen and Steiner 1996), and for the retention or loss of a byssus in the adult. Here, in the absence of anatomical data for the presence of pedal gland, we simply coded for the presence or absence of a byssus. Coding is restricted to bivalves with a foot in the adult.
70 Ontogenetic loss of foot immediately after settlement: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 116).
71 Pedal reversal: (0) absent); (1) present. All known extant limoids have a unique foot that is rotated 180 degrees, affecting the pedal nerves (e.g. Seydel 1909; Stuardo 1968; Gilmour 1990).
72 Cruciform muscle: (0) absent; (1) present. The cruciform muscle is a cross-shaped muscle at the base of the incurrent siphon in Tellinoidea (and recognised as a synapomorphy of that superfamily; Yonge 1949) that absorbs the physical strain experienced when the siphons extend and retract. New character.
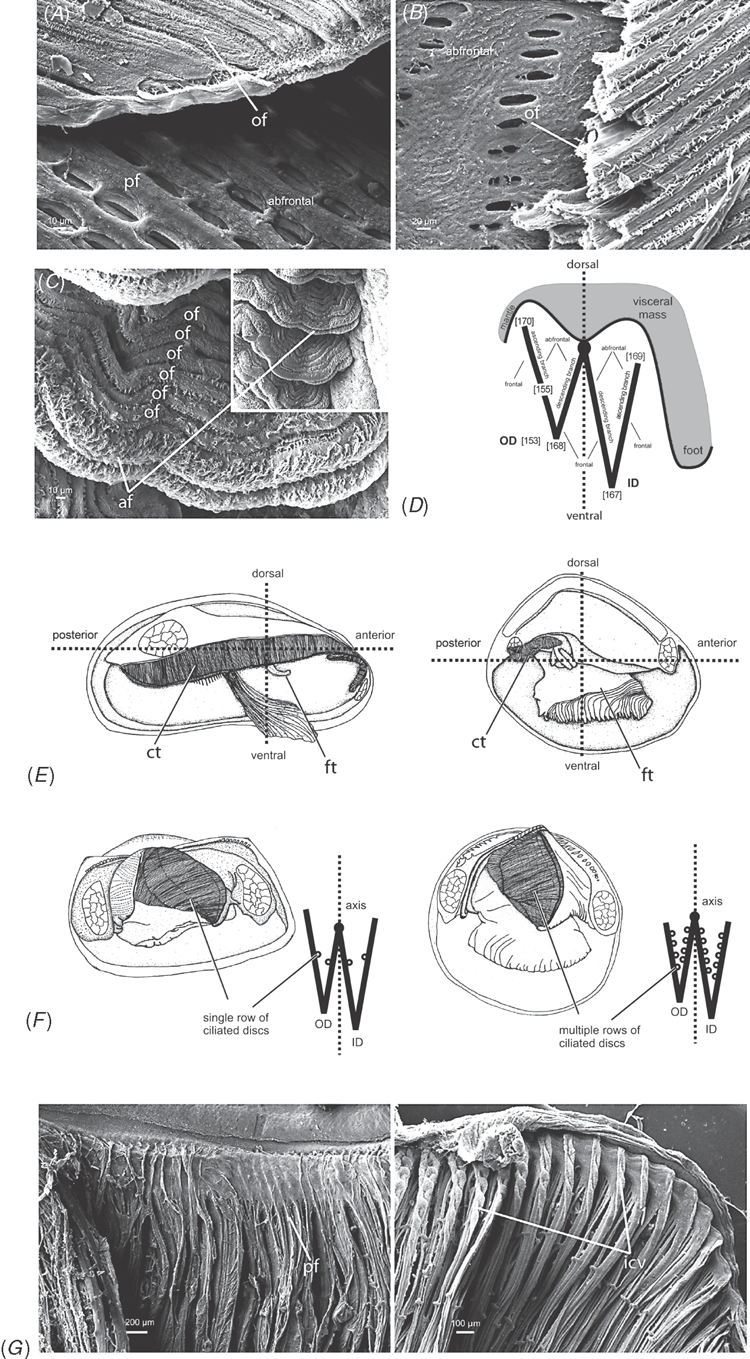
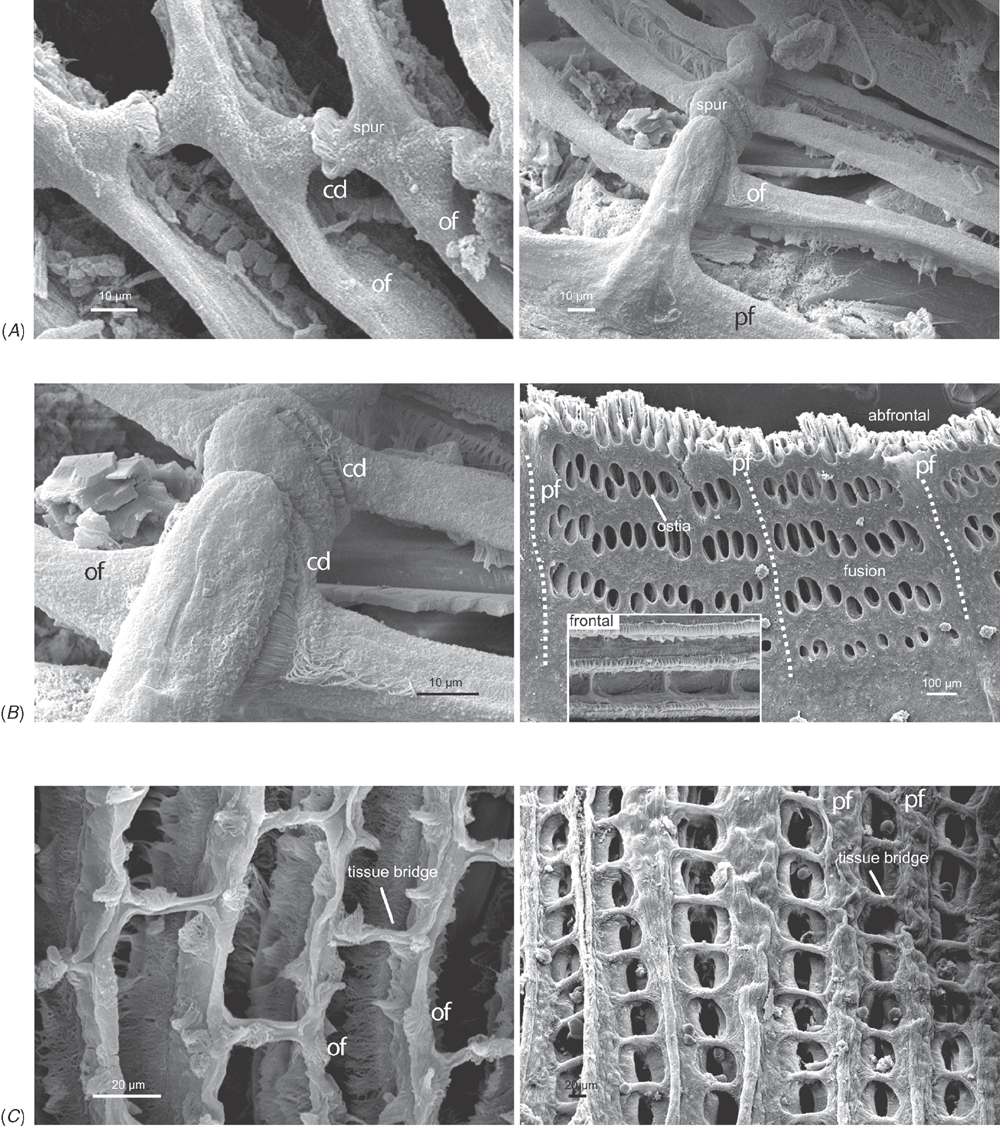
Alimentary system characters (Figs 16–18)
|
The gastric chamber (‘stomach’) is one of the most complex and character-rich organs in bivalves and has been extensively described (e.g. Graham 1949; Owen 1955, 1956; Reid 1965; Dinamani 1967). These early studies were greatly expanded by R. D. Purchon in a series of publications that established five, now ‘classic,’ stomach types (Purchon 1956, 1957, 1958, 1959, 1960, 1985, 1987a, 1990). The stomach types are idealised categories characterised by presumably unique combinations of several key features on the inner surface of the gastric chamber: (1) the number of ducts of digestive diverticula and the degree of their consolidation; (2) the course of the major and minor typhlosoles, the intestinal groove, and their association with the ducts of the digestive diverticula; (3) the presence and position of sorting areas; and (4) the presence of specialised compartments (e.g. dorsal hood, left pouch, food-sorting caecum). The stomach types largely reflect the diversity of feeding modes in bivalves: deposit-feeding (Type I), carnivory (Type II) and filter-feeding (Types III, IV and V).
Most prior phylogenetic analyses based on morphological data subsumed gastric chamber characters under Purchon’s stomach types and coded them as such (e.g. Purchon 1978; Waller 1978; Lee 2004; Graf and Cummings 2006); other studies either used a relatively small set of alimentary system characters (e.g. Hoagland and Turner 1981; Schneider 1995; Korniushin and Glaubrecht 2002; Simone and Chichvarkhin 2004; Tëmkin 2006; Simone and Wilkinson 2008) or included both a stomach type character with finer-grained characters in a single matrix (e.g. Schneider 1992; Dreher Mansur and Meier-Brook 2000; Harper et al. 2000; Giribet and Wheeler 2002). In this analysis we have attempted to re-evaluate homologies of the complex internal modifications of the gastric chamber based on more stringent and consistent positional criteria. As a result, several previously defined characters were revised and novel characters were defined.
The intestine is not as character-rich a system as the gastric chamber. Relevant characters primarily concern aspects of intestinal coiling, and the relationship of the intestine with the style sac and the pericardial cavity. In addition, several characters pertain to the distal-most part of the intestine: its association with the posterior adductor muscle and the presence of the anal funnel. Only adults were used for coding so any co-variation of some alimentary system characters with size would not reflect ontogenetic variation.
73 Digestive tract: (0) present; (1) reduced. The digestive tract is primitively present in Bivalvia but greatly reduced in complexity in species of Solemyidae (Reid 1980; Reid and Bernard 1980), some Nucinellidae (but see discussion in Oliver and Taylor 2012) and some cardiids. This character was also coded by Giribet and Wheeler (2002: char. 103).
74 Radular apparatus: (0) absent; (1) present. Salvini-Plawen (1988) considered loss of the radular apparatus and associated glands and ganglia to be a synapomorphy of the Bivalvia. This character was also coded, in part, by Giribet and Wheeler (2002: char. 85).
75 Style: (0) absent; (1) protostyle; (2) crystalline style. This character was also coded by Giribet and Wheeler (2002: char. 89) and, in part, by Simone (2009: char. 45).
76 Gastric chamber muscular envelope: (0) absent; (1) present. The muscular envelope is a modified wall of the gastric chamber that is considerably thickened and internally longitudinally folded. The muscular envelope is a characteristic feature of carnivorous bivalves (Yonge 1928; Purchon 1956, 1987a). New character.
77 Antechamber: (0) absent; (1) present. The antechamber is a compartment at the anterior part of the stomach formed by the expansion of the stomach lumen anterior to the gastric shield into which the oesophagus opens. In bivalves in which an antechamber is absent, the gastric shield is found immediately proximal to the oesophageal opening. Purchon (1987a) applied this term to describe this structure in Chamidae, in which it is particularly prominent. The antechamber has been referred to as the ‘anterior chamber’ in oysters (Galtsoff 1964) and ‘capacious vestibule’ in Chamidae (Allen 1976). New character.
78 Antechamber position: (0) dorsal to main chamber; (1) anterior to main chamber. When the antechamber is present, it is positioned either dorsally or anteriorly relative to the main stomach chamber. The axes of the main stomach chamber are defined by the stable, relative arrangement of: the entry of the intestine/style sac, and the position of the gastric shield and the dorsal hood. New character.
79 Oesophageal longitudinal folds: (0) absent; (1) present. A character signifying the presence of oesophageal ridges was used in a higher-level analysis of the Bivalvia by Salvini-Plawen and Steiner (1996), but the structure and its variations were neither illustrated nor discussed. This character was also coded by Giribet and Wheeler (2002: char. 86) and Tëmkin (2006: char. 31).
80 Dorsal hood position relative to the gastric shield: (0) anterodorsal; (1) posterodorsal; (2) dorsal. The dorsal hood extends either anteriorly or posteriorly relative to the gastric shield in right lateral view. New character.
81 Oesophageal entry into gastric chamber lumen: (0) dorsal to the gastric shield; (1) ventral to the gastric shield. The oesophagus can open to the gastric chamber lumen either dorsal or ventral to the gastric shield. This character is applicable to taxa with the gastric shield is displaced anteriorly, so that it originates from an extension of the left wall that is continuous with the anterior wall of the gastric chamber. New character.
82 Oesophageal lip: (0) absent; (1) present. The oesophageal lip is a conspicuous thickened rim sharply demarcating the entry of the oesophagus into the stomach lumen. New character.
83 Fold-and-ridge area of food-sorting caecum: (0) absent; (1) present. This character corresponds to the Sorting Area 1 (‘SA1’) in Purchon’s (1956) system. This character was also coded, in part, by Giribet and Wheeler (2002: char. 96).
84 Ventral extension of food-sorting caecum: (0) absent; (1) present. A ventral expansion of the food-sorting caecum can form an extensive, posteriorly directed or conspicuous, coiled diverticulum. It has previously been described as ‘ventral diverticulum’ (Sabatier 1877; White 1937). The possible function of this ‘blind sac’ was discussed by Nelson (1918). New character.
85 Posterior wall of food-sorting caecum (fleshy fold): (0) not expanded; (1) with posterior slender process; (2) widening posteriorly; (3) widening posteriorly producing digitate folds. The fleshy fold (Purchon 1957) has been variously described by other authors as ‘the left wall of the caecum’ (Graham 1949), an internally projecting ‘longitudinal ridge’ (Nakazima 1958), and ‘the axial fold’ (Dinamani 1967). Purchon (1985, 1987a) later substituted his earlier term ‘fleshy fold’ for ‘(fleshy) buttress’. This character was also coded by Tëmkin (2006: chars 33 and 34).
86 Dorsal hood groove: (0) absent; (1) present. Based on drawings by Reid (1965: 162–163, figs 3, 4), the structure as defined here is a groove passing from the food sorting caecum to the oesophagus and joining a groove on the right wall leading over the stomach roof into the dorsal hood. It apparently links the apex of the major typhlosole with the dorsal hood (Purchon 1987a: 194, fig. 3). New character.
87 Food-sorting caecum: (0) absent; (1) present. The food-sorting caecum (referred to simply as ‘caecum’ by Graham 1949), is a dorsoventrally expanded pocket in the anterior part on the left wall of the stomach. It is typically bordered posteriorly by the fleshy fold and is invested by the major typhlosole. New character.
88 Left pouch: (0) absent; (1) present. The left pouch is an embayment from the left stomach wall that is partly invested by a lobe of the gastric shield and typically receives one or more ducts of the digestive diverticula (Purchon 1954: 29). New character.
89 Left pouch sorting area: (0) absent; (1) present. This character corresponds to the Sorting Area 6 (‘SA6’) in Purchon’s system. It is located either entirely inside the left pouch or immediately proximal to it. New character.
90 Fold(s)/groove(s) connecting left pouch and dorsal hood: (0) absent; (1) present. The left pouch and the dorsal hood can communicate via fold(s)/groove(s) that extend along the left wall of the gastric chamber. New character.
91 Blind symmetrical shallow pockets lateral to the oesophagus: (0) absent; (1) present. Hemispheric blind pockets located more or less symmetrically to the left and right of the oesophageal opening were reported in Protobranchia (Purchon 1956: 514–515). New character.
92 Left pouch duct field: (0) absent; (1) present. The left pouch duct field is a single duct or multiple ducts of the digestive diverticula opening to the interior of the left pouch. New character.
93 Dorsal hood: (0) absent; (1) present. The dorsal hood is a distinct, blind, frequently grooved pouch that opens mid-dorsally or dorsolaterally to the left from the roof of the stomach (Purchon, 1954: 28). It corresponds to the ‘dorsal pouch’ of Graham (1949). This character was also coded by Giribet and Wheeler (2002: char. 104), and Simone and Wilkinson (2008: char. 35).
94 Right posterodorsal appendix: (0) absent; (1) present. The appendix is a narrow outpocketing extending from the posterodorsal region of the stomach roof; it presumably functions as a temporary storage compartment for food (Purchon, 1955: 886, 887; Reid, 1965: 160). New character.
95 Right wall forked embayment: (0) absent; (1) present. The forked embayment, with a complex sorting area, is formed by two extensive, interconnected, blind, conical pockets on the right stomach wall in some Unionida (Purchon 1987a: 216–219, fig. 8). New character.
96 Longitudinal folds and grooves of the dorsal hood: (0) absent; (1) present. This character refers to regular, longitudinal folds and grooves on the interior surface of the dorsal hood. New character.
97 Transverse folds and grooves of the dorsal hood: (0) absent; (1) present. This character refers to regular, transverse folds and grooves on the interior surface of the dorsal hood. New character.
98 Typhlosoles: (0) absent; (1) present. Typhlosoles are two longitudinal ridges that separate the intestine from the style sac but allow communication between them throughout their extent (Nelson 1918). When present, these structures vary in size and position among species, with one typhlosole (the major typhlosole) typically larger than the other (the minor typhlosole), and may extend into the lumen of the gastric chamber to different degrees. New character.
99 Major typhlosole projecting into gastric chamber lumen: (0) not projecting; (1) projecting. This character was also coded by Giribet and Wheeler (2002: char. 91).
100 Major typhlosole and anterior duct field: (0) curving posterior to duct field; (1) spiraling posterior to duct field; (2) terminating immediately proximal to duct field; (3) entering left vestibule; (4) entering left and right vestibules; (5) entering food-sorting caecum. This character was also coded, in part, by Schneider (1992: char. 16; 1998: char. 14), Dreher Mansur and Meier-Brook (2000: char. 25), Giribet and Wheeler (2002: chars 92, 93, 100) and Korniushin and Glaubrecht (2002: char. 42).
101 Major typhlosole path inside left vestibule: (0) terminating upon entry; (1) penetrating deeply; (2) recurving (but not exiting left vestibule); (3) spiraling; (4) recurving and terminating upon exit of left vestibule; (5) undulating. This character was also coded, in part, by Giribet and Wheeler (2002: chars 92, 93, 100).
102 Intestinal groove: (0) consisting of single groove; (1) consisting of multiple parallel grooves. The intestinal groove is a ciliated rejection tract bounded by the typhlosoles of the style sac. The groove can project along the major typhlosole into the gastric chamber. New character.
103 Tongue of major typhlosole diving into ducts of digestive diverticula: (0) absent; (1) present. This character was also coded by Tëmkin (2006: char. 36).
104 Path of major typhlosole inside food-sorting caecum: (0) major typhlosole not reaching apex of food-sorting caecum; (1) major typhlosole reaching apex of food-sorting caecum. When entering the food-sorting caecum, the major typhlosole either reaches or does not reach its apex. New character.
105 Low ridges circumnavigating major typhlosole: (0) absent; (1) present. The low ridges circumnavigating the major typhlosole within the food-sorting caecum comprise the Type B sorting mechanism of Reid (1965). New character.
106 Major typhlosole and embayment of right anterior stomach gastric chamber wall: (0) typhlosole not entering embayment; (1) typhlosole entering embayment. When the right wall embayment is present, the major typhlosole either invades the embayment or by-passes it. New character.
107 Minor typhlosole path in stomach gastric chamber lumen: (0) terminating upon entry; (1) extending deep into lumen. The minor typhlosole typically terminates upon entry into the lumen of the gastric chamber but in some species extends deeply into the lumen forming a distinct ridge or ledge. New character.
108 Inter-typhlosolar sorting area: (0) absent; (1) present. The principal sorting area in the Protobranchia is situated on the stomach floor between the major and minor typhlosoles. This feature corresponds to an unnumbered Sorting Area (‘SA’) in Purchon’s (1957) system. New character.
109 Right wall sorting area: (0) absent; (1) present. This feature corresponds to the Sorting Area 3 (‘SA3’) in Purchon’s (1956, 1957) system. This character was also coded, in part, by Korniushin and Glaubrecht (2002: char. 40).
110 Right wall sorting area position: (0) extending from right wall into dorsal hood; (1) restricted to dorsal hood; (2) restricted to right wall. The right wall sorting area varies in extent from occupying a large surface on the right wall of the gastric chamber and extending dorsally over the gastric chamber roof into the dorsal hood, to being restricted to either the dorsal hood or right wall. New character.
111 Pattern of transverse ridges of right wall sorting area: (0) simple transverse ridges; (1) alternating long and short ridges. The right wall sorting area is typically equipped with relatively evenly spaced, transverse, parallel ridges of uniform size. In some taxa, ridges of the sorting area are not uniform in length resulting in a pattern of alternating long and short ridges. New character.
112 Ridge posteriorly bordering right wall sorting area: (0) absent; (1) present. This character refers to a ridge on the right wall of the gastric chamber extending along the posterior border of the right wall sorting area. New character.
113 Ridge anteriorly bordering right wall sorting area: (0) absent; (1) present. This character refers to a ridge on the right wall of the gastric chamber extending along the anterior border of the right wall sorting area. New character.
114 Accessory right wall sorting area: (0) absent; (1) present. This feature corresponds to the Sorting Area 8 (‘SA8’) in Purchon’s (1960) system. New character.
115 Ridge anteriorly bordering accessory right wall sorting area: (0) absent; (1) present. This character refers to a ridge on the right wall of the gastric chamber extending along the anterior border of the accessory right wall sorting area. New character.
116 Right wall acceptance tract: (0) absent; (1) smooth; (2) with longitudinal grooves and ridges. This feature was defined as the acceptance tract by Owen (1953), corresponds to the anterodorsal tract of Reid (1965), and is possibly homologous to Sorting Area 5 (‘SA5’) of Purchon (1960). New character.
117 Anterior duct field: (0) absent; (1) with individual duct(s); (2) with ducts partially condensed into vestibules; (3) with ducts fused into vestibules. Ducts of digestive diverticula can either open into the stomach lumen directly or fuse, opening to the lumen via a large duct. In the most extreme condition, none of the ducts open into the stomach directly but condense, forming large caeca, here termed ‘vestibules’ to avoid confusion with other stomach outpockets that have been referred to as ‘caeca’. The vestibules generally correspond to the left and right caeca defined by Purchon (1955). This character was also coded, in part, by Giribet and Wheeler (2002: chars 97, 99, 101), Korniushin and Glaubrecht (2002: char. 42) and Tëmkin (2006: char. 37).
118 Posterior duct field: (0) absent; (1) with ducts scattered along right side; (2) with ducts scattered on both sides; (3) with ducts condensed on right side; (4) with ducts scattered along left side. This character was also coded, in part, by Giribet and Wheeler (2002: chars 97, 99, 101).
119 Right wall duct field: (0) absent; (1) present. The right wall duct field is a collection of ducts of the digestive diverticula that can be present on the right lateral and/or lateroventral surface of the gastric chamber. New character.
120 Gastric shield: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 90).
121 Gastric shield teeth: (0) absent; (1) present. The gastric shield tooth is a distinct projection of the gastric shield that presumably receives the projecting tip of the crystalline style. Gastric shield teeth are variable in shape and size. The gastric shield can be equipped with none, one, or several teeth. New character.
122 Cuticularisation of gastric chamber inner surface: (0) absent; (1) cuticle restricted to gastric shield area; (2) cuticle lining most of left side; (3) cuticle lining nearly entire inner surface. This character was also coded, in part, by Giribet and Wheeler (2002: char. 90).
123 Style sac and proximal intestine relationship: (0) merged; (1) confluent; (2) separated. The style sac and proximal intestine are considered merged when only a single opening leading to a common tract is present. The style sac and proximal intestine are considered confluent when two distinct tracts are present that communicate, at least partially, along their lengths. The style sac and proximal intestine are separated when they form completely independent, non-communicating tracts. New character.
124 Groove joining style sac and intestinal openings: (0) absent; (1) present. A groove on the floor of the gastric chamber can connect the openings of the style sac and the proximal intestine in those taxa where the orifices are separate. New character.
125 Style sac caecum: (0) absent; (1) present. The style sac caecum is a blind pocket at the junction of the style sac and the descending arm of the intestine. Coding is restricted to bivalves in which the style sac and intestine are confluent or merged. This character was also coded by Tëmkin (2006: char. 39).
126 Intestinal coiling: (0) no coiling; (1) loose (loop-like) coil; (2) tight (knot-like) coil. This character refers to relative degree of coiling rather than to the topology of coiling (see characters 127–128). Characters pertaining to different aspects of intestinal coiling were coded by Hoagland and Turner (1981: char. 109), Schneider (1998: char. 13), Hoeh et al. (2001: char. 5), Giribet and Wheeler (2002: char. 95), Korniushin and Glaubrecht (2002: char. 46), Graf and Cummings (2006: char. 41), Tëmkin (2006: chars 41–43), and Simone and Wilkinson (2008: char. 41).
127 Intestinal coil position relative to gastric chamber: (0) anteroventral; (1) posterior/posterodorsal. The intestinal coil typically occupies the visceral mass ventral to the gastric chamber but can be displaced posterior to the gastric chamber relative to the anteroposterior axis (as defined by Jackson 1890). New character.
128 Intestine looping over gastric chamber dorsally: (0) absent; (1) present. This character refers to the passage of the intestine dorsal to the gastric chamber roof. New character.
129 Style sac position: (0) produced anteriorly; (1) produced posteriorly; (2) style sac very short. This character was also coded, in part, by Schneider (1992: char. 15; 1998: char. 18).
130 Relative positions of rectum and pericardial cavity: (0) rectum penetrating ventricle; (1) rectum passing dorsal to ventricle; (2) rectum passing ventral to ventricle; (3) rectum passing lateral to ventricle. This character was also coded, in part, by Hoagland and Turner (1981: char. 75), Harper et al. (2000: char. 35) and Tëmkin (2006: char. 54).
131 Rectum terminus: (0) attached to posterior adductor muscle; (1) free-hanging papilla. This character was also coded by Tëmkin (2006: char. 45).
132 Anal funnel: (0) absent; (1) present. The anal funnel (Herdman 1904) has been referred to as an ‘anal process’ (Herdman 1904), ‘anal membrane’ (Ranson 1961) and ‘anal flag or ear’ (Pelseneer 1911). Its functional and systematic significance were reviewed by Tëmkin (2006). This character was also coded by Tëmkin (2006: char. 47).
Endosymbiont characters
133 Intracellular ctenidial bacteria constituting sulphide-oxidising symbiosis: (0) absent; (1) present.-The phenomenon of chemosymbiosis in bivalves was recently reviewed by Taylor and Glover (2010; see also Oliver and Taylor 2012; Oliver et al. 2013). Chemosymbiotic bivalves have been reported from seven distinct families: Solemyidae, Nucinellidae, Mytilidae, Lucinidae, Thyasiridae, Vesicomyidae and Montacutidae. The symbiosis has been identified in all species of Lucinidae, Solemyidae and Vesicomyidae studied so far and is likely obligatory (groundplan coding for these families is used herein), whereas in Thyasiridae, many species possess symbionts but others lack them. In Mytilidae, chemosymbiosis is confined to members of the subfamily Bathymodiolinae, whereas other mytilids are asymbiotic (Taylor and Glover 2010). This character was also coded by Giribet and Wheeler (2002: char. 146).
134 Symbiotic zooxanthellae: (0) absent; (1) present. Symbiotic zooxanthellae of the genus Symbiodinium have been found in all members of Tridacninae (Yonge 1981; Trench et al. 1981) and other members of Cardiidae (Kawaguti 1950; Ohno et al. 1995; Morton 2000; Kirkendale 2009), including Fragum unedo (see Carlos et al. 1999), but not in Cerastoderma edule. Zooxanthellae have also been found in Fluviolanatus subtortus, a species tentatively placed in Trapezidae (Morton 1982). This character was also coded by Giribet and Wheeler (2002: char. 147), and although uninformative in this matrix, it should be of value when more cardiids are added in the future. Another character, referring exclusively to Tridacninae (Giribet and Wheeler 2002: char. 148), was omitted here because our matrix does not include any members of this clade. Likewise, coding for the zooxanthella tube system (Giribet and Wheeler 2002: char. 149) was omitted, because it refers to a putative clade of Fragiinae + Tridacninae, but the observations of this system (Morton 2000) have not been made for any of our terminals.
135 Gland of Deshayes containing cellulolytic nitrogen-fixing bacteria: (0) absent; (1) present. Cellulolytic nitrogen-fixing bacteria have been isolated from the gland of Deshayes in numerous Pholadoidea, including Xylophaga (Popham and Dickson 1973; Waterbury et al. 1983; Distel and Roberts 1997) and are known to be transmitted vertically (Sipe et al. 2000). This character was also coded by Giribet and Wheeler (2002: char. 150), and although uninformative in the current matrix (only coded for Teredo clappi), we have retained it for future reference.
Gill & palp characters (Figs 19–22)
Gills (ctenidia) are the main bivalve respiratory organ and, in most cases, are also suspension-feeding organs. These complicated, often extensive, structures have been long and broadly studied, and formed the basis of early higher-level classifications (Pelseneer 1891; Ridewood 1903). Bivalve gills have been categorised into several useful functional types (protobranch, lamellibranch, filibranch, septibranch). Other ‘condition’ terms in use to define gills are homorhabdic/heterorhabdic (having one or more than one type of filament, respectively) and eutherorhabdic/synaptorhabdic (having filaments connected by ciliated disks or tissue junctions, respectively). Stasek (1963) categorised the kinds of associations of labial palps with the anterior gill filaments. Although useful in general descriptions, such categories and ‘types’ can be formed of multiple, potentially useful, individual phylogenetic characters, and emphasis has been placed here on breaking these down into their presumably homologous parts. Ciliation on the gill filaments can be complex and has also been studied extensively (e.g. Atkins 1937; Owen 1978); four main types of cilia (frontal, lateral, laterofrontal, abfrontal) have been defined.
136 Labial palps: (0) absent; (1) present. This character describes the presence of bipartite lateral expansions of the mouth, termed labial palps. Labial palps are generally symmetrical (excluding several anomalodesmatans; see char. 137), and consist of an inner and an outer labial palp that each bear ridges on their inner surfaces. Labial palps sort food particles delivered by the palp proboscides (Protobranchia) or the gills (Autobranchia) and transport the food particles to the mouth. Bivalve labial palps differ from other molluscan mouth extensions, such as labial tentacles, labial lobes or the gastropod lip organ, in their complex ciliated ridge structure. This character was also coded by Giribet and Wheeler (2002: char. 79).
137 Symmetry of labial palps: (0) symmetrical; (1) asymmetrical. Left and right labial palps are symmetrical in most bivalve species, with exceptions within the anomalodesmatans in which the palps often differ in size and shape. This character does not refer to symmetry between the inner and outer palps on a single side. New character.
138 Relative position of labial palps (Fig. 19A): (0) lateral to visceral mass; (1) close to anterior region of visceral mass. In protobranchs that use palp proboscides (and not the gills) for particle delivery, the labial palps are larger than the gills. A lateral position supports the function of the palp proboscides and might be necessary due to the overall large size of the palps (Yonge 1939; Zardus 2002). New character.
139 Palp proboscides: (0) absent; (1) present. A palp proboscis is an elongated tentacle-like structure situated dorsoposteriorly near the labial palps. It provides the labial palps with food particles by transporting them from the surrounding substratum to the palp pouch or the palps directly. Palp proboscides only occur in the Protobranchia. This character was also coded (as palp appendages) by Giribet and Wheeler (2002: char. 81).
140 Palp pouch: (0) absent; (1) present. This is an unpaired concave structure, also called palp caecum, at the posterior end of the labial palps that receives food particles from the palp proboscides. The food particles are transported from the palp pouch to the labial palps for sorting. This character was also coded by Giribet and Wheeler (2002: char. 82).
141 Labial palp shape (Fig. 19B): (0) triangular; (1) semicircular; (2) falciform. Coding is restricted to the Autobranchia, and describes the general geometric shape of the labial palps. Differentiation between ‘triangular’ and ‘falciform’ is based on labial palp length–width ratio; a labial palp more than three times longer than its maximum width is considered falciform. This character was coded (in modified form) by Hoeh et al. (2001: char. 26), Roe and Hoeh (2003: chars 51–53) and Graf and Cummings (2006: char. 26).
142 Anterior/posterior differentiation of the labial palps (Fig. 19C): (0) absent; (1) present. This character describes the subdivision of the labial palps into a posterior expanded area (with prominent ridges) and an anterior grooved narrow area (with less prominent or no ridges) that connects the posterior part to the mouth opening. The differentiation is often present within the Pteriomorpha. New character.
143 Outer labial palp tip (Fig. 19D): (0) free; (1) not free. This character describes whether the dorsoposterior tip of the outer labial palp is free within the mantle cavity. Because the homology of protobranch palp tips remains unclear, coding is restricted to the Autobranchia. A similar character, but also including size, was also coded by Monari (2009: char. 46).
144 Outer labial palp tip shape: (0) angled; (1) rounded. This character describes the free edge of the outer labial palp, as described by Thiele (1886) and also coded by Tëmkin (2006: char. 24). Because homology of the palp tips in protobranchs is unclear, coding is restricted to the Autobranchia.
145 Hood formed by labial palps: (0) absent; (1) present. This character decribes the formation of a hood-like structure by the labial palps by a ventral, non-anatomical connection of the left and right labial palps. This character is present only in the Autobranchia. New character.
146 Gill–palp association (insertion): (0) absent; (1) present. This character describes the insertion of the ventral tips of the anterior filaments of the inner demibranchs into the distal oral groove. It was used by Tëmkin (2006: char. 26) to distinguish between gill–palp association categories I/II (present) and III (absent) of Stasek (1963).
147 Gill–palp association (fusion) (Fig. 19E): (0) not fused; (1) fused. This character accommodates the fused insertion of the ventral tips of the anterior filaments of the inner demibranchs. Coding is restricted to those bivalves exhibiting the former categories I (not fused) and II (fused) of Stasek (1963). This character has not been used as a separate character (from char 152) in previous phylogenetic analyses.
148 Palp lamella division ridge (Fig. 20A): (0) absent; (1) present. This character accommodates the presence of a ridge on the inner surface of the inner and outer labial palps, dividing the inner surface into dorsal and ventral partitions. The palp lamella division ridge as described by Morse and Zardus (1997) for Acila castrensis is a very prominent structure that has only been found among protobranchs. New character.
149 Ctenidia: (0) absent; (1) present. Gills with alternating leaflets or filaments occur in all molluscan classes except Scaphopoda and Solenogastres (Waller 1998; Reynolds and Okusu 1999). Numerous modifications and loss of one or both ctenidia have occurred within the Gastropoda. A modified version of this character was coded by Giribet and Wheeler (2002: char. 64).
150 Ctenidial structure: (0) leaflets; (1) filaments. This character is the first in a series (chars 150–156) replacing the ‘gill types’ postulated by Ridewood (1903), based primarily on the relationship of the outer and inner demibranchs (see Fig. 20C). This series of characters has not been used in this configuration in previous phylogenetic analyses.
151 Ctenidial leaflet orientation: (0) horizontal; (1) vertical. See character 150.
152 Demibranch relations (Fig. 20B): (0) outer demibranch not overlaying inner demibranch; (1) outer demibranch partially overlaying inner; (2) outer demibranch completely overlaying inner. See character 150.
153 Outer demibranch: (0) present; (1) absent. See character 150.
154 Inner demibranch reflection: (0) reflected; (1) not reflected. See character 150. The character of demibranch reflection was also coded by Giribet and Wheeler (2002: char. 66), but without the distinction between outer and inner demibranch.
155 Outer demibranch reflection: (0) reflected; (1) not reflected. See characters 150 and 161.
156 Outer demibranch (relative size of lamellae) (Fig. 20B): (0) ascending lamella less than descending lamella; (1) ascending lamella greater than descending lamella. See character 150.
157 Outer demibranch (keel) (Fig. 20B): (0) present; (1) absent. This character describes the presence of a keel on the ascending lamella of the outer demibranch. This character has not been used in previous phylogenetic analyses. New character.
158 Demibranch length: (0) both demibranchs of subequal length; (1) outer demibranch distinctly shorter than inner demibranch. This character describes the relative lengths of the inner and outer demibranchs. This character has not been used in previous phylogenetic analyses. New character.
159 Ctenidial shape: (0) short and small; (1) short and wide; (2) elongate. This character has not been used in this configuration in previous phylogenetic analyses. Used (in modified form) by Graf and Cummings (2006: char. 26).
160 Ctenidial symmetry: (0) symmetrical; (1) asymmetrical. The ctenidia of the left and right side are symmetrical in most bivalves, with exceptions within some of the anomalodesmatans, in which they differ in size and shape.
161 Demibranch origin (Fig. 20B, D): (0) at approximately the same dorsoventral level; (1) outer demibranch originating more dorsally than inner demibranch. The origin of the demibranch is separated by a membrane (see Fig. 20D). New character.
162 Inner demibranch anatomical fusion: (0) not fused; (1) fused. This character accommodates the partial anatomical (tissue) fusion of the right and left ascending lamellae of the inner demibranch at the inner (dorsal) margin. Because the foot interferes with this connection, this character is determined within the area posterior to the foot. This character has not been used in previous phylogenetic analyses. New character.
163 Ctenidial leaflet shape: (0) square-like; (1) V-like. In square-like leaflets, the dorsal, lateral and ventral margins of each leaflet are of similar length. In V-like leaflets, the ventral and dorsal margins are significantly longer than the lateral margin. New character.
164 Leaflets laterally broadened (Fig. 20C): (0) present; (1) absent. In some bivalves, the sides of the gill leaflets are distinctly broadened and lack frontal cilia, with ciliary junctions between the leaflets. New character.
165 Gill position: (0) anterior to posterior adductor muscle; (1) extending posterior of posterior adductor muscle. This character was coded in modified form by Monari (2009: char. 40).
166 Posterior inner demibranch connection: (0) absent; (1) ciliary; (2) anatomical (tissue). This character describes the connection of the posteriormost filaments of the right and left ctenidia to each other. If the posterior tips of the ctenidia are completely separated, the character is coded as absent; this can be observed in most of the Protobranchia and several Pteriomorpha. The second character state describes a posterior ciliary joining of the ctenidia (e.g. within the Solemyidae), and the third state codes a tissue connection. This character was defined and also coded by Tëmkin (2006: char. 10).
167 Inner demibranch food groove: (0) absent; (1) present. This character accommodates the presence of a food groove on the ventral margin of the inner demibranch. This character has never been used separate from character 168 in previous phylogenetic analyses. New character.
168 Outer demibranch food groove: (0) absent; (1) present. This character accommodates the presence of a food groove on the ventral margin of the outer demibranch. This character was also coded by Tëmkin (2006: char. 14).
169 Inner demibranch/visceral mass connection: (0) absent; (1) ciliary; (2) anatomical (tissue). This character has not been used in this configuration in previous phylogenetic analyses. Used (in modified form) by Graf and Cummings (2006: char. 24).
170 Outer demibranch/mantle connection: (0) absent; (1) ciliary; (2) anatomical (tissue). This character was defined and also coded by Tëmkin (2006: char. 13).
171 Interlamellar junctions: (0) absent; (1) multiple cross bars; (2) dorsal septa; (3) complete septa. This character was defined and also coded by Tëmkin (2006: char. 15).
172 Separation between frontal and laterofrontal cilia (Fig. 20D): (0) present; (1) absent. This character accommodates a clear separation of the frontal cilia and the laterofrontal cilia on the front edge of leaflets. Coding was restricted to members of Protobranchia. New character.
173 Laterofrontal gill cilia: (0) absent; (1) present. This character was also coded by Giribet and Wheeler (2002: char. 75).
174 Laterofrontal cilia composition: (0) eulaterofrontal with prolaterofrontal cilia; (1) microlaterofrontal; (2) anomalous with paralaterofrontal cilia. This character was also coded by Giribet and Wheeler (2002: char. 76).
175 Abfrontal cilia: (0) present; (1) absent. Abfrontal cilia are those on the inner surfaces of the gill filaments. This character was also coded by Giribet and Wheeler (2002: char. 77).
176 Interlocking cilia: (0) absent; (1) present. This character accommodates the presence of elongated cilia on both sides of the lateral edge of gill leaflets, interconnecting the leaflets. Yonge (1939) postulated that the interlocking cilia were modified lateral cilia. Coding is restricted to members of Protobranchia. This character has not been used in this configuration for previous phylogenetic analyses. New character.
177 Chitinous rods: (0) absent; (1) present. These are chitinous filament rods with structural enlargement at the base of the filament. This character was also coded by Giribet and Wheeler (2002: char. 72).
178 Principal filaments (Fig. 21A): (0) absent; (1) present. This character, together with character 179, represents different states of filament differentiation within bivalve gills (an additional character, presence of ordinary filaments, will become relevant with larger taxon sampling). These characters have been used in phylogenetic analyses, but with a different definition, for example as ‘homorhabdic’ or ‘heterorhabdic’ states of ctenidial filament morphology (Graf and Cummings 2006: char. 18), and as states of a filament differentiation character (Tëmkin 2006: char. 17). These characters have not been used in this configuration in previous analyses.
179 Apical filaments: (0) absent; (1) present. See character 178.
180 Principal filament distribution (Fig. 20E): (0) on entire gill; (1) on posterior part of gill only. This character describes the presence of principal filaments over the full length of the gill. In character state 1, principal filaments are only present in the posterior two-thirds of the gill and are lacking in the anterior third. New character.
181 Mantle gills: (0) absent; (1) present. Mantle (pallial) gills were described in detail by Taylor and Glover (2000). These gill-like structures are located on the inner mantle surface ventral to the anterior adductor muscle. New character.
182 Septum: (0) absent; (1) present. This character accommodates the incorporation of the gills into a muscular branchial septum (in septibranchs). This character has been used in similar configuration by Giribet and Wheeler (2002: char. 61).
183 Portion of the mantle cavity occupied by gills (Fig. 21E): (0) both lateral and posterior to the foot; (1) posterior to the foot only. This character was also coded by Giribet and Wheeler (2002: char. 63). Coding is restricted to molluscs with one pair of gills.
184 Interconnecting vessels (Fig. 21G): (0) absent; (1) present. This character describes the presence of interconnecting vessels on the abfrontal side of the principal filaments of the descending branches of the inner and outer demibranchs. These structures were described in detail by Veniot et al. (2003) for Placopecten magellanicus, but have never been used in a phylogenetic analysis. During this study, we found interconnecting vessels in an additional pteriomorph species (Spondylus ambiguus), allowing this character’s use in a phylogenetic analysis for the first time. New character.
185 Filamental spurs (Fig. 22A): (0) absent; (1) present. Filamental spurs are tissue extensions on the filaments on which the ciliated discs are located. This character is one of a series describing the interfilamental junctions within the Bivalvia (chars 185–190). These structures were described in detail by Veniot et al. (2003) for Placopecten magellanicus, and were also coded in modified form by Tëmkin (2006: char. 19).
186 Ciliated discs (Fig. 22B): (0) absent; (1) present. See character 185. Ciliated discs serve as one kind of interfilamental junctions. Because homology of the ciliated discs of the Protobranchia is questionable because of their location on the leaflets, coding is restricted to the Autobranchia. This character was also coded by Tëmkin (2006: char. 18).
187 Ciliated disc configuration (Fig. 21F): (0) single; (1) multiple. See character 185. This character distinguishes between the more common state of multiple rows of ciliated discs along the ctenidium (e.g. Glycymeris glycymeris) and only a single row on each branch of the inner and outer demibranchs (e.g. Arcopsis adamsi). New character.
188 Abfrontal filamental fusion (Fig. 22B): (0) absent; (1) present. See character 185. This character describes the presence of a membrane on the abfrontal side of the gill lamellae, creating a permanent tissue connection between filaments. The membrane bears openings (ostia) to allow water flow. This membrane is not correlated with the absence or presence of principal filaments. New character.
189 Interfilamental tissue bridges (ordinary filaments) (Fig. 22B, C): (0) absent; (1) present. See character 185. This character describes conditions of interfilamental junctions between ordinary and prinicipal filaments. Although the presence of tissue bridges does not preclude the presence of an abfrontal filamental fusion (char. 188), the co-occurrence of tissue bridges and ciliated discs (char. 186) has not been observed. Although interfilamental tissue bridges were used in phylogenetic analyses to specify gill types by, for example, Giribet and Wheeler (2002: char. 68), this character as defined here has not been used in previous phylogenetic analyses.
190 Interfilamental tissue bridges (principal filaments): (0) absent; (1) present. See character 185. This character describes the connection between principal filaments. Although interfilamental tissue bridges were used in phylogenetic analyses to specify gill types by, for example, Giribet and Wheeler (2002: char. 68), this character as defined has not been used in previous phylogenetic analyses.
Oxygen transport character
191 Intracellular haemoglobin: (0) absent; (1) present. Haemoglobin is present in erythrocytes in Arcidae and Glycymeridae (Boyd 1998; Morton et al. 1998), Carditidae and Crassatellidae (see citations in Slack-Smith 1998b, 1998c; Taylor et al. 2005) and Calyptogena magnifica (see Terwilliger et al. 1983); see also Booth and Mangum (1978).
Sperm characters (Figs 23–25)
Although the taxonomic and phylogenetic importance of sperm ultrastructure in Bivalvia is widely appreciated (for general review, see Healy 1996 and literature cited therein), only one phylogenetic study of bivalves has included sperm characters in a cladistic analysis (Giribet and Wheeler 2002: chars 154–169). The characters selected for this study attempt to cover as much of the observable sperm ultrastructure as possible, but acknowledge that this dataset has limitations imposed upon it by missing information for several target species, the influence of fertilisation biology (possible convergences) and, in certain taxa, fixation quality of available material.
The environment of fertilisation and other factors, such as the type and longevity of sperm storage conditions before fertilisation, have an important influence on sperm morphology (Franzén 1955, 1956), thereby creating difficulties when comparing results for taxa of widely disparate reproductive modes in a phylogenetic context. Coding of sperm characters presents a special problem in that there are essentially two competing hypotheses concerning the fertilisation biology and sperm structure of ancestral molluscs: (1) fertilisation in seawater using spermatozoa of the ect-aquasperm or ‘primitive’ type (see Franzén 1955, 1956; an idea originating with Gustaf Retzius (his contributions recently reviewed by Afzelius 1995) for animals in general); (2) fully internal fertilisation using spermatozoa of the introsperm or ‘modified’ type (Buckland-Nicks and Scheltema 1995; developed by them for Bilateria in general but using primarily molluscan examples) (For formal definitions of ‘ect-aquasperm’ and ‘introsperm’, see Jamieson and Rouse (1989)). Thus, the first hypothesis holds that introsperm and fully internal fertilisation are secondary, whereas the second hypothesis holds that external fertilisation and ect-aquasperm (at least in molluscs) are secondary (although Buckland-Nicks and Scheltema (1995) accepted that basal metazoans (sponges and cnidarians) have external fertilisation and ect-aquasperm). Healy (2000) evaluated both of these scenarios and concluded that good cases for both hypotheses could be put forward and that both, in fact, could be operating within molluscs. Buckland-Nicks and Scheltema (1995) based their hypothesis on the occurrence of introsperm in Solenogastres (spermatozoa of Epimenia australis (Thiele, 1897), which show mature and/or developmental similarities to those of some neritimorphs, caenogastropods and the polychaete genus Protodrilus). They argued that through suppression of spermiogenesis at the ‘ect-aquasperm’ phase, it would be possible to generate aquasperm secondarily from molluscs (or other bilaterians) possessing introsperm. Although this is a very attractive concept, we believe that, at least for the earliest Conchifera, an ect-aquasperm starting point is much more consistent with available data for the following reasons: (1) ect-aquasperm of the same basic structure occur in all Bivalvia and Scaphopoda and most basal Gastropoda, as well as basal Polyplacophora and Caudofoveata; (2) there are no known incidences of ect-aquasperm (and external fertilisation) arising in Neritimorpha, Caenogastropoda or Heterobranchia, even in sedentary or cemented taxa in which the dispersive advantage for free-spawning would seem considerable; (3) the absence of introsperm in Bivalvia, Scaphopoda, Polyplacophora and, as far as is known, Monoplacophora (this is a corollary to (1) but just as significant); and (4) marked differences in sperm features between introsperm of neritimorphs, vent taxa, apogastropods and various trochoidean (skeneiform) taxa are suggestive of independent origins for some or possibly all of these groups from lineages possessing ect-aquasperm. For these reasons, we have coded sperm characters here from the standpoint of ‘ect-aquasperm’ being basal for Conchifera and at least plausibly for molluscs in general. This contrasts with Buckland-Nicks (2008), who coded nine sperm characters for his study of polyplacophoran phylogeny from the standpoint of introsperm (as exemplified by E. australis) being basal to the Placophora and molluscs in general. Thus, the polarity of some of Buckland-Nicks’ characters relating to centriolar position, mitochondrial number, shape and position differ markedly from that applied here. Fig. 25 summarises all sperm characters and states for Bivalvia and the ingroups. Micrographs demonstrating several character states for Bivalvia are indicated below. (Sources for sperm data and additional literature references are provided in the accompanying MorphoBank Project 790.)
192 Acrosomal vesicle (Fig. 25): (0) composed of one vesicle only (Fig. 23A, B, G); (1) composed of multiple small vesicles (Fig. 23E). This character describes the number of vesicle components in the acrosomal complex. All Mollusca except Palaeoheterodonta have a single acrosomal vesicle. This was coded by Giribet and Wheeler (2002: char. 155).
193 Acrosomal complex (vesicle + subacrosomal material) position (Fig. 25): (0) anterior, seated vertically on nuclear apex (Fig. 23A, B, G); (1) posterior, associated with midpiece (Figs 23C, D, 24H). All Mollusca except the non-cuspidariid anomalodesmatans have the acrosomal complex positioned at the nuclear apex. This was coded by Giribet and Wheeler (2002: char. 158).
194 Acrosomal vesicle longitudinal profile (Fig. 25): (0) cap/low dome-shaped (Fig. 23C); (1) high dome-shaped (Fig. 23A, B); (2) elongate–conical, with wide base (Fig. 23F); (3) elongate–conical, narrow throughout; (4) flat, discoidal (Fig. 23E); (5) spherical. This character describes the shape of the acrosomal vesicle in longitudinal section. The cap/low cone-shaped acrosomal vesicle is seen in Laevipilina antarctica, scaphopods, some basal gastropods and in many ingroup taxa (mostly pteriomorphians and anomalodesmatans). New character.
195 Acrosomal vesicle contents (Fig. 25): (0) homogeneous (or apparently so); (1) with radial structures peripherally (Fig. 23I); (2) with internally structured (reticulate) basal ring and one or more other zones (Fig. 23G, H); (3) with dense internal layers (Fig. 23C); (4) with electron-lucent periphery (Fig. 23E); (5) with differentiated anterior and posterior zones. This character describes the ultrastructural appearance of the acrosomal vesicle contents. Developmentally, the contents of proacrosomal vesicles in molluscs are initially homogeneous and thus we consider this state plesiomorphic. Because these vesicles fuse to form the acrosomal vesicle, their contents might (or might not) differentiate into various layers/elements, depending on the taxon. New character.
196 Acrosomal vesicle apex (Fig. 25): (0) simple (Fig. 23G); (1) elaborated as a rod-shaped or filiform extension (Fig. 23D). This character describes the upper surface of the acrosomal vesicle. All Mollusca except certain non-cuspidariid anomalodesmatans have a simple acrosomal vesicle apex. New character.
197 Subacrosomal material (Fig. 25): (0) granular deposit (Fig. 23C, G); (1) granular deposit with electron-lucent layer; (2) granular deposit + rod; (3) rod only (Fig. 23F); (4) rod + subacrosomal plate on nuclear apex; (5) solid, columnar body. This character describes the appearance of subacrosomal material (i.e. material associated with the base of the acrosomal vesicle). Subacrosomal material present as a simple granular deposit is seen in Laevipilina antarctica, scaphopods, most basal gastropods, polyplacophorans, cephalopods, and many of the ingroup taxa. Some partial overlap with the coding of Buckland-Nicks (2008: char. 2) exists, although his states specifically relate to features of the solenogastre Epimenia australis and Polyplacophora. New character.
198 Nucleus shape (Fig. 25): (0) barrel-shaped (Fig. 23N); (1) rod-shaped (Fig. 23L); (2) spheroidal (Fig. 23M); (3) filiform; (4) rod-shaped with helical twists; (5) teardrop-shaped. This character describes the overall shape of the sperm nucleus. Laevipilina antarctica, scaphopods, the majority of vetigastropods, many patellogastopods and a large proportion of the ingroup have a barrel-shaped nucleus. Although there can be difficulties in assigning a sperm nucleus shape for some taxa, in general the states listed above are distinctive and accurately reflect the range of morphologies encountered here. Franzén (1983) showed a positive correlation between nuclear elongation and increased size and yolk content of eggs, but further work is required to test the extent and consistency of this correlation. New character.
199 Nucleus curvature (Fig. 25): (0) absent (Fig. 23L); (1) present (Fig. 23K). This character relates to whether or not the sperm nucleus is curved. No curvature is seen in Laevipilina antarctica, scaphopods, most basal gastropods, nautiloid and octopodomorph cephalopods, polyplacophorans and the majority of the ingroup. New character.
200 Nuclear contents (mature) (Fig. 25): (0) coarse granulation visible (Figs 23A, 24G); (1) coarse granulation not visible (Fig. 24F). This character describes the ultrastructural appearance of the condensed nuclear contents. Retention of coarse granulation of the nucleus is seen in protobranch and pteriomorphian bivalves. The condition in monoplacophorans is unknown. All other molluscs appear to have more complex nuclear condensation, leaving no trace of the coarse granulation at maturity. Buckland-Nicks (2008: char. 8), in his study of solenogastre and polyplacophoran taxa, coded nuclear condensation (as chromatin pattern) during spermiogenesis and not (as in this study) for mature spermatozoa. New character.
201 Nuclear apex (Fig. 25): (0) flat to almost so (Fig. 23G); (1) strongly convex; (2) concave and/or with deep fossa (Fig. 23F, M); (3) with projecting ridge or rod (Fig. 23J). This character describes the shape of the distal tip of the nucleus. New character.
202 Nuclear basal fossa (Fig. 25): (0) short, with centrioles largely or wholly excluded (Fig. 24A); (1) short, containing centrioles or derivatives thereof; (2) very shallow or absent, with centrioles wholly excluded (Fig. 23C). This character describes the invagination of the nuclear base. The nuclear fossa is short with centrioles largely or wholly excluded in scaphopods, most basal gastropods, polyplacophorans and most of the ingroup. The condition in Laevipilina antarctica is inferred from available (incomplete) data. Buckland-Nicks (2008: char. 9), in his study of solenogastre and polyplacophoran taxa, interpreted the fossa containing the centriolar derivative (‘basal body’) of Epimenia australis as being the plesiomorphic condition (in our study this state is coded as derived). New character.
203 Centriolar complex (Fig. 25): (0) proximal and distal centrioles present, both short and orthogonally arranged (Figs 23A, 24A); (1) proximal centriole lateral (not anterior) to distal centriole; (2) proximal centriole wholly or partially modified into banded rod (Fig. 24E,F); (3) both centrioles parallel, each acting as a distal centriole (each with separate flagellum) (Fig. 24J); (4) present as centriolar derivative only; (5) proximal centriole lateral to distal, but distal very elongate (Fig. 24H). This character describes the ultrastructure of the centrioles. The centrioles are short and orthogonally arranged in Laevipilina antarctica, scaphopods, basal gastropods, basal polyplacophorans and the majority of the ingroup. Buckland-Nicks (2008: char. 9), in his study of solenogaster and polyplacophoran taxa, coded a different series of character states relating specifically to his ingroup. New character.
204 Centriolar rootlet (Fig. 25): (0) without periodically banded component; (1) with periodically banded component (Fig. 24G); (2) poorly developed or absent (Fig. 24A). This character describes the material connecting the centriole (usually the proximal centriole, if present) to the nucleus. The centriolar rootlet lacks any periodically banded component in Laevipilina antarctica, scaphopods, basal gastropods, basal polyplacophorans and the majority of the ingroup taxa. New character.
205 Pericentriolar material (Fig. 25): (0) diffuse or absent (Figs 23A, 24A); (1) well developed, often sheathing both centrioles (Fig. 24K). This character describes the presence or absence of granular material around the centrioles. Pericentriolar material appears to be diffuse or absent in Laevipilina antarctica, scaphopods, basal gastropods, basal polyplacophorans and a large proportion of the ingroup. New character.
206 Prevailing (usual) mitochondrial number (Fig. 25): (0) 5 (sometimes 4) (Fig. 24B); (1) 4 (sometimes 5) (Fig. 24C); (2) 6 (sometimes 5); (3) greater than 6 (Fig. 24D, E); (4) 1 (a continuous cylindrical sheath). This character describes the usual number of mitochondria present. Despite the fact that there is some variation in mitochondrial number within most bivalve species, it is often possible to identify a ‘prevailing number’ (i.e. in a particular species, most spermatozoa might show five mitochondria, but some might only have four – in this case, the ‘prevailing number’ would be five). The prevailing mitochondrial number in the monoplacophoran Laevipilina antarctica, scaphopods, basal gastropods and basal polyplacophorans is five, as is also the case in a large proportion of the ingroup. Buckland-Nicks (2008: char. 4), in his study of aplacophoran and polyplacophoran taxa, coded the lowest number of mitochondria (1 or 2) as (0) (in Epimenia australis) and polyplacophoran taxa with more mitochondria as apomorphic states (3 or 4 as (1); 5 or 6 as (2); 7–9 as (3)). This is based on the idea of introsperm rather than aquasperm being plesiomorphic in molluscs. This character is an extension of Giribet and Wheeler’s (2002: char. 167) coding for the eight mitochondria of Archiheterodonta.
207 Mitochondrial shape (Fig. 25): (0) spherical, uncompressed (Figs 23A, B, 24A–D); (1) subspherical with marked lateral compression (Fig. 24E, I); (2) elongate, straight; (3) extremely elongate, components helical. Spherical, uncompressed mitochondria are seen in Laevipilina antarctica, scaphopods, most basal gastropods, basal polyplacophorans and a large proportion of the ingroup. Buckland-Nicks (2008: char. 5), in his study of aplacophoran and polyplacophoran taxa, coded ‘fused spiral’ (partially equivalent to our state (3)) as (0), with ‘all spherical’ (our state (0)) as (1), and ‘not all spherical’ (partially equivalent to our states (1) and (2)) as (2). This is based on the idea of introsperm rather than aquasperm being plesiomorphic in molluscs. New character.
208 Mitochondrial arrangement in relation to centriolar complex (Fig. 25): (0) radial (Fig. 24B–E); (1) with marked asymmetry (semi-radial) (Fig. 24I); (2) no close spatial relationship to centriolar complex. This character relates centriolar and mitochondrial positioning. All of the ingroup, with the exception of certain non-cuspidariid anomalodesmatans, exhibit radial arrangement of the mitochondria in relation to the centrioles, as do Laevipilina antarctica, scaphopods, basal gastropods, basal polyplacophorans (mostly Lepidopleurida) and the majority of the ingroup. Buckland-Nicks (2008: char 3), in his study of aplacophoran and polyplacophoran taxa, coded the radial arrangement (our state (0)) as (1), and the partial equivalent of our state (2) as (0). This is also similar to character 166 of Giribet and Wheeler (2002).
209 Mitochondrial arrangement in relation to nucleus (Fig. 25): (0) minimal or no overlap with nucleus (Figs 23A, 24A, F); (1) anterior region of one or all mitochondria surrounding the base of the nucleus (Fig. 24J, K). This character relates nuclear and mitochondrial positioning. Most of Mollusca show no or minimal overlap of the mitochondria (or their derivatives) with the nucleus. New character.
210 Axoneme (Fig. 25): (0) in a simple flagellum (axoneme sheathed by a plasma membrane) (Fig. 24L); (1) in a simple flagellum, but with the plasma membrane elaborated posteriorly as two paddles (Fig. 24M); (2) sheathed proximally by mitochondrial elements and posteriorly by glycogen. This character describes the structural elements associated with the sperm axoneme. A simple flagellum is seen in Laevipilina antarctica, scaphopods, basal gastropods, polyplacophorans and most of the ingroup. Buckland-Nicks (2008: char 7), in his study of aplacophoran and polyplacophoran taxa, coded ‘flagellum reinforcement’ features that do not apply in the present analysis. New character.
Molecular sequence data
Amplification of the five molecular markers resulted in sequences of the following lengths: between 1761 bp for Huxleya munita and 1990 bp for Frenamya elongata for 18S rRNA (amplified in three amplicons); between 643 bp for Pododesmus patelliformis and 865 bp for Cerastoderma edule for the first amplicon of 28S rRNA; between 496 bp for H. munita and 561 bp for F. elongata and Myochama anomioides for the second amplicon of 28S rRNA; between 594 bp for Lyonsia floridana and 706 bp for Bathyneaera demistriata for the third amplicon of 28S rRNA; between 423 bp for L. floridana and 842 bp for Chama macerophylla for 16S rRNA; between 659 bp for H. munita, Isognomon alatus, Malleus albus, Crassostrea virginica and Cardita calyculata and 661 bp for the Arcida species, Mytilus edulis, Pecten maximus, Kelliella sp., Calyptogena magnifica and Mya arenaria for COI (all outgroups and most species had a COI amplicon of 658 bp); and 328 bp for histone H3.
Phylogenetic analyses – dynamic homology under parsimony
Parsimony analysis of morphological data resulted in a single tree of 1410 steps. After multiple rounds of tree fusing following the protocols described above, tree lengths for the molecular data analyses stabilised after 5–10 searches. The optimal lengths for the six-parameter sets after stabilisation, and the results of the individual partitions (nuclear ribosomal, 16S rRNA, COI and histone H3) after two rounds of fusing, are shown in Table 3. The WILD identified parameter set 3221 as the optimal one for the molecular analyses, with a value of 0.02343, followed closely by parameter set 3211 (WILD = 0.02348).
The optimal parameter set yielded a single most parsimonious tree at 92 811 weighted steps (Fig. 26). This parameter set did not recover monophyly of Bivalvia, because the scaphopod sequence nested within the protobranchs. Monophyly of Bivalvia and Protobranchia were, however, found under the next two best parameter sets, 3211 and 111 (Table 3). Nearly all parameter sets recovered monophyly of Autobranchia (except 221), Pteriomorphia, Heteroconchia (except 221), Archiheterodonta, Palaeoheterodonta, Euheterodonta and Anomalodesmata. The non-anomalodesmatan euheterodonts were recovered in analyses under the three optimal parameter sets (3221, 3211 and 111). All parameter sets except 3211 found trees supporting a more basal position of Archiheterodonta than Palaeoheterodonta, which is sister-group to Euheterodonta. Neoheterodontei (Taylor et al. 2007b) is problematic due to the unstable position of Chama among analyses.
Because the difference between the two optimal parameter sets was minimal in terms of WILD scores (0.02343 for 3221 versus 0.02348 for 3211), and because this difference could be due to search heuristics (Wheeler et al. 2005), we conducted a total evidence analysis with morphology for both parameter sets. The combined analysis of the five genes and morphology for parameter set 3221 resulted in a single tree of 97 613 weighted steps (Fig. 27), and for parameter set 3211, resulted in a single tree of 77 343 weighted steps (Fig. 28).

|
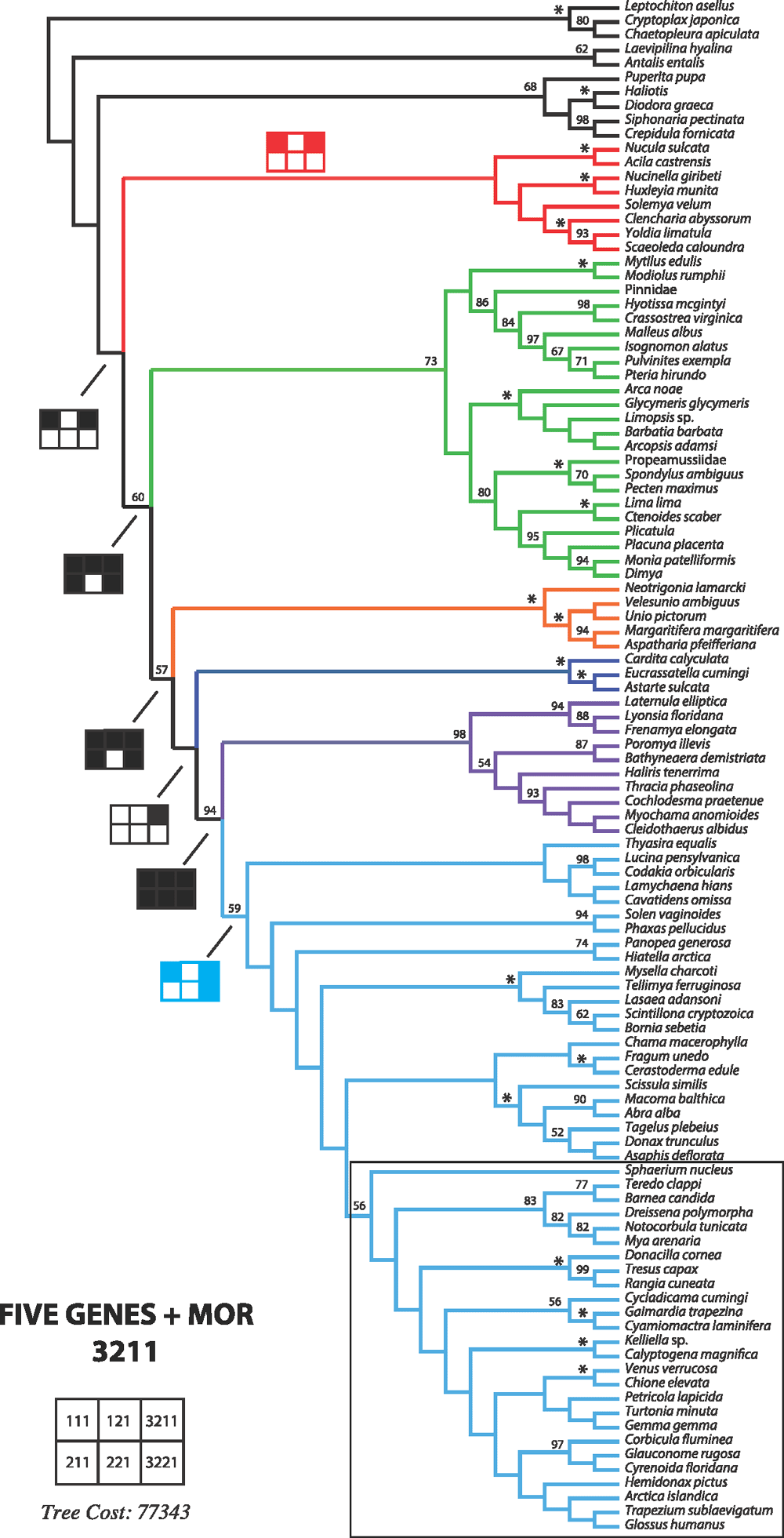
|
Monophyly of Protobranchia is found in one of the total evidence trees, but it is paraphyletic in the other, and nodal support (jackknife support; JS hereafter) is low for either hypothesis. Nucinellidae (JS = 100%), Nuculoidea (JS = 100%), and Nuculanoidea (JS = 100%) are well supported clades. Solemya does not form a clade with Nucinella and Huxleya in either tree. Within Pteriomorphia, Arcida is well supported (JS = 100%), as are Mytilidae (JS = 100%) and two other clades, one including (Pinnidae + (Ostreoidea + Pteroidea)) (JS = 82–86%), and another including Limidae + Pectinoidea + Anomioidea (including Dimyoidea) (JS = 76–86%). Support for Pteroidea (JS = 94–97%), Limidae (JS = 100%), Pectinoidea (JS = 99–100%) and Anomioidea (JS = 100%) is high. However, the deepest splits within Pteriomorphia are unsupported and unstable to parameter set variation.
Heteroconchia appears with the relationship (Archiheterodonta + (Palaeoheterodonta + Euheterodonta)) under most parameter sets (Fig. 26), but support for this particular configuration of the three heteroconch lineages is low. However, this result is stable to parameter set variation, with most parameter sets supporting a sister-group relationship of Palaeoheterodonta to Euheterodonta. Only parameter set 3211, with or without morphology, supports the more traditional Heterodonta (Fig. 28). Within palaeoheterodonts, both the monophyly of Unionida and its sister-group relationship to Trigoniidae are supported in all analyses (JS = 100%)
Euheterodonta’s main division into Anomalodesmata and remaining species is well supported and stable to parameter set variation. Anomalodesmatan relationships receive moderate to high support. Laternulidae + Lyonsiidae + Pandoridae form a well supported clade (JS = 96–100%), as do the latter two families (JS = 93–98%). The three septibranch families (Verticordiidae, Poromyidae and Cuspidariidae) form a clade under parameter set 3221 (JS < 50%), which is sister to the remaining non-septibranch families, but again, this relationship is not well supported (JS < 50%). The septibranch families appear as a sister-group to Myochamidae, Cleidothaeridae, Thraciidae and Periplomatidae, under parameter set 3211. A clade of these four families is well supported in our analyses (JS = 92%).
Relationships among the remaining euheterodonts (JS = 68–72% for the monophyly of Euheterodonta) generally are not well supported, and neither Venerida nor Myida, in the traditional sense (e.g. Beesley et al. 1998), are monophyletic (Figs 26–28). Among suprafamilial clades represented by multiple species, Galeommatoidea, Mactroidea and Tellinoidea appear well supported (JS = 100%), as is the sister-group of Kelliellidae and Vesicomyidae (JS = 100%); Pholadoidea receives moderate support (JS = 75%). A clade of Cyrenidae, Glauconomidae and Cyrenoididae is also well supported (JS = 98%), as is the clade containing Dreisseniidae, Corbulidae and Myidae (JS = 80–84%). A sister-group relationship of Pholadidae to the latter clade (JS = 89%) is supported under parameter set 3211, suggesting a relationship of multiple myoidan families, but including Dreisseniidae and excluding Hiatellidae and Gastrochaenidae. Gaimardia trapezina (Gaimardioidea) and Cyamiomactra laminifera (Cyamioidea) are united with strong support (JS = 99–100%) and group with Cycladicama cumingi (Ungulinoidea) (JS = 53–56%).
Several heterodont families are not monophyletic or receive little support/stability, including Veneridae, Lucinidae, Montacutidae and Tellinidae. Kelliellidae + Vesicomyidae are not closely related to Glossidae. Several species appear in spurious positions, especially those with the longest ribosomal sequences, such as Chama macerophylla, Cavitidens omissa, Lamychaena hians and the cardiids.
Phylogenetic analyses – probabilistic approaches
Five-gene analyses
The ML analysis of the five-gene dataset yielded a tree of -ln L = 78776.58 (Fig. 29). Runs of MrBayes v.3.1.2 for the same dataset reached stationarity after 4 million generations; 5 million generations (25%) were thus discarded as burn-in. The two topologies were remarkably similar (Fig. 29) and support monophyly of Bivalvia albeit with equivocal nodal support (Bremer support (BS) = 42%; posterior probability (PP) = 0.99), as well as monophyly of Autobranchia (BS < 50%; PP = 0.97). Other clades recovered are Pteriomorphia (BS = 86%; PP = 1.00), Heteroconchia (BS = 100%; PP = 1.00), Archiheterodonta (BS = 100%; PP = 1.00), Palaeoheterodonta (BS = 100%; PP = 1.00), Palaeoheterodonta + Euheterodonta (BS = 71%; PP = 0.78), Euheterodonta (BS = 100%; PP = 1.00), and a division of the latter into Anomalodesmata (BS = 100%; PP = 1.00) and the rest of euheterodonts (BS = 85%; PP = 1.00). Archiheterodonta is thus the sister-group to Palaeoheterodonta + Euheterodonta, as supported under most parameter sets in the POY analyses.
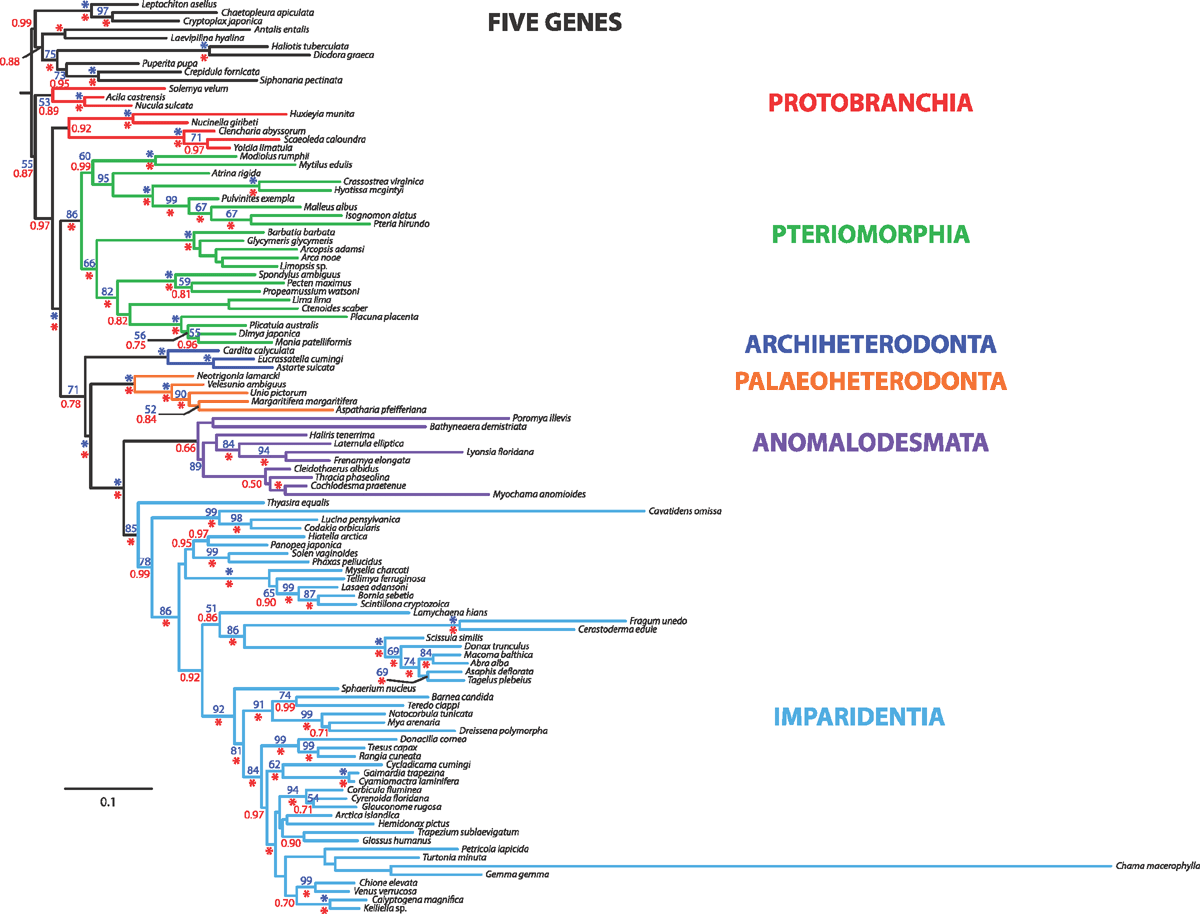
|
Protobranchia is paraphyletic with respect to Autobranchia, with Solemyidae (Solemya velum) being the sister-group to Nuculidae (BS = 53%; PP = 0.89), whereas Nucinellidae is the sister-group to Nuculanoidea, although without nodal support (BS < 50%; PP = 0.92). Nuculidae, Nucinellidae and Nuculanoidea each receive BS = 100% and PP = 1.00.
Relationships within Pteriomorphia are similar to those found under some parameter sets for the POY analyses, and only differ from the optimal parameter set in the position of Mytiloidea, which is the sister-group of (Pinnidae + (Ostreoidea + Pterioidea)), as in the 3211 POY analysis. Support for this relationship is modest (BS = 60%; PP = 0.99), whereas the Pinnidae + Ostreoidea + Pterioidea clade is well supported (BS = 95%; PP = 1.00). Arcida is well supported (BS = 100%; PP = 1.00) and sister to (Pectinoidea + (Limidae + Anomioidea (including Dimyoidea))) (BS = 82%; PP = 1.00). Limopsis sp. (Limopsoidea) and Glycymeris glycymeris and Arcopsis adamsi appear well nested within Arcidae in the ML topology. There are minor topological differences between the ML and BI trees with respect to relationships within Arcida and its sister-clade, but these are not supported.
Resolution within Archiheterodonta and Palaeoheterodonta is similar to that of the POY analyses. Resolution of the internal relationships of Anomalodesmata finds little support, with the exception of a clade that includes all of the non-septibranch species plus Haliris tenerrima (BS = 89%), and a clade with (Laternulidae (Lyonsiidae + Pandoridae)) (BS = 94%; PP = 1.00).
Euheterodont relationships remain poorly supported with a few notable exceptions. Thyasiridae and Lucinidae, the latter well supported as monophyletic (BS = 99%; PP = 1.00), constitute the basalmost nodes, the remaining families grouping with BS = 86% and PP = 1.00. Galeommatoidea (BS = 100%; PP = 1.00) groups with Hiatelloidea + Solenoidea, with low support. Lamychaena hians (Gastrochaenidae) is supported as the sister-group (BS = 51%; PP = 0.86) to a clade composed of Cardiidae + Tellinoidea (BS = 86%; PP = 1.00); both Cardiidae and Tellinoidea each receive high nodal support (BS = 100%; PP = 1.00). Its sister-clade, Neoheterodontei (BS = 92%; PP = 1.00), places Sphaerium nucleus (Cyrenidae) as sister-group to all other members (BS = 81%; PP = 1.00), followed by a clade containing Barnea candida, Mya arenaria, Notocorbula tunicata and Dreissena polymorpha (BS = 91%; PP = 1.00). Mactroidea (BS = 99%; PP = 1.00), Cyamioidea (BS = 100%; PP = 1.00), Kelliellidae + Vesicomyidae (BS = 100%; PP = 1.00), Cyrenidae + Glauconomidae + Cyrenoididae (BS = 94%; PP = 1.00), Corbulidae + Myidae + Dreissenidae (BS = 99%; PP = 1.00), Pholadoidea (BS = 74%; PP = 0.99) and Pholadoidea + Corbulidae + Myidae + Dreissenidae (BS = 91%; PP = 1.00), are among the suprafamilial supported clades. However, families such as Tellinidae, Veneridae and Montacutidae are paraphyletic or polyphyletic. As in the previous analyses, the position of some long-branch taxa, especially Chama macerophylla, appears problematic. As with Pteriomorphia, minor topological differences between ML and BI trees occur within Euheterodonta, and these are not supported.
Nine-gene analyses
Two nine-gene analyses were conducted, one with the 42 taxa that overlapped with those of Sharma et al. (2012) and one with all 108 taxa. The combined nine-gene alignment consisted of 6446 characters. In the ML analysis of the 42-taxon dataset (–ln L = –80409.47; Fig. S1, available as Supplementary Material on the journal website), Bivalvia receives little support and Protobranchia is paraphyletic. Autobranchia (BS = 100%), Pteriomorphia (BS = 100%), Heteroconchia (BS = 100%), Palaeoheterodonta (BS = 100%), Archiheterodonta (BS = 100%), Euheterodonta (BS < 50%), Anomalodesmata (BS = 100%) and the remaining Euheterodonta (BS < 50%) are monophyletic. A difference in tree topology with respect to previous analyses is that a clade composed of Palaeoheterodonta + Archiheterodonta is recovered (BS = 87%).
The results of the nine-gene analysis including all taxa (Fig. 30) are comparable with those of the 42-taxon analysis, but also include the monophyly of Bivalvia (BS < 50%; PP = 0.91) and Protobranchia (BS < 50%; PP = 0.81). As in the previous analysis, Autobranchia (BS = 81%; PP = 1.00), Pteriomorphia (BS = 99%; PP = 1.00), Heteroconchia (BS = 100%; PP = 1.00), Palaeoheterodonta (BS = 100%; PP = 1.00), Archiheterodonta (BS = 100%; PP = 1.00), Palaeoheterodonta + Archiheterodonta (BS = 81%; PP = 0.99), Euheterodonta (BS = 100%; PP = 1.00), Anomalodesmata (BS = 100%; PP = 1.00), and the remaining Euheterodonta (BS = 91%; PP = 1.00) are monophyletic. Internal resolution within each of these clades is similar to the five-gene analysis, with some notable differences in the internal resolution of Pteriomorphia, especially in the position of Arcida and Mytilida.
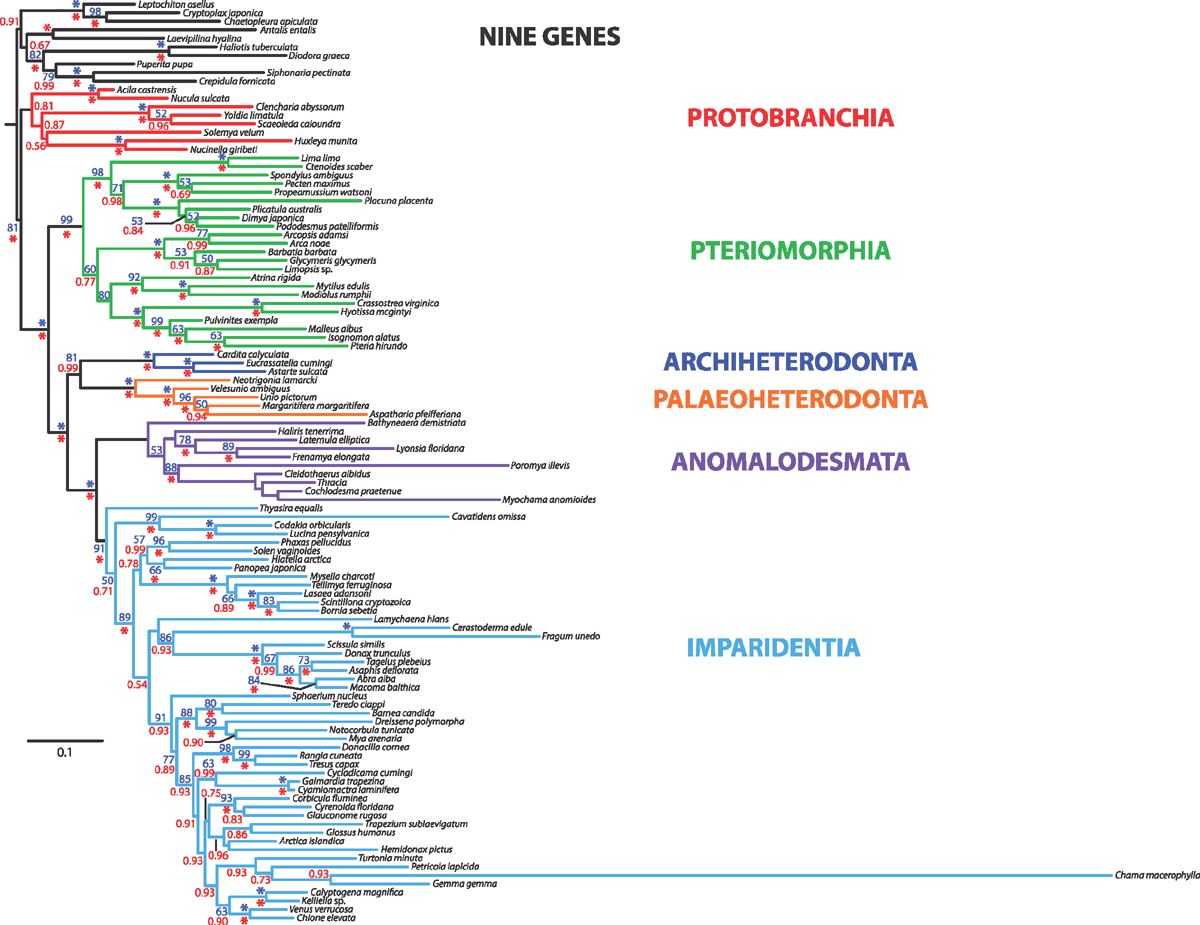
|
Total evidence Bayesian inference analysis
The total evidence analysis of the nine genes plus morphology using a Bayesian approach (Fig. 31) is highly congruent with the nine-gene analyses, again with some differences in the internal topology of Pteriomorphia. High posterior probabilities are recovered for most major clades, including Bivalvia (PP = 1.00), Protobranchia (PP = 1.00), Autobranchia (PP = 1.00), Pteriomorphia (PP = 1.00), Heteroconchia (PP = 1.00), Palaeoheterodonta (PP = 1.00), Archiheterodonta (PP = 1.00), Euheterodonta (PP = 1.00), Anomalodesmata (PP = 1.00) and the non-anomalodesmatan Euheterodonta (PP = 1.00). As in all analyses with nine genes, Palaeoheterodonta and Archiheterodonta form a clade (PP = 0.98), in sister relationship to Euheterodonta.
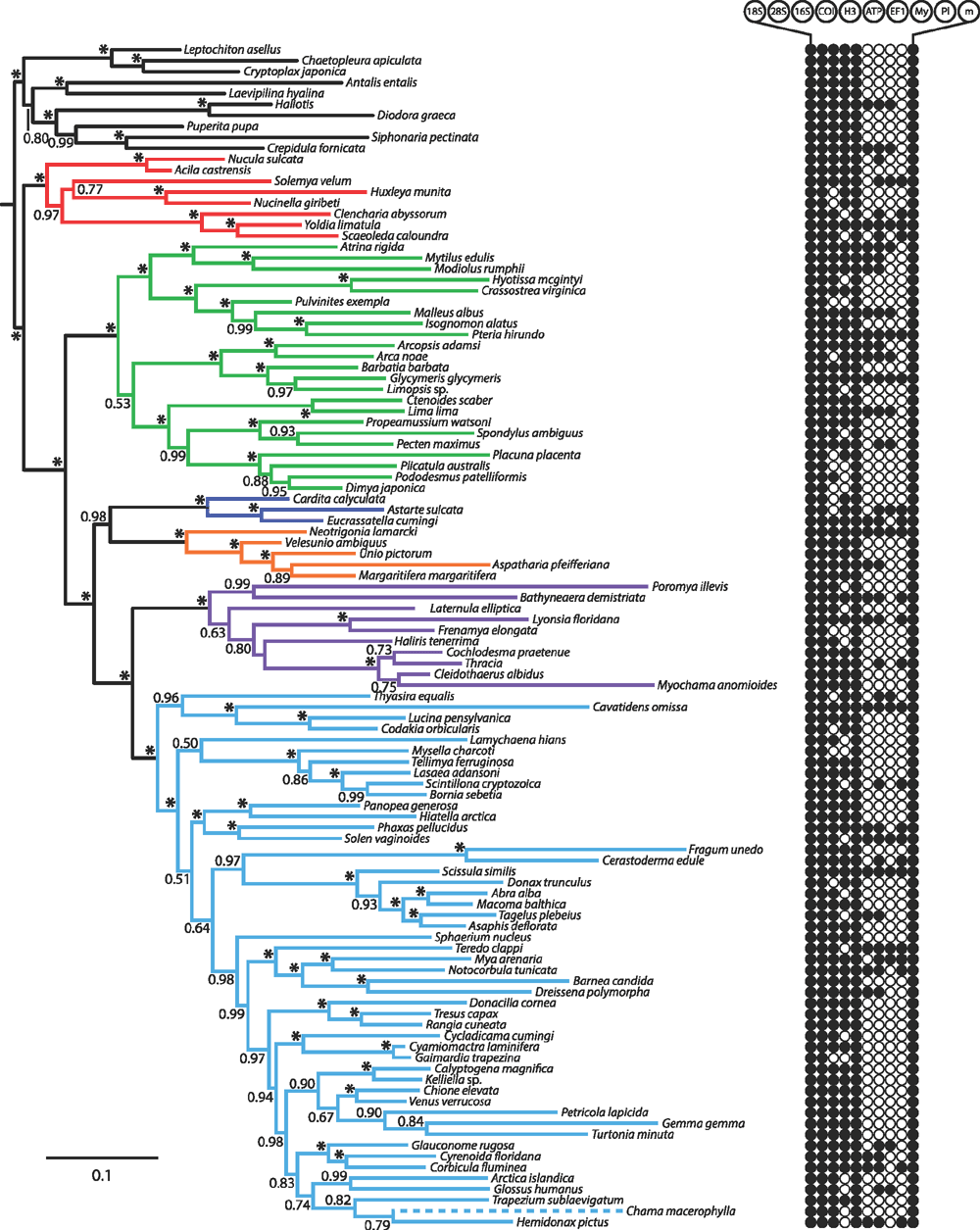
|
Protobranchia is well resolved, with Nuculida (PP = 1.00), as the sister-group to Nuculanida + Solemyida (PP = 0.97), but Solemyida receives no support (PP = 0.77). Pteriomorphia lacks resolution for the position of a monophyletic Arcida (PP = 1.00), but both Arcidae and Arcoidea are paraphyletic with respect to Noetiidae, Glycymerididae and Limpsoidea. Mytilida appears well supported as sister-group to Pinnidae (PP = 1.00), nested within Pterida (PP = 1.00). Limida appears well supported as sister to Pectinida (PP = 0.99). Anomioidea is not monophyletic, but the clade including them with Plicatuloidea and Dimyoidea is well supported (PP = 1.00). Support within Archiheterodonta is high for the separation between Trigoniida and Unionida (PP = 1.00), but the unionoid superfamilies are not supported by the analyses.
Support within Anomalodesmata is low for most nodes, with Poromyidae + Cuspidariidae forming a clade (PP = 0.99), but not Septibranchia, because Verticordiidae (Haliris tenerrima) appears nested within the non-septibranch families, although without support. Pandoridae + Lyonsiidae (PP = 1.00) and Periplomatidae + Thraciidae + Cleidothaeridae + Myochamidae (PP = 1.00) are the only supported anomalodesmatan clades.
Within the remaining Euheterodonta, a basal split separates a clade containing Thyasiridae + Lucinidae (PP = 0.96) from the rest of the families (PP = 1.00); deep resolution within this clade receives low support. Well supported suprafamilial relationships include: Galeommatoidea (PP = 1.00); Hiatelloidea + Solenoidea (PP = 1.00); Tellinoidea (PP = 1.00); Mactroidea (PP = 1.00); Cyamioidea (PP = 1.00); Cyamioidea + Ungulinidae (PP = 1.00); Kelliellidae + Vesicomyidae (PP = 1.00); Glauconomidae + Cyrenoididae + Cyrenidae (PP = 1.00); Cyrenoididae + Cyrenidae (PP = 1.00); and (Teredinidae (Myidae + Corbulidae) (Dreissenidae + Pholadidae)) (all nodes with PP = 1.00). Many deep nodes, including Neoheterodontei, receive modest support (PP = 0.98). Some clades that are clearly not monophyletic include Glossoidea, because Glossus humanus groups with Arctica islandica (PP = 0.99), instead of with Kelliellidae + Vesicomyidae. Veneroidea is also diphyletic, due to the position of Glauconomidae, and Veneridae requires the inclusion of Turtonia and Petricola to be monophyletic. Cardioidea, Arcticoidea and Pholadoidea are not monophyletic. Chamidae and Hemidonacidae appear as an unsupported sister-group nested deep within the tree.
Estimation of divergence times
Runs of BEAST v.1.7.4 achieved stationarity after 2 × 107 generations; 2.5 × 107 generations (25%) were discarded as burn-in. The tree topology recovered is comparable in all major aspects with the nine-gene and Bayesian total evidence topologies (Fig. 32). Estimated dates of diversification for major clades of bivalves are inferred as follows: Protobranchia: 436 Ma (highest posterior density interval (HPD): 372–492 Ma; Autobranchia: HPD: 529 Ma (527–530 Ma); Pteriomorphia: 516 Ma (HPD: 511–521 Ma); Heteroconchia: 516 Ma (HPD: 509–522 Ma); Archiheterodonta + Palaeoheterodonta: 501 Ma (HPD: 491–510 Ma); and non-anomalodesmatan Euheterodonta: 459 Ma (HPD: 431–483 Ma). Dates for all other nodes are available in Fig. S2. The estimated timing of the split between Protobranchia and Autobranchia almost coincides with the timing of bivalve diversification, which in turn was estimated to occur close to the upper bound of the prior in all a posteriori trees.
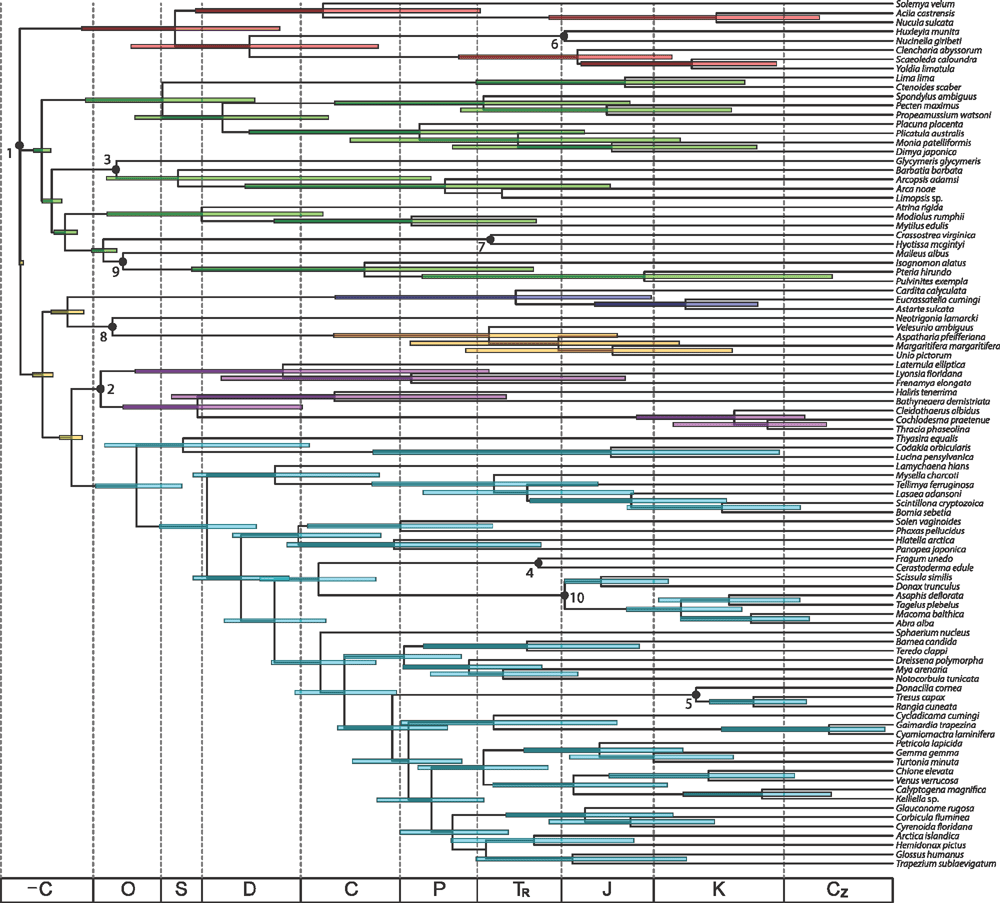
|
The log-lineage through time (LTT) plot showed a characteristic increase in lineage accumulation immediately subsequent to the end-Permian for Palaeoheterodonta and Archiheterodonta, with almost no cladogenesis before the Mesozoic, comparably with Protobranchia (Fig. 33). In contrast, Imparidentia new clade shows little evidence for a lower rate of cladogenesis immediately before the Mesozoic. The LTT plot of Pteriomorphia could be construed as anti-sigmoidal, but is not statistically distinguishable from a density-dependent process (i.e. decelerating diversification rate). Anomalodesmata, which diversified early in the Palaeozoic, is insufficiently sampled taxonomically for this analysis, and its lineage accumulation curve is therefore not dispositive of either hypothesis (Fig. 33).
Analysis of phylogenetic signal
Of the 210 characters in the morphological matrix (with 16.6% missing data), 99 had significant phylogenetic signal (see Supplementary Material). We analysed these data to observe the distribution of informative characters by character system (Fig. 34). On average, 49.5% of characters in each class bore phylogenetic signal, but the number of characters per class is not evenly distributed, and thus character systems with few characters and/or more missing data (e.g. oxygen transport, sensory, endosymbiont) can differ in actual distribution of informative characters. Of those character systems with more definable characters, sperm ultrastructure and external shell classes included a greater proportion of informative characters (≥50%). The larval class bore some of the fewest informative characters (27.3%), although this set is the least completely sampled character system (69.5% missing data).
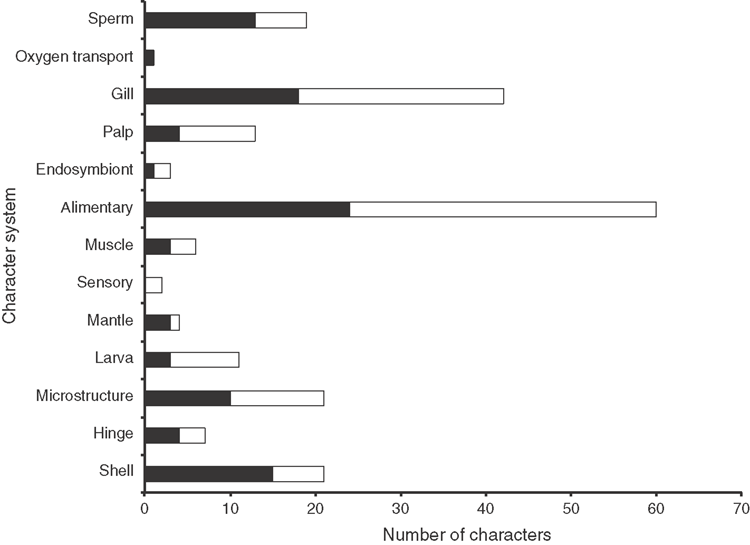
|
To determine the parts of bivalve phylogeny elucidated by the 99 significantly informative morphological characters, this smaller dataset was analysed using the same methods as for the 210-character dataset (above). The resulting most parsimonious tree of 911 unweighted steps is shown in Fig. 35. Notable aspects of this topology include the monophyly of Protobranchia, Archiheterodonta and Euheterodonta, but the paraphyly of Pteriomorphia, Palaeoheterodonta, Imparidentia and Anomalodesmata.
|
Taxonomic actions
We propose the name Imparidentia Bieler, Mikkelsen & Giribet, new clade for a clade of Euheterodonta excluding Anomalodesmata, as a sister-group relationship between these two groups is a recurrent theme in bivalve molecular phylogenetics (e.g. Giribet and Wheeler 2002; Taylor et al. 2009; Sharma et al. 2012; but see Dreyer et al. 2003; Taylor et al. 2007b); although Imparidentia new clade was supported in the analysis by Plazzi et al. (2011), Euheterodonta was not recovered. The name (from the Latin adjective impar, unequal, and the neuter plural of the Latin noun dens, tooth) refers to the unequal teeth that predominate in the hinges of the members of this clade. This is thus essentially the ‘traditional’ Heterodonta (as understood in many pre-molecular studies before including Anomalodesmata in Heterodonta; e.g. Newell 1965; Waller 1990, 1998; Cope 1997), but excluding Archiheterodonta.
Discussion
Phylogenetic analysis of the molecular data using more than 5 Kb of DNA sequence data (including ~3.7 Kb of conserved positions after culling of ambiguous nucleotides in the static alignments) per complete taxon yields comparable results between analytical approaches, with some differences. First, it is important to note that the direct optimisation analyses used all sequence information, whereas probabilistic approaches based on static alignments discarded highly variable regions. This can make the direct optimisation analysis more sensitive to long or idiosyncratic sequences, while providing more support for shallower branches (e.g. Lindgren and Daly 2007), as is the case with the Septibranchia and the better resolution of anomalodesmatan relationships in the POY analyses than in the probabilistic ones. Examples of the former are the unstable positions of Lamychaena hians and Chama macerophylla in the different POY analyses, although their position is also unsupported in the probabilistic analyses. Alternatively, clades like Arcida, with extremely low levels of variation in the nuclear ribosomal markers, can suffer from removal of the few variable sites in the probabilistic analyses. The ML analysis finds generally higher support for the deepest nodes. Both analyses, however, agree in the low support for bivalve monophyly, lack of support for protobranch monophyly and the high support for the monophyly of Euheterodonta. These receive higher support in the nine-gene Bayesian analyses, especially when combined with morphology (Fig. 31), wherein both Bivalvia and Protobranchia receive high support (PP = 1.00). All analyses generally agree in deep branching patterns, with monophyly of Autobranchia, Pteriomorphia, Heteroconchia, Palaeoheterodonta, Archiheterodonta, Euheterodonta, Anomalodesmata and Imparidentia. Resolution within many of these clades is also compatible between analyses with some notable exceptions involving internal relationships, such as the position of Arcida, Mytilida, or the monophyly of Lucinidae and the position of Thyasiridae.
In comparison with recent bivalve analyses, Giribet and Wheeler (2002) also combined morphology and molecular data, although that analysis included fewer species and only a subset of the molecular data explored here. Taylor et al. (2009) explored the largest heteroconch dataset amassed to date, but again for a subset of the molecular data included here and without morphology. For a comparable amount of molecular data, the analyses of Giribet et al. (2006) and Wilson et al. (2010) included only a handful of bivalve lineages and no morphology. More recent studies have analysed comprehensive datasets including largely different types of data, such as mitochondrial genomes (Plazzi and Passamonti 2010; Plazzi et al. 2011) and a novel set of nuclear protein-encoding genes (Sharma et al. 2012). Finally, the most fundamental question of bivalve monophyly has been explored with large transcriptomic datasets (Kocot et al. 2011; Smith et al. 2011; see a recent review in Kocot 2013).
In contrast to these earlier studies, our combined molecular and morphological dataset has resolving power at the base of the tree and unambiguously supports bivalve monophyly when all data are considered (POY combined analyses and molecular analyses under parameter sets 111 and 3211; probabilistic analyses). Molluscan monophyly has only received high support (other than in Bayesian phylogenetics) in the two recent phylogenomic analyses (Kocot et al. 2011; Smith et al. 2011), which also found support for the monophyly of Protobranchia, a clade only recovered in some of our analyses, and often with marginal support.
Inferring a classification
Protobranchia (red)
The protobranchiate bivalves are confirmed as a monophyletic group comprising Nuculida (Nuculoidea), Solemyida (Solemyoidea + Manzanelloidea) and Nuculanida (Nuculanoidea), much in agreement with early morphology-based classifications (e.g. Thiele (1934) who placed these groups together in ‘Stirps Nuculacea’). Earlier studies focussing on molecular data found non-monophyly of Protobranchia and proposed a split into Opponobranchia (a clade uniting Nuculida and Solemyida) and a clade of the mutually monophyletic (Nuculanida + Autobranchia) (see Giribet 2008). That hypothesis was not supported in phylogenomic studies (Kocot et al. 2011; Smith et al. 2011) and in analyses of nuclear protein-encoding genes (Sharma et al. 2012), but is recovered (without jackknife support) in our POY combined analysis (Fig. 27). Protobranch monophyly is supported in the suboptimal POY combined analysis tree as well as in two of the molecular-only parameter sets analysed in POY (Fig. 28). Alternative resolutions rejecting protobranch monophyly are recovered in different probabilistic trees (Fig. 29, Fig. S1), and protobranch monophyly is found in the nine-gene probabilistic analyses (Fig. 30) but with negligible support. Only when combining large amounts of molecular data with morphology is support found for the monophyly of Protobranchia.
Resolution within Protobranchia is analysis-dependent, and our data only unambiguously support Nuculida, Nuculanida and Nucinellidae, but not the monophyly of the Solemyida ( = Nucinellidae + Solemyidae), which was recovered only in the nine-gene analyses and without significant support. This is broadly consistent with the results of a recent molecular phylogeny sampling all extant families of Protobranchia (Sharma et al. 2013). In that study, based on the same five workhorse genes employed herein, Solemyida formed a grade at the base of Protobranchia, with Solemyoidea sister to the remaining protobranchs, and a clade of the mutually monophyletic Nuculida and Nuculanida (the traditional Palaeotaxodonta hypothesis). Adequately preserved protobranch material can be difficult to obtain for many lineages and the present analyses, although based on up to nine-genes and morphology, still have limited taxonomic representation, specifically for the four protein-encoding genes analysed by Sharma et al. (2012). Future work focussing on the internal relationships of protobranchs might have to depend upon much more fragmentary datasets because many miniaturised and deep-sea samples are available only from formalin-fixed specimens or from specimens collected occasionally and not fixed properly (see Boyle et al. 2004; Zardus et al. 2006; Etter et al. 2011; Sharma et al. 2013).
Though morphological and molecular phylogenetic work on this early clade of bivalves has appeared sporadically in the literature, Protobranchia have nevertheless figured prominently in studies on speciation in the deep sea, and recent efforts have focussed on discovery of species from extreme environments (e.g. Oliver et al. 2011; Oliver and Taylor 2012) or on the characterisation of endosymbiosis (e.g. Taylor and Glover 2010; Oliver and Taylor 2012). Although the presence of chemosymbiosis in Nucinellidae has been inferred (Reid 1998; Taylor and Glover 2010), and confirmed through anatomical data (Oliver and Taylor 2012), no molecular confirmation is yet available. Here we report the sequencing of a Gamma proteobacteria COI from the DNA extraction of Huxleyia munita, which is consistent with the presence of chemosynthesis in this deep-sea family.
The relationships of Nucinellidae and Solemyidae were recently reviewed by Oliver and Taylor (2012; see also Pojeta 1988), and a small analysis of Solemyidae was published by Taylor et al. (2008). Albeit with limited taxon sampling, Taylor et al. (2008) addressed the taxonomy of Solemyidae and considerably advances our knowledge of these bivalves, supporting the reciprocal monophyly of Acharax and Solemya, and the monophyly of the subgenus Solemyarina. Systematic treatment awaits for many genera and families of Nuculida and Nuculanidae recovered as non-monophyletic by Sharma et al. (2013).
Autobranchia
Autobranchia is confirmed as the monophyletic sister-group of Protobranchia, containing all remaining bivalve groups. Autobranchia is characterised by the presence of enlarged ctenidia with a filtering function – although the ctenidia have been highly modified in the septibranchs – and comprises two sister-taxa: Pteriomorphia and Heteroconchia.
Pteriomorphia (green)
Pteriomorphia is monophyletic, as has been found in virtually all prior phylogenetic analyses of bivalves (e.g. Steiner and Hammer 2000; Giribet and Wheeler 2002; Giribet and Distel 2003; Matsumoto 2003; see Xue et al. 2012 for a recent analysis based on 28S rRNA). It contains four well supported clades: Arcida, Mytilida, Pectinida and Ostreida + Pinnida (represented by a single species in our analyses).
A clade consisting of Pinnida (Pinnidae) + Mytilida (Mytiloidea) is supported in some of the probabilistic analyses (Figs 30–31; Fig. S1; but not in Fig. 29), and it is sister to Eupteriomorphia, consisting of Ostreida (Ostreoidea + Pterioidea). The placement of Pinnidae outside Eupteriomorphia, however, is contradicted in the direct optimisation analyses (Figs 26–28) and in the five-gene probabilistic analyses (Fig. 29), which place Pinnidae in a more traditional position, sister to the two pteriomorphian superfamilies Ostreoidea and Pterioidea (see also Tëmkin 2010). The conflict between the two datasets could be due to the protein-coding genes of Sharma et al. (2012), and we prefer to leave this unresolved. The direct optimisation trees are also compatible with the recent results by Xue et al. (2012) based on probabilistic analyses of a large sample of 28S rRNA data, although they decided to include Pinnoidea in their order Pteriina ( = Ostreida herein).
The clade Arcida (Arcoidea) is well supported in all analyses (Figs 26–31). The position of the representative of Limopsidae does not support the hypothesis of a separate superfamily Limopsoidea (see also Xue et al. 2012). The family Arcidae as currently understood does not appear to be monophyletic, because multiple relationships between Noetiidae and some members of Arcidae appear in the different analyses. Arca, for example, groups with Arcopsis (Noetiidae) and Barbatia with Glycymeris + Limopsis (Glycymerididae + Limopsidae) in the nine-gene + morphology Bayesian analysis (Fig. 31). Glycymerididae + Limopsidae appears supported in different analyses, as in Limopsoidea sensu Beesley et al. (1998). Inclusion of members of the family Philobryidae should further test monophyly of Limopsoidea, which, however, appears nested within the family Arcidae. Resolution of Arcida affinities of its members remain some of the most contentious issues in bivalve taxonomy, as discussed in earlier reviews (Giribet and Distel 2003; Giribet 2008). The constituent families are supported by very few synapomorphic characters (Oliver and Holmes 2006; Xue et al. 2012), although they are similar in shell microstructure consisting of an outer crossed-lamellar and an inner crossed-lamellar shell layers only, both of which are penetrated by tubules passing through the entire thickness of the shell. This microstructural arrangement is quite distinct from the other pteriomorph taxa that show either some combination of calcitic prisms and nacre (with many Mytilida only nacre) or calcite prisms and foliated calcite with crossed-lamellar aragonite, and all of which lack shell tubules.
A clade of Limida (Limidae) + Pectinida is supported, the latter composed of two well supported clades, Pectinoidea (with Pectinidae, Spondylidae and Propeamussiidae) and a group formed by Anomiidae, Dimyidae, Plicatulidae and Placunidae (see also Xue et al. 2012). The latter four families are currently classified in three superfamilies, Anomioidea, Plicatuloidea and Dimyoidea, the latter two with a single family each. Anomioidea is sister to Pectinoidea (or Limoidea, in some analyses) and includes Anomiidae and Placunidae, but the monophyly of the superfamily is rejected by most analyses. Thus it might be more appropriate to group these four families into a single superfamily. Xue et al. (2012) proposed to use the clade Pectinoida (Pectinida here) for all members of Limida + Pectinida, maintaining the superfamilies Limoidea, Pectinoidea, Anomioidea, Dimyoidea and Plicatuloidea. However, their sampling included a single genus each for Anomioidea, Dimyoidea and Plicatuloidea, and therefore precluded testing the validity of these entities. The system that we propose maintains Limida and Pectinida as orders, and the superfamilies Limoidea, Pectinoidea and Anomioidea, the latter including the families Anomiidae, Dimyidae, Placunidae and Plicatulidae.
Heteroconchia
Heteroconchia is confirmed as monophyletic and composed of three clades, Archiheterodonta, Palaeoheteterodonta and Euheterodonta. To some degree, the resolution of these three clades depends on the analysis and method used, but most analyses contradict the traditional monophyly of Heterodonta ( = Archiheterodonta + Euheterodonta) as the sister-group to Palaeoheterodonta, and instead place Archiheterodonta in a more basal position in most of the five-gene analyses (e.g. Figs 26, 27, 29), or as sister to Palaeoheterodonta in the nine-gene analyses (Figs 30, 31), as also found in other recent studies (Wilson et al. 2010; Sharma et al. 2012). Potential synapomorphies of this clade include the hind end of the ctenidia unattached to the mantle (Purchon 1990), and the presence of Atkin’s type-D ciliary currents (see Sharma et al. 2012). Nevertheless, the shell microstructures of our exemplars from Archiheterodonta and Palaeoheterodonta are quite distinct, with the former having layers comprising combinations of crossed-lamellar, homogeneous and complex crossed-lamellar structures, whereas in the latter, the layers consist of aragonitic prismatic and nacreous structures.
Archiheterodonta (dark blue)
The well supported Archiheterodonta comprises Carditoidea (Carditidae) and Crassatelloidea (Crassatellidae + Astartidae), a result corroborated in all analyses, with the caveat that our sampling is currently limited to three archiheterodont species and lacks Condylocardiidae. Detailed work addressing the relationships within Archiheterodonta is currently in progress (V. L. González and G. Giribet). Within this analysis, the members of this clade present a unique sperm morphology featuring more than six mitochondria (char. 206), which are laterally compressed (char. 207) and arranged around a modified centriolar complex (char. 203) and possses shells with a very distinctive dentition pattern.
Palaeoheterodonta (orange)
The likewise well supported clade Palaeoheterodonta consists of Trigoniida (Trigoniidae; represented by a single species in our analyses) and Unionida, represented by four species: one Etherioidea (Iridinidae: Aspatharia pfeifferiana); one Hyrioidea (Hyriidae: Velesunio ambiguus); and two Unionoidea (Margaritiferidae: Margaritifera margaritifera; Unionidae: Unio pictorum). Unionoidea is not monophyletic in any analysis and although support for the internal Unionida relationships is low, most analyses place Hyriidae in a basal position and Iridinidae as sister-group to Margaritiferidae. These results are at odds with recent analyses of unionoid relationships (e.g. Graf 2000; Graf and Cummings 2006, 2007; Whelan et al. 2011), and could be due to our limited sampling.
Both Archiheterodonta and Palaeoheterodonta have a long fossil record dating back to the Ordovician (e.g. Cope 2004). Throughout this long history, trigoniids and large crassatellids display remarkably conservative and unique hinges with prominent primary dentition of very large teeth involved in aligning the valves, and secondary dentition running transversely on the primary hinge teeth.
Euheterodonta
Euheterodonta consists of two well supported clades, Anomalodesmata (purple) and the remaining Heterodonta, to which we apply the name Imparidentia (light blue). Anomalodesmata was nested within the latter in the study of Giribet and Distel (2003), which introduced the name Euheterodonta. Here we find further support for a sister-group relationship between the Anomalodesmata and the newly defined Imparidentia.
Anomalodesmata
Anomalodesmata is confirmed as monophyletic and well supported in all analyses (Figs 26–31), as already shown in nearly all previous studies based on morphology (Morton 1981; Harper et al. 2000), molecules (Dreyer et al. 2003; Giribet and Distel 2003; Harper et al. 2006; Taylor et al. 2009; Plazzi et al. 2011; Sharma et al. 2012), or both (Harper et al. 2006). Unambiguous synapomorphies of the clade include the presence of a lithodesma (char. 15), arenophilic gland secretions (char. 46), and the presence of an acrosomal complex (vesicle + subacrosomal material) positioned posteriorly, associated with the midpiece (char. 193). One of the most interesting questions of anomalodesmatan evolution is the origin of the carnivorous septibranch mode of life, which was specifically addressed with denser taxon sampling by Harper et al. (2006), but left unresolved. Likewise, we find conflicting results. The optimal POY analyses of molecular data (parameter set 3221) in isolation (Fig. 26), or combined with morphology (Fig. 27), recovers the Septibranchia as the sister-group to the other anomalodesmatans, albeit with low jackknife support (<50%). The latter analysis strongly supports two other clades, one including the families (Laternulidae (Pandoridae + Lyonsiidae)) (JF = 96% and 93%, respectively) and another including Periplomatidae + Thraciidae + Cleidothaeridae + Myochamidae (JF = 92%). This analysis thus supports a deep division of Anomalodesmata into three clades, Septibranchia, and two lineages corresponding to the ‘lyonsiid’ and largely to the ‘thraciid’ lineages of Harper et al. (2006). Recognising the limitations of our study due to the lack of representation of Clavagellidae, Clistoconchidae, Euciroidae, Halonymphidae, Lyonsiellidae, Parilimyidae, Penicillidae, Pholadomyidae, Protocuspidaridae and Spheniopsidae (see Morton 2012), these relationships appear better resolved in the POY analyses than in the probabilistic ones (Figs 29–31), which reject Septibranchia and the ‘lyonsiid’ lineage, although without support for the alternative positions of Laternulidae and Verticordiidae. As discussed earlier, this lack of resolution might be due to the exclusion of the variable regions in the static alignments (see Lindgren and Daly 2007), especially rich in informative characters in anomalodesmatans. Thus, our results are largely congruent with those of Harper et al. (2006), and we add Periplomatidae, unsampled in their analysis, to their ‘thraciid’ lineage. Periploma had only been analysed molecularly in an earlier study on bivalve phylogeny (Adamkewicz et al. 1997), but that analysis only incorporated a short fragment of 18S rRNA, and Periploma appeared as sister-group to Solemya. Our study has potential implications for the classification of Anomalodesmata, which currently includes numerous superfamilies consisting of one to just a few families (e.g. Bieler et al. 2010). Myochamoidea is rejected and only Pandoroidea, from among the superfamilies represented by more than one family, survives in all analyses, suggesting that reassessment of the suprafamilial taxonomy of Anomalodesmata is warranted with denser family-level sampling.
Imparidentia (light blue)
Imparidentia new clade consists of several clades, the relationships of which are not fully resolved. These include:
(1) Lucinida has been a much discussed clade, currently considered to encompass the extant superfamilies Thyasiroidea and Lucinoidea (Bieler et al. 2010). Alternatively, Lucinoidea has been treated as encompassing the families Lucinidae, Fimbriidae, Thyasiridae, Ungulinidae and the extinct Mactromyidae (in Beesley et al. 1998). Others have also included the amphi-Atlantic freshwater family Cyrenoididae. As summarised by Taylor and Glover (2006) in their revision of Lucinoidea, species of the Recent genus Bathycorbis Iredale, 1930 have been claimed as living representatives of Mactromyidae (Cox et al. 1969) – a family with otherwise with no post-Cretaceous records. However, the affinities of these small (~5 mm) offshore bivalves from Australia, known only from shells, are uncertain. Fimbriinae nests within Lucinidae (Taylor and Glover 2006) and is now considered a subfamily of Lucinidae (Bieler et al. 2010; Taylor et al. 2011). Ungulinidae appears in a well supported clade as the sister-group of Cyamiidae + Gaimardiidae (see discussion below), and corroborates Taylor and Glover’s (2006) removal of Ungulinidae from Lucinoidea. Likewise, our results corroborate the removal of Cyrenoididae by Taylor et al. (2009), which appears in both studies in a well supported clade with the freshwater families Glauconomidae and Cyrenidae (see also Sharma et al. 2012). In addition, Taylor and Glover (2006), based on their results and those from prior analyses (Williams et al. 2004; see also Taylor et al. 2007a, 2007b), rejected monophyly of Lucinoidea (here as Lucinida) and removed Thyasiridae. This result implies that the specific chemosymbiosis with sulfide-oxidising bacteria housed in the ctenidia found in Thyasiridae and Lucinidae (e.g. Taylor and Glover 2006, 2010) had independent origins. With a much more comprehensive taxon sampling of Lucinidae and Thyasiridae, Taylor et al. (2007b) found strong support for a clade of non-thyasirid imparidentians, but Thyasiridae appeared as the most-basal euheterodont clade, with Anomalodesmata as more derived. Other studies using multiple molecular markers have found support for a clade of Thyasiridae + Lucinidae (e.g. Giribet and Distel 2003; Sharma et al. 2012), as found in several of our analyses. Only a single thyasirid species was analysed here, but direct optimisation analyses always support this sister-group relationship (Fig. 27), although with low support. In some cases, Gastrochaenidae nests within this clade (Figs 26, 28), a result clearly driven by some sort of systematic error. The probabilistic analyses, whether using five or nine genes (Figs 29, 30), favour Taylor and Glover’s (2006) strongly supported result, but with low support. This can be explained by some sort of conflict between the ‘workhorses’ of bivalve molecular phylogenetics (the mitochondrial genes 16S rRNA and COI; the nuclear ribosomal genes 18S rRNA and 28S rRNA; and the nuclear protein-encoding gene histone H3), and the nuclear protein-encoding genes used by Sharma et al. (2012). However, those same ‘workhorses’ support Lucinoidea in the POY analyses (Fig. 27), as does the total evidence probabilistic analysis (Fig. 31), and thus we favour the hypothesis of Lucinida monophyly. This has important implications for the origins of chemosymbiosis in lucinids and thyasirids. Work in progress using a larger taxon sampling (J.T. and E.G., unpubl. results), but with the same markers and methodologies, favours the previous result. Differences in ctenidial structure between the two families, differences in the symbioses (location and bacterial groups) and the absence of a thyasirid fossil record in the Early Mesozoic and Palaeozoic lend support to the non-monophyly of Lucinida, but our combined analyses support this clade and we consider the issue unresolved.
(2) Galeommatoidea is a well supported clade, but there is little known structure in this species-rich group. More than a dozen available family-group names have been introduced, and only recently a comprehensive analysis examining galeommatoid relationships has been published (Goto et al. 2012) that would allow dividing them into supported clades. Recent classifications (Bieler et al. 2010; Carter et al. 2011; Coan and Valentich-Scott 2012) used a preliminary arrangement that subdivided members of the superfamily into only two families, Galeommatidae and Lasaeidae; the former characterised by small or absent cardinal hinge teeth, the latter by cardinal hinge teeth developed in at least one valve. This arrangement does not reflect actual clade diversity (Goto et al. 2012). In the present analyses, Scintillona, our single representative of Galeommatidae, groups as sister-taxon to Bornia (Lasaeidae or Kelliidae of authors) within other members of Lasaeidae (or Montacutidae of authors), but in the analysis of Goto et al. (2012), Scintillona groups with several montacutids. Our sample size is too small to address the substructure of Galeommatoidea and is intended only to test its phylogenetic placement within Imparidentia.
(3) Adapedonta, a well supported clade consisting of Hiatellidae + Solenoidea ( = Solenidae + Pharidae), is found in all of the probabilistic analyses and in the optimal POY combined tree (Fig. 27), although Hiatellidae and Solenoidea form a grade in other direct optimisation analyses. This grouping has been found in previous analyses of ribosomal genes (e.g. Giribet and Distel 2003; Taylor et al. 2007b, 2009; Yuan et al. 2012a, 2012b). The name Adapedonta Cossmann & Peyrot, 1909, is here applied to this putative clade. The original concept of this grouping also included Myidae and Gastrochaenidae, but was subsequently modified by various authors (e.g. Thiele (1934) who used it at the same rank as Anomalodesmata). The position of Gastrochaenidae (Lamychaena) was poorly resolved in this study, but Myidae appears well resolved as a member of Neoheterodontei (see below). Characters tentatively uniting Adapedonta are: (1) the anterodorsal position of the dorsal hood with respect to the gastric shield (char. 80); (2) the absence of the oesophageal lip (char. 82); (3) simple transverse ridge pattern of the right wall sorting area (char. 111); and (4) the presence of a ridge posteriorly bordering the right wall sorting area (char. 112). Some combination (but not all four) of these character states also occurs in unrelated lineages, such as Thracia, Tellinoidea and some Neoheterodontei (e.g. Chione, Gaimardia, Gemma). Although ancestral state reconstructions suggest that these four characters in concert could constitute adapedont synapomorphies, the incidence of missing data in non-adapedont lineages for the characters of interest precludes conclusive morphological definition of this group.
(4) Cardiidae + Tellinoidea appears as a clade in all probabilistic analyses, albeit inconsistently supported, whereas the direct optimisation analyses support placement of Chamidae within this clade as sister-group to the Cardiidae. Monophyly of Tellinoidea (including Tellinidae, Donacidae, Semelidae, Solecurtidae, and Psammobiidae) is confirmed, but monophyly of Tellinidae (Macoma and Scissula) is not, with Macoma grouping instead with Abra. Resolving the internal relationships of Tellinoidea has proven difficult (Taylor et al. 2007b; see also a recent mitogenomic analysis by Yuan et al. 2012b) and is beyond the scope of these analyses and will require much denser taxon sampling and detailed anatomical analysis. A conspicuous character supporting this clade is the presence of the cruciform muscle (char. 72). Cardiidae is represented by two species, which form a clade in all analyses. Hemidonacidae, which had been grouped with Cardiidae into Cardioidea in earlier classifications (e.g. Beesley et al. 1998), does not group with Cardiidae in any analysis (see also Taylor et al. 2007b; Sharma et al. 2012), a result supported by studies of sperm ultrastructure (Healy et al. 2008b). Chamidae has appeared as the sister-group of Cardiidae in previous analyses mostly using parsimony (Giribet and Wheeler 2002; Giribet and Distel 2003), and this relationship appears in the present parsimony analyses as a stable hypothesis, although with low support (Figs 26–28). However, given the unstable position of Chamidae and its long branch in the probabilistic analyses (Figs 29–31), the affinities of this family remain unresolved. Regardless, Cardiidae + Tellinoidea (with or without Chamidae and Gastrochaenidae) constitutes the sister-group of Neoheterodontei (discussed below).
(5) Neoheterodontei was erected by Taylor et al. (2007b) as an unranked clade including a large diversity of imparidentians, i.e. Sphaerioidea (without Cyrenidae), Myoidea (Myidae and Corbulidae), Pholadoidea, Dreissenoidea, Gaimardioidea, Mactroidea, Ungulinoidea, Cyrenoidea (without Sphaeriidae), Chamoidea, Veneroidea (Veneridae plus Petricolidae), Glossidae, Hemidonacidae, Glauconomidae, Trapezidae, Arcticidae, Vesicomyidae and Kelliellidae. This clade had been recovered in earlier analyses (e.g. Adamkewicz et al. 1997; Giribet and Wheeler 2002; Giribet and Distel 2003; Taylor et al. 2005) for the most part. Our probabilistic analyses all recover Neoheterodontei (with moderate to high support) (Figs 29–31), as proposed originally, but the direct optimisation analyses exclude Chamidae (see above) (Figs 26–28).
(5.1) Sphaeriidae (Sphaerium) appears as the most basal offshoot of Neoheterodontei in all analyses with moderate support.
(5.2) Myida is a well supported clade in all analyses, and appears as the sister-group to all non-sphaeriid Neoheterodontei. The circumscription of Myida has changed drastically over time. For example, Newell (1965: 20) included Myoidea, Gastrochaenoidea and Hiatelloidea, all of which now group in different branches of Imparidentia. The current composition, including Myidae, Corbulidae, Pholadidae and Teredinidae, agrees with the proposed arrangement by Bieler et al. (2010: 131), to which Dreissenidae is added here following the results of virtually all previous molecular phylogenetic analyses of heterodonts (e.g. ribosomal data of Giribet and Distel 2003; Taylor et al. 2007b). However, relationships within this clade differ among analyses. The direct optimisation analyses all support monophyly of Pholadoidea and of Myoidea, and a sister-group relationship between Myoidea and Dreissenidae (as in Taylor et al. 2007b), but the probabilistic analyses vary, some recovering this configuration (e.g. nine-gene analysis), while others do not support monophyly of Pholadoidea nor of Myoidea.
(5.3) Mactroidea (Mactridae + Mesodesmatidae) and Mactridae (with two species) are well supported in all analyses, and Mactroidea appears as the sister-group to all remaining Neoheterodontei, excluding Sphaeriidae and Myida.
(5.4) A clade composed of Ungulinidae + Cyamioidea ( = Cyamiidae + Gaimardiidae) is found in all analyses, further corroborating the clade Ungulinidae + Gaimardiidae recovered by Taylor et al. (2007b, 2009), and the removal of Ungulinidae from Lucinoidea (Williams et al. 2004; Taylor et al. 2007b; Taylor et al. 2009).
(5.5) Venerida remains a poorly resolved clade that includes several components:
(5.5.1) A clade including Vesicomyidae + Kelliellidae as the sister-group of Veneridae sensu lato (with Petricolidae and Turtoniidae) is found in all of the direct optimisation analyses (Figs 26–28) and in the Bayesian combined analysis of nine genes + morphology (Fig. 31). This clade receives moderate to low support, and is contradicted by the probabilistic analyses, which artificially nest Chamidae within Veneridae (Figs 29, 30). Vesicomyidae + Kelliellidae finds strong support in all analyses (see Krylova and Sahling 2010; Janssen and Krylova 2012), but contradicts the presumed composition of Glossoidea (e.g. Carter et al. 2011), because Glossidae often appears closely related to Arcticidae or Arcticidae + Trapezidae ( = Arcticoidea). Veneridae appears supported in all of the direct optimisation analyses and some of the probabilistic ones, although in some cases (e.g. nine-gene analysis), the well supported Chioninae + Venerinae clade appears as sister-group to Vesicomyidae + Kelliellidae. The internal phylogeny of Veneridae has been examined and discussed earlier using much denser taxon sampling (Kappner and Bieler 2006; Mikkelsen et al. 2006; Chen et al. 2011), and our results do not allow us to explore this problem further.
(5.5.2) A well supported clade of the freshwater–brackish water families Glauconomidae, Cyrenoididae, and Cyrenidae has been previously reported (Taylor et al. 2009; Sharma et al. 2012) and is also found in all of our analyses. This clade, encompassing the superfamilies Cyrenoidea and Cyrenoidoidea, was used to justify the further dismantling of Lucinoidea by Taylor et al. (2009), the previous position of Cyrenoididae. Members of Glauconome are deep burrowers with long siphons, while the members of the other two familes are shallow burrowers with short siphons. Glauconome and Corbicula both have a vacuolated layer in the periostracum, but this is also found in Arctica. Purchon (1987a: 267) highlighted a set of six stomach characters shared between Glauconome and Geloina (Cyrenidae), but Healy et al. (2006) rejected a relationship with Cyrenoidea based on sperm morphology, and suggested a possible connection to Arctica.
(5.5.3) Arcticidae, Glossidae, Trapezidae and Hemidonacidae are four families of medium- to large-sized bivalves that form a clade (e.g. Figs 28–30) or a grade (Fig. 27) in most analyses. In the Bayesian total evidence tree, Chamidae also nests within a clade formed by these four families (Fig. 31), but this relationship is never found in the direct optimisation analyses. The direct optimisation analyses consistenly retrieve a topology of (Arctica (Trapezium + Glossus)), but this is not found in the probabilistic ones, which place Glossus with Arctica (PP = 0.99 in the total evidence tree) or with Trapezium, but without support. Stability in the direct optimisation trees (sensu Giribet 2003) probably best reflects the phylogeny of these families.
The position of Hemidonacidae is highly unstable, and multiple options emerge from the different analyses, including as a sister-group to Arcticidae (Figs 29, 30), as sister to a clade containing three other families (Fig. 28), as sister to all other Venerida (Fig. 27), or as sister to Chamidae in the Bayesian total evidence tree (Fig. 31). Hemidonacidae has appeared as sister to a clade including Veneridae sensu lato and Chamidae in previous analyses (Taylor et al. 2007b) and has been difficult to position even with sperm ulstrastructure (Healy et al. 2008b).
(6) Chamidae occurs on a very long branch and its placement is highly unstable among analyses, supported within Neoheterodontei, and typically associated to Venerida, in the probabilistic analyses (Figs 29, 30), or outside of Neoheterodontei, with Cardiidae + Tellinoidea, as sister-group to Cardiidae, in the direct optimisation analyses (Figs 26–28). Morphologically, Chamidae are highly autapomorphic, and the spermatozoa of Chama macerophylla is of the primitive type present in many heterodont groups, including Cardiidae and Venerida, two of the alternative positions indicated here. The position of Chamidae has been long debated (see discussion in Taylor et al. 2007b), and is not resolved here, even when we conducted a probabilistic analysis excluding 18S rRNA sequences, which placed Chama as the sister-group to Hemidonax, although without support (BS = 30%; tree not shown).
The tempo of bivalve diversification
The fossil record of bivalves extends back to the Cambrian, and many modern lineages diversified during the Ordovician (e.g. Cope 2000, 2004). Our calibrated phylogeny (Fig. 32) suggests a Cambrian diversification of Autobranchia, Pteriomorphia, Heteroconchia, Archiheterodonta, Paleoheoterodonta and Euheterodonta. Diversification of Protobranchia is more recent (Silurian), but its highest posterior density interval spans the Late Cambrian through the Upper Devonian (but see Sharma et al. 2013 for a Mid- to Early Cambrian estimate with denser taxonomic sampling within protobranchiate bivalves). The major diversification of Pteriomorphia occurred during the early Paleozoic, whereas the diversification of the major heteroconch clades Archiheterodonta and Paleoheoterodonta is Mesozoic. Anomalodesmata mostly diversified during the Ordovician to the Devonian, although the ‘thraciid’ lineage did so in the Cretaceous. Imparidentia seems to have diversified steadily since its Ordovician origin, and continued generating higher diversity until the Cretaceous. This contrasts with other groups that have long branches separating their origin and the diversification of the modern forms likely indicative of major-lineage extinctions.
As demonstrated previously by Sharma et al. (2013), the chronogram of Protobranchia is unusual among marine invertebrates in that it captures the signature of the end-Permian mass extinction (Sharma et al. 2013: fig. 8). Drastic mass extinctions are visualised as a statistically significant depression in the accumulation of linages before the timing of extinction, engendering an anti-sigmoidal curve. To compare effects of the end-Permian mass extinction on major bivalve lineages, we visualised the log-lineage through time (LTT) curves derived from the dated tree topology’s branch lengths (Fig. 33). As with Protobranchia (Sharma et al. 2013), Palaeoheterodonta shows evidence of strongly depressed cladogenetic rate immediately before the Mesozoic. The shape of the Palaeoheterodonta curve is the consequence of the prolonged period between the diversification of Unionida (dated as 245.1 Ma) and its divergence from Trigoniida (473.5 Ma). But in contrast to Protobranchia, the absence of multiple relictual lineages of Palaeoheterodonta (like Trigonioida) diverging in the early Palaeozoic precludes the observation of a strong anti-sigmoidal curve. For example, the LTT curve of Palaeoheterodonta can also be reconciled with an exponential distribution, which corresponds to a density-dependent cladogenetic process. At present, Protobranchia constitutes an unusual case among bivalves (and indeed, other marine invertebrates) of a Cambrian crown group that has captured the signal of the most severe mass extinction in the history of animal life on Earth (Sharma et al. 2013).
The intriguing observation of accelerated diversification in Unionida and Archiheterodonta immediately subsequent to the end-Permian is comparable to such invertebrate lineages as Crinoidea, as all crown-group crinoids have been shown to stem from a single lineage surviving at the end-Permian (Rouse et al. 2013). However, these observations should be tempered with caution, due to limited taxonomic sampling for both lineages, which hinders both accurate measurement of the post-Palaeozoic net diversification rate, and possibly inference of actual diversification age.
By contrast, the accumulation curves for the better-sampled Imparidentia is inconsistent with an anti-sigmoidal curve with a point of inflection at the end-Permian. These data are consistent with Imparidentia, the most diverse ramus of the bivalve tree of life, weathering the end-Permian mass extinction better than other bivalve lineages. The accumulation curve of Pteriomorphia lies somewhere in between the two extremes; a roughly antisigmoidal curve is observed, but cannot be statistically distinguished from a density-dependent cladogenetic process (data not shown). This result is probably not due to actual deceleration of diversification, but to a taxonomic sampling artefact, as LTT plots with poor sampling of extant lineages will mimic a diversification rate slowdown (Cusimano and Renner 2010). The sampling of Anomalodesmata is not dispositive of either steady diversification or depressed cladogenesis due to pronounced limitations in taxonomic representation.
Future investigations of the effect of the end-Permian mass extinction on extant bivalve phylogeny should therefore focus on denser sampling of all bivalve lineages excepting Imparidentia. Molecular dating of a published two-gene Palaeoheterodonta dataset that sampled major lineages of Unionida (Graf and Cummings 2006), in concert with palaeontological data, may facilitate investigation of whether Unionida constitutes a true revenant clade (sensu Sharma and Wheeler 2013).
Phylogenetic signal in morphological characters
A significant portion of our efforts was dedicated towards construction of a morphological matrix, including characters from major character systems traditionally used to infer bivalve classification and phylogeny. The juxtaposition of such a matrix with a total evidence phylogeny is required to test which characters bear coherent phylogenetic signal. We observed that only the majority of characters derived from the shell and hinge, and those of sperm ultrastructure, are reliable indicators of phylogenetic relationships, even though the sperm character submatrix includes significant amounts of missing data (Fig. 34). This result accords with our total evidence topologies, which largely validate the traditional, higher-level classification of bivalves. Furthermore, phylogenies constructed upon isolation of phylogenetically informative characters demonstrate that these characters inform higher-level relationships, such as the monophyly of Pteriomorphia, Arcida, Palaeoheterodonta, Unionida, Anomalodesmata and Heteroconchia (Fig. 35). However, even isolating the phylogenetically informative characters from their noisy counterparts does not elucidate key relationships within Imparidentia, a result comparable to phylogenetic inference based upon molecular sequence data alone.
That many character systems thought to be of potential utility bear numerous noisy characters was an unexpected result. One potential concern for this analysis was the possibility of a correlation between the amount of missing data and the presence of phylogenetic signal. We tested for such a correlation using the mean proportion of missing data and the proportion of significantly informative phylogenetic characters in each character system. Little evidence for such a confounding correlation was obtained (Spearman rank correlation coefficient ρ = 0.0962). We similarly tested for correlation between the number of matrix cells with missing data and whether the character was scored as phylogenetically informative, for all characters and regardless of character system. As before, no evidence for correlation between these variables (ρ = -0.101) was recovered, and distributions of proportion of missing data are largely comparable between phylogenetically informative and noisy characters (Fig. 36). These observations suggest that missing data do not strongly influence the inference of phylogenetic informativeness in our dataset.
Our results are advocative of careful reconsideration of morphological characters used for inferring phylogenetic relationships. Of all character systems evaluated, exploration of sperm ultrastructural characters could prove the most fruitful for forthcoming morphological studies of extant taxa. Our data also offer promising prospects for the use of chararacters that can be readily inspected in fossil taxa for phylogenetic placement (namely, gross shell characters and shell microstructure).
Conclusions
The inferred relationships discussed above, especially those within the Heteroconchia, differ considerably from previous hypotheses and in particular from the recently proposed synoptic classification of Carter et al. (2011: 4 ff.). The extant taxa in that classification that are relevant to the current discussion are arranged in Table 5.

|
We provide a new analysis of bivalve relationships integrating novel morphological characters and a combination of up to nine molecular markers. Although some persistent problems in bivalve systematics are not completely eliminated (e.g. some deep relationships of Pteriomorphia and Imparidentia), we have made significant progress in resolving previously uncertain relationships and postulating novel hypotheses that now appear well supported. This has allowed us to refine the higher order bivalve classification (Table 6) reflecting the results of our varied analyses and datasets, resulting in the recognition of six major monophyletic lineages of modern Bivalvia: Protobranchia, Pteriomorphia, Palaeoheterodonta, Archiheterodonta, Anomalodesmata and Imparidentia. Among the most difficult issues remaining – and a high-value target for forthcoming phylogenetic efforts – is the position of Chamidae, which is highly sensitive to choice of optimality criterion.
Acknowledgements
The Bivalve Assembling the Tree-of-Life project (http://www.bivatol.org) is supported by the USA National Science Foundation (NSF) Assembling the Tree of Life (AToL) program (DEB-0732854 / 0732903 / 0732860). Fieldwork and specimen acquisition in Florida was partly supported by the Comer Science and Education Foundation, the Negaunee Foundation Ltd, the Grainger Foundation, and Field Museum’s (FMNH) Department of Zoology’s Marshall Field Fund. Specimen collecting in the protected waters of the Florida Keys was conducted under Florida Keys National Marine Sanctuary Research Permit FKNMS-2009–024 and USA Fish & Wildlife Service Special Use Permit 41580–2010–20 (and earlier issues of these permits) for work in the National Wildlife Refuges. Collecting in the Moreton Bay Marine Park was permitted under Permit number QS2005/CVL588 from Queensland Parks and Wildlife Service. Collecting in Spain was primarily supported by a sabbatical fellowship from the Ministerio de Educación y Ciencia, Spain (SAB2006–0124) to G. Giribet in 2007 to work in the Centre d’Estudis Avançats de Blanes (CEAB, CSIC), sponsored by Iosune Uriz, to whom we are indebted. Collecting was done under a general collecting permit from the CEAB. Additional specimens came from the Protostome Tree of Life project (NSF EF-0334932 and EF-0531757 to GG). Work on Cardiidae and Veneridae was supported, in part, by NSF award DEB-0918982 to RB and PMM. We acknowledge a grant from UC Ship Funds Panel to N.G. Wilson for a collecting trip with the R/V Robert Gordon Sproul, in the Santa Rosa-Cortes Ridge of the southern California continental borderland at depths to ca. 400 m. Nucinella specimens were obtained with the help of the Panglao Marine Biodiversity Project, a joint project of Muséum National d’Histoire Naturelle, Paris (Philippe Bouchet, PI) and University of San Carlos, Cebu City (Danilo Largo, PI), funded by grants from the Total Foundation, the French Ministry of Foreign Affairs, and the Asean Regional Center for Biodiversity Conservation (ARCBC), and operating under a permit from the Philippine Bureau of Fisheries and Aquatic Resources (BFAR). We thank the Captain and crew of the R/V Endeavour, George Hampson and Steve Aubrey for help in collecting deep-sea specimens from the Gay Head–Bermuda transect and for the invitation to participate from PI Ron Etter (University of Massachusetts, Boston), supported by the National Science Foundation (OCE-0726382). The specimens from deep-water off of Mozambique were collected by R/V Vizconde de Eza during the MAINBAZA cruise in April 2009. The cruise, under PI Philippe Bouchet, was operated by Muséum National d’Histoire Naturelle (MNHN) and Instituto Espanol de Oceanografia (IOE), as part of a cluster of Mozambique-Madagascar expeditions funded by the Total Foundation, Prince Albert II of Monaco Foundation, and Stavros Niarchos Foundation, and conducted by MNHN and Pro-Natura International (PNI). Dredging off of the coast of western Scotland aboard the R/V Prince Madog was jointly sponsored by Bangor University and the National Museum of Wales and organised by Chris Richardson and Graham Oliver. Collecting in Salcombe-Kingsbridge estuary, a local nature preserve and SSI (Site of Special Scientific Interest) in Devon, UK, was arranged under permit 1/2009. Collecting at Tjärnö, Sweden, was carried out during a marine biological workshop organised by Per Sundberg, Malin Strand, and Christer Erseus for the Swedish Taxonomy Initiative. Bill Anderson (South Carolina Department of Natural Resources, Charleston) made possible and provided assistance collecting coastal species in South Carolina. Logistical support was kindly provided by the Mote Marine Laboratory’s Tropical Research Laboratory (Summerland Key, Florida), the Smithsonian Marine Station (Ft. Pierce, Florida), the Moreton Bay Research Station (Stradbroke Island, Queensland), Swire Institute of Marine Sciences (Hong Kong), Sven Lovén Centre for Marine Sciences, Fiskebäckskil and Tjärnö (Sweden). Freshwater sampling in Australia was supported by NSF-0542575, facilitated by Hugh Jones of the New South Wales Department of Environment, Climate Change and Water, and exported under permit WT2010–8037. Freshwater sampling in Zambia was supported by NSF-0542575, facilitated by Alex Chilala of the Zambian Department of Fisheries, Ministry of Agriculture and Cooperatives, and collected under permit DFH/8/3/3. For assistance during fieldwork, we thank Brian Gollands (Paleontological Research Institution, Ithaca, NY), Sherry Reed (Smithsonian Marine Station, Ft. Pierce, FL), Petra Sierwald and Jochen Gerber (FMNH), Tan Koh Siang (University of Singapore), Martin Taylor (Bangor University), Peter Middelfart (Australian Museum, Sydney), Lisa Kirkendale (Western Australian Museum, Perth), and Anthony Geneva (Academy of Natural Sciences of Philadelphia). Although the majority of the material studied here was collected anew, we greatly appreciate the help of numerous colleagues and friends who provided additional specimens and tissues. These include David Duggins (Friday Harbor Laboratories, University of Washington, WA, USA), Anders Warén (Naturhistoriska Riksmuseet, Stockholm, Sweden), Lloyd Peck and Melody Clark (British Antarctic Survey, Cambridge, UK), Paul Valentich-Scott (Santa Barbara Museum of Natural History, CA, USA), Janet Voight (FMNH), Stacy Galleher (Oregon Department of Fish and Wildlife, Salem, OR, USA), Antonio Checa (Universidad de Granada, Spain), David Roberts (Queens University, Belfast, UK), Chris Richardson (School of Ocean Sciences, Bangor University, UK), Tan Koh Siang (University of Singapore, Singapore), Serge Gofas (Universidad de Málaga, Spain), Judith Fuchs (University of Göteborg, Sweden), and Alexandra Zieritz (University of Cambridge, UK). Thomas Waller (National Museum of Natural History, Washington, DC) is thanked for his identification of Propeamussiidae species, and Cleo Oliviera (Universidade Federal do Rio de Janeiro) advised on cuspidariid taxonomy. All processing, sectioning and photography for sperm ultrastructure (TEM and SEM) was carried out by BivAToL EM technician Erica Lovas (Queensland Museum) at the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, University of Queensland (staff here thanked for access to facilities and technical assistance). The Department of Earth Sciences, University of Cambridge and the Natural History Museum (London) are thanked for the free use of their SEM facilities for shell microstructural work. Janeen Jones (FMNH) helped with specimen and data management. Participants in Field Museum’s REU student internship program (supported by NSF DBI-084995 to Petra Sierwald), Emily Rudick (Temple University) and Hannah Wirtshafter (Carnegie Mellon University), assisted with SEM investigations. BivAToL illustrator Lisa Kanellos and research assistant Gracen Brilmyer (both FMNH) assisted with photography and artwork. The MorphoBank team, especially Maureen O’Leary, facilitated the morphological research, helped develop tools, and accommodated our many requests to incorporate new features. PPS was supported by NSF Postdoctoral Research Fellowship in Biology Grant No. DBI-1202751. The very constructive input on an earlier draft by Carmen Salas (University of Málaga) and an anonymous reviewer are greatly appreciated.
References
Adamkewicz, S. L., Harasewych, M. G., Blake, J., Saudek, D., and Bult, C. J. (1997). A molecular phylogeny of the bivalve mollusks. Molecular Biology and Evolution 14, 619–629.| A molecular phylogeny of the bivalve mollusks.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2szjtl2nsw%3D%3D&md5=5f4fbed845259a4209b321fd018cb12aCAS | 9190063PubMed |
Afzelius, B. A. (1995). Gustaf Retzius and spermatology. The International Journal of Developmental Biology 39, 675–685.
| 1:STN:280:DyaK283gvVOmsA%3D%3D&md5=76f88391e31e76a3a1ce4230132a9818CAS | 8645550PubMed |
Allen, J. A. (1976). On the biology and functional morphology of Chama gryphoides Linne (Bivalvia; Chamidae). Vie et Milieu 26A, 243–260.
Allen, J. A. (2000). An unusual suctorial montacutid bivalve from the deep Atlantic. Journal of the Marine Biological Association of the United Kingdom 80, 827–834.
| An unusual suctorial montacutid bivalve from the deep Atlantic.Crossref | GoogleScholarGoogle Scholar |
Allen, J. A., and Hannah, F. J. (1989). Studies on the deep sea Protobranchia: the subfamily Ledellinae (Nuculanidae). Bulletin of the British Museum Natural History. Zoology 55, 123–171.
Allen, J. A., and Sanders, H. L. (1982). Studies on the deep-sea Protobranchia (Bivalvia); the subfamily Spinulinae (family Nuculanidae). Bulletin of the Museum of Comparative Zoology 150, 1–30.
Allen, J. A., and Sanders, H. L. (1996). Studies on the deep-sea Protobranchia (Bivalvia): the family Neilonellidae and the family Nuculanidae. Bulletin of the Natural History Museum Zoology Series 62, 101–132.
Amler, M. R. W. (1999). Synoptical classification of fossil and Recent Bivalvia. Geologica et Palaeontologica 33, 237–248.
Amler, M., Fischer, R., and Rogalla, N. (2000). ‘Muscheln’. (Enke/Thieme Verlag: Stuttgart.)
Atkins, D. (1937). On the ciliary mechanisms and interrelationships of Lamellibranchs: Part III: Types of lamellibranch gills and their food currents. The Quarterly Journal of Microscopical Science 79, 375–421.
Babin, C. (1982). Mollusques bivalves et rostroconches. In ‘Brachiopodes (articules) et mollusques (bivalves, rostroconches, monoplacophores, gastropodes) de l’Ordovicien inférieur (Trémadocien-Arenigien) de la Montagne Noire (France méridionale)’. pp. 37–49. (Société des Études Scientifiques de l’Aude: Carcassone.)
Barrande, J. (1881). Système Silurien de Centre de la Bohême. Volume VI: Acéphales. Système Silurien de Centre de la Bohême. Volume VI: Acéphales 1–342.
Bassler, R. S. (1915). Bibliographic index of American Ordovician and Silurian fossils, volume 2. United States National Museum Bulletin 92, 719–1521.
Beesley, P. L., Ross, G. J. B., and Wells, A. (Eds) (1998). ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5.’ (CSIRO Publishing: Melbourne.)
Bieler, R., Carter, J. G., and Coan, E. V. (2010). Classification of bivalve families. Malacologia 52, 113–133.
Bieler, R., Mikkelsen, P. M., and Giribet, G. (2013). Bivalvia—a discussion of known unknowns. American Malacological Bulletin 31, 123–133.
| Bivalvia—a discussion of known unknowns.Crossref | GoogleScholarGoogle Scholar |
Bøggild, O. B. (1930). The shell structure of the mollusks. Det Kongelige Danske Videnskabernes Selskab, Skrifter, Naturvidenskabelige og Mathematiske Afhandlinger ser. 9 2, 231–326.
Bonetto, A. A., and Ezcurra, I. D. (1965). Notas malacologicas. III. 5) la escultura del periostraco en el género Anodontites, 6) el lasidium de Anodontites trapezeus (Spix), 7) el lasidium de Mycetopoda siliquosa (Spix). Physis Rio de Janeiro, Brazil 25, 197–204.
Booth, C. E., and Mangum, C. P. (1978). Oxygen uptake and transport in the lamellibranch mollusc Modiolus demissus. Physiological Zoology 51, 17–32.
Boss, K. J. (1982). Mollusca. In ‘Synopsis and Classification of Living Organisms’. (Ed. S. P. Parker.) pp. 1092–1166. (McGraw-Hill Book Company: New York.)
Boyd, S. E. (1998). Order Arcoida. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5’. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 253–261. (CSIRO Publishing: Melbourne.)
Boyle, E. E., Zardus, J. D., Chase, M. R., Etter, R. J., and Rex, M. A. (2004). Strategies for molecular genetic studies of preserved deep-sea macrofauna. Deep-sea Research. Part I, Oceanographic Research Papers 51, 1319–1336.
| Strategies for molecular genetic studies of preserved deep-sea macrofauna.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmsl2itLo%3D&md5=7ef0ed893e8ef3515cc496f7ac655bcdCAS |
Brooks, W. K. (1875). An organ of special sense in the lamellibranchiate genus Yoldia. Proceedings of the American Association for the Advancement of Science, 23rd meeting, Hartford, Connecticut 1874, 79–82.
Buckland-Nicks, J. (2008). Fertilization biology and the evolution of chitons. American Malacological Bulletin 25, 97–111.
| Fertilization biology and the evolution of chitons.Crossref | GoogleScholarGoogle Scholar |
Buckland-Nicks, J., and Scheltema, A. (1995). Was internal fertilization an innovation of early Bilateria? Evidence from sperm structure of a mollusc. Proceedings. Biological Sciences 261, 11–18.
| Was internal fertilization an innovation of early Bilateria? Evidence from sperm structure of a mollusc.Crossref | GoogleScholarGoogle Scholar |
Buckland-Nicks, J., Gibson, G., and Koss, R. (2002). Phylum Mollusca: Polyplacophora, Aplacophora, Scaphopoda. In ‘Atlas of Marine Invertebrate Larvae’. pp. 245-259. (Academic Press: San Diego, CA.)
Campbell, D. C. (2000). Molecular evidence on the evolution of the Bivalvia. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 31–46. (The Geological Society of London: London.)
Campbell, D. C., Hoekstra, K. J., and Carter, J. G. (1998). 18S ribosomal DNA and evolutionary relationships within the Bivalvia. In ‘Bivalves: An Eon of evolution—Palaeobiological studies honoring Norman D. Newell’. (Eds P. A. Johnston and J. W. Haggart.) pp. 75–85. (University of Calgary Press: Calgary.)
Carlos, A. A., Baillie, B. K., Kawachi, M., and Maruyama, T. (1999). Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain. Journal of Phycology 35, 1054–1062.
| Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXns1Cqt70%3D&md5=57fd17689ad9e01c6732ea8f3471f739CAS |
Caron, J.-B., Scheltema, A., Schander, C., and Rudkin, D. (2007a). Odontogriphus: earliest mollusk. In ‘McGraw-Hill Yearbook of Science & Technology 2008’. (McGraw-Hill Companies: New York.)
Caron, J.-B., Scheltema, A., Schander, C., and Rudkin, D. (2007b). Reply to Butterfield on stem-group “worms”: fossil lophotrochozoans in the Burgess Shale. BioEssays 29, 200–202.
| Reply to Butterfield on stem-group “worms”: fossil lophotrochozoans in the Burgess Shale.Crossref | GoogleScholarGoogle Scholar | 17226817PubMed |
Carter, J. G. (1978). Ecology and evolution of the Gastrochaenacea (Mollusca, Bivalvia) with notes on the evolution of the endolithic habitat. Peabody Museum of Natural History, Yale University, Bulletin 41, vi + 92 pp.
Carter, J. G. (1990a). Evolutionary significance of shell microstructure in the Palaeotaxodonta, Pteriomorphia and Isofilibranchia (Bivalvia: Mollusca). In ‘Skeletal Biomineralization: Patterns, Processes and Evolutionary Trends, v. 1’. (Ed. J. G. Carter.) pp. 135–296. (Van Nostrand Reinhold: New York.)
Carter, J. G. (1990b). Shell microstructural data for the Bivalvia. In ‘Skeletal Biomineralization: Patterns, Processes and Evolutionary Trends, v. 1’. (Ed. J. G. Carter) pp. 297–411. (Van Nostrand Reinhold: New York.)
Carter, J. G., Campbell, D. C., and Campbell, M. R. (2000). Cladistic perspectives on early bivalve evolution. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 47–79. (The Geological Society of London: London.)
Carter, J. G., Altaba, C. R., Anderson, L. C., Araujo, R., Biakov, A. S., Bogan, A. E., Campbell, D. C., Campbell, M., Jin-hua, C., Cope, J. C. W., Delvene, G., Dijkstra, H. H., Zong-jie, F., Gardner, R. N., Gavrilova, V. A., Goncharova, I. A., Harries, P. J., Hartman, J. H., Hautmann, M., Hoeh, W. R., Hylleberg, J., Bao-yu, J., Johnston, P., Kirkendale, L., Kleeman, K., Hoppka, J., Kriz, J., Machado, D., Malchus, N., Márquez-Aliaga, A., Masse, J.-P., McRoberts, C. A., Middelfart, P. U., Mitchell, S., Nevesskaja, L. A., Özer, S., Pojeta, J., Polubotko, I. V., Pons, J. M., Popov, S., Sánchez, T., Sartori, A. F., Scott, R. W., Sey, I. I., Signorelli, J. H., Silantiev, V. V., Skelton, P. W., Thomas, S., Waterhouse, J. B., Wingard, G. L., and Yancey, T. (2011). A synoptical classification of the Bivalvia (Mollusca). Paleontological Contributions 4, 1–47.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552.
| Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXisVSgt7g%3D&md5=b9e72f4f43e92e5aaa6c2696c6b069dbCAS | 10742046PubMed |
Chanley, P. E. (1968). Larval development in the Class Bivalvia. Symposium on Mollusca: Series 3, Part II. Cochin, India: Marine Biological Association of India, Marine Fisheries P.O., Mandapam Camp, India, 475–481 + IV pls.
Chanley, P. E., and Andrews, J. D. (1971). Aids for identification of bivalve larvae of Virginia. Malacologia 11, 45–119.
Checa, A. G., and Harper, E. M. (2010). Spikey bivalves: intra-periostracal crystal growth in anomalodesmatans. The Biological Bulletin 219, 231–248.
| 21183444PubMed |
Checa, A. G., Ramírez-Rico, J., González-Segura, A., and Sánchez-Navas, A. (2009). Nacre and false nacre (foliated aragonite) in extant monoplacophorans (= Tryblidiidae: Mollusca). Naturwissenschaften 96, 111–122.
| Nacre and false nacre (foliated aragonite) in extant monoplacophorans (= Tryblidiidae: Mollusca).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsV2qu77P&md5=df6536628c282a960f26e68a779f6526CAS | 18843476PubMed |
Checa, A. G., Harper, E. M., and Willinger, M. (2012). Aragonitic dendritic prismatic microstructure of the anomalodesmatan bivalve Thracia. Invertebrate Biology 131, 19–29.
| Aragonitic dendritic prismatic microstructure of the anomalodesmatan bivalve Thracia.Crossref | GoogleScholarGoogle Scholar |
Chen, J., Li, Q., Kong, L. F., and Zheng, X. D. (2011). Molecular phylogeny of venus clams (Mollusca, Bivalvia, Veneridae) with emphasis on the systematic position of taxa along the coast of mainland China. Zoologica Scripta 40, 260–271.
| Molecular phylogeny of venus clams (Mollusca, Bivalvia, Veneridae) with emphasis on the systematic position of taxa along the coast of mainland China.Crossref | GoogleScholarGoogle Scholar |
Clark, M. S., Thorne, M. A. S., Vieira, F. A., Cardoso, J. C. R., Power, D. M., and Peck, L. S. (2010). Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics 11, 362.
| Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing.Crossref | GoogleScholarGoogle Scholar | 20529341PubMed |
Coan, E. V., and Valentich-Scott, P. (2012). Bivalve seashells of tropical West America. Marine bivalve mollusks from Baja California to northern Peru. 2 vols, 1258 pp. (Santa Barbara Museum of Natural History: Santa Barbara, CA.)
Colgan, D. J., McLauchlan, A., Wilson, G. D. F., Livingston, S. P., Edgecombe, G. D., Macaranas, J., Cassis, G., and Gray, M. R. (1998). Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Australian Journal of Zoology 46, 419–437.
| Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution.Crossref | GoogleScholarGoogle Scholar |
Conti, S. (1954). Stratigrafi e paleontologia della Val Solda (Lago di Lugano). Memorie Descrittive della Carta Geologica d’Italia 30, 1–248.
Cope, J. C. W. (1996). The early evolution of the Bivalvia, In ‘Origin and Evolutionary Radiation of the Mollusca’. (Ed. J. D. Taylor.) pp. 361–371. (Oxford University Press: Oxford.)
Cope, J. C. W. (1997). The early phylogeny of the class Bivalvia. Palaeontology 40, 713–746.
Cope, J. C. W. (2000). A new look at early bivalve phylogeny. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 81–95. (The Geological Society of London: London.)
Cope, J. C. W. (2004). Bivalve and rostroconch mollusks. In ‘The Great Ordovician Biodiversification Event’. (Eds B. D. Webby and M. L. Droser.) pp. 196–208. (Columbia University Press: New York.)
Coppe, A., Bortoluzzi, S., Murari, G., Marino, I. A., Zane, L., and Papetti, C. (2012). Sequencing and characterization of striped Venus transcriptome expand resources for clam fishery genetics. PLoS ONE 7, e44185.
| Sequencing and characterization of striped Venus transcriptome expand resources for clam fishery genetics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVWlsbnJ&md5=7c3156ffd8b3dbf560332d91ded58a0cCAS | 23028497PubMed |
Cox, L. R., Newell, N. D., Boyd, D. W., Branson, C. C., Casey, R., Chavan, A., Coogan, A. H., Dechaseaux, C., Fleming, C. A., Haas, F., Hertlein, L. G., Kauffman, E. G., Myra Keen, A., LaRocque, A., McAlester, A. L., Moore, R. C., Nuttall, C. P., Perkins, B. F., Puri, H. S., Smith, L. A., Soot-Ryen, T., Stenzel, H. B., Trueman, E. R., Turner, R. D., and Weir, J. (1969). Part N. Bivalvia. In ‘Treatise on Invertebrate Paleontology, Part N, Mollusca 6’. (Ed. R. C. Moore.) pp. N2–N129. (The Geological Society of America and The University of Kansas: Lawrence, KS.)
Cragg, S. M. (1989). The ciliated rim of the velum of larvae of Pecten maximus (Bivalvia: Pectinidae). The Journal of Molluscan Studies 55, 497–508.
| The ciliated rim of the velum of larvae of Pecten maximus (Bivalvia: Pectinidae).Crossref | GoogleScholarGoogle Scholar |
Cusimano, N., and Renner, S. S. (2010). Slowdowns in diversification rates from real phylogenies may not be real. Systematic Biology 59, 458–464.
| Slowdowns in diversification rates from real phylogenies may not be real.Crossref | GoogleScholarGoogle Scholar | 20547781PubMed |
Dan, J. C., and Wada, S. K. (1955). Studies of the acrosome. IV. The acrosome reaction in some bivalve spermatozoa. The Biological Bulletin 109, 40–55.
| Studies of the acrosome. IV. The acrosome reaction in some bivalve spermatozoa.Crossref | GoogleScholarGoogle Scholar |
De Laet, J. (2010). A problem in POY tree searches (and its work-around) when some sequences are observed to be absent in some terminals. Cladistics 26, 453–455.
| A problem in POY tree searches (and its work-around) when some sequences are observed to be absent in some terminals.Crossref | GoogleScholarGoogle Scholar |
Dinamani, P. (1967). Variation in the stomach structure of the Bivalvia. Malacologia 5, 225–268.
Distel, D. L., and Roberts, S. J. (1997). Bacterial endosymbionts in the gills of the deep-sea wood-boring bivalves Xylophaga atlantica and Xylophaga washingtona. The Biological Bulletin 192, 253–261.
| Bacterial endosymbionts in the gills of the deep-sea wood-boring bivalves Xylophaga atlantica and Xylophaga washingtona.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2s3ptVymug%3D%3D&md5=4dbc3c99f43bdb80c69c4a090818dde7CAS | 9145497PubMed |
Dreher Mansur, M. C., and Meier-Brook, C. (2000). Morphology of Eupera Bourguignat, 1854 and Byssanodonta Orbigny, 1846 with contributions to the phylogenetic systematics of Sphaeriidae and Corbiculidae (Bivalvia: Veneroida). Archiv fuer Molluskenkunde 128, 1–59.
Drew, G. A. (1899). Some observations on the habits, anatomy and embryology of members of the Protobranchia. Anatomischer Anzeiger 15, 493–519.
Drew, G. A. (1901). The life-history of Nucula delphinodonta (Mighels). The Quarterly Journal of Microscopical Science 44, 313–392.
Dreyer, H., Steiner, G., and Harper, E. M. (2003). Molecular phylogeny of Anomalodesmata (Mollusca: Bivalvia) inferred from 18S rRNA sequences. Zoological Journal of the Linnean Society 139, 229–246.
| Molecular phylogeny of Anomalodesmata (Mollusca: Bivalvia) inferred from 18S rRNA sequences.Crossref | GoogleScholarGoogle Scholar |
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7, 214.
| BEAST: Bayesian evolutionary analysis by sampling trees.Crossref | GoogleScholarGoogle Scholar | 17996036PubMed |
Dunachie, J. F. (1963). The periostracum of Mytilus edulis. Royal Society of Edinburgh Transactions 65, 383–411.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797.
| MUSCLE: multiple sequence alignment with high accuracy and high throughput.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXisF2ks7w%3D&md5=0f644003e0450eab42e285b06371fbb5CAS | 15034147PubMed |
Edgecombe, G. D., and Giribet, G. (2006). A century later—a total evidence re-evaluation of the phylogeny of scutigeromorph centipedes (Myriapoda: Chilopoda). Invertebrate Systematics 20, 503–525.
| A century later—a total evidence re-evaluation of the phylogeny of scutigeromorph centipedes (Myriapoda: Chilopoda).Crossref | GoogleScholarGoogle Scholar |
Edgecombe, G. D., Giribet, G., and Wheeler, W. C. (2002). Phylogeny of Henicopidae (Chilopoda: Lithobiomorpha): a combined analysis of morphology and five molecular loci. Systematic Entomology 27, 31–64.
| Phylogeny of Henicopidae (Chilopoda: Lithobiomorpha): a combined analysis of morphology and five molecular loci.Crossref | GoogleScholarGoogle Scholar |
Elshahawi, S. I., Trindade-Silva, A. E., Hanora, A., Han, A. W., Flores, M. S., Vizzoni, V., Schrago, C. G., Soares, C. A., Concepcion, G. P., Distel, D. L., Schmidt, E. W., and Haygood, M. G. (2013). Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proceedings of the National Academy of Sciences of the United States of America 110, E295–E304.
| Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvFCgtL8%3D&md5=6651e9298d43015dd24ba6eb9f8fe55dCAS | 23288898PubMed |
Etter, R. J., Rex, M. A., Chase, M. R., and Quattro, J. M. (2005). Population differentiation decreases with depth in deep-sea bivalves. Evolution 59, 1479–1491.
| 16153033PubMed |
Etter, R. J., Boyle, E. E., Glazier, A., Jennings, R. M., Dutra, E., and Chase, M. R. (2011). Phylogeography of a pan-Atlantic abyssal protobranch bivalve: implications for evolution in the Deep Atlantic. Molecular Ecology 20, 829–843.
| Phylogeography of a pan-Atlantic abyssal protobranch bivalve: implications for evolution in the Deep Atlantic.Crossref | GoogleScholarGoogle Scholar | 21199034PubMed |
Eyster, L. S., and Morse, M. P. (1984). Early shell formation during molluscan embryogenesis, with new studies on the surf clam, Spisula solidissima. American Zoologist 24, 871–882.
Farris, J. S., Albert, V. A., Källersjö, M., Lipscomb, D., and Kluge, A. G. (1996). Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124.
| Parsimony jackknifing outperforms neighbor-joining.Crossref | GoogleScholarGoogle Scholar |
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. C. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3, 294–299.
| 1:CAS:528:DyaK2MXjt12gtLs%3D&md5=41891c10f444b637826cbf6001a4954cCAS | 7881515PubMed |
Franzén, Ä. (1955). Comparative morphological investigations into the spermiogenesis among Mollusca. Zoologiska Bidrag fran Uppsala 30, 399–456, + 2 pls.
Franzén, Ä. (1956). On spermiogenesis, morphology of the spermatozoon, and biology of fertilization among invertebrates. Zoologiska Bidrag fran Uppsala 31, 355–482, + 6 pls.
Franzén, Ä. (1983). Ultrastructural studies of spermatozoa in three bivalve species with notes on evolution of elongated sperm nucleus in primitive spermatozoa. Gamete Research 7, 199–214.
| Ultrastructural studies of spermatozoa in three bivalve species with notes on evolution of elongated sperm nucleus in primitive spermatozoa.Crossref | GoogleScholarGoogle Scholar |
Galtsoff, P. S. (1964). ‘The American Oyster Crassostrea virginica Gmelin’. (United States Government Printing Office: Washington, DC.)
Garcia-March, J. R., Márquez-Aliaga, A., and Carter, J. G. (2008). The duplivincular ligament of Recent Pinna nobilis L., 1758: further evidence for pterineid ancestry of the Pinnoidea. Journal of Paleontology 82, 621–627.
| The duplivincular ligament of Recent Pinna nobilis L., 1758: further evidence for pterineid ancestry of the Pinnoidea.Crossref | GoogleScholarGoogle Scholar |
Gilmour, T. H. J. (1990). The adaptive significance of foot reversal in the Limoida. In ‘The Bivalvia—Proceedings of a Memorial Symposium in Honour of Sir Charles Maurice Yonge, Edinburgh, 1986’. (Ed. B. Morton.) pp. 247–263 (Hong Kong University Press: Hong Kong.)
Giribet, G. (2003). Stability in phylogenetic formulations and its relationship to nodal support. Systematic Biology 52, 554–564.
| Stability in phylogenetic formulations and its relationship to nodal support.Crossref | GoogleScholarGoogle Scholar | 12857646PubMed |
Giribet, G. (2007). Efficient tree searches with available algorithms. Evolutionary Bioinformatics 3, 1–16.
Giribet, G. (2008). Bivalvia. In ‘Phylogeny and Evolution of the Mollusca’. (Eds W. F. Ponder and D. R. Lindberg.) pp. 105–141. (University of California Press: Berkeley, CA.)
Giribet, G., and Carranza, S. (1999). What can 18S rDNA do for bivalve phylogeny? Journal of Molecular Evolution 48, 256–258.
| What can 18S rDNA do for bivalve phylogeny?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXhs1yms74%3D&md5=e9babeb89152ea2d6e16ff3fde120f31CAS | 10627192PubMed |
Giribet, G., and Distel, D. L. (2003). Bivalve phylogeny and molecular data. In ‘Molecular Systematics and Phylogeography of Mollusks’. (Eds C. Lydeard and D. R. Lindberg.) pp. 45–90. (Smithsonian Books: Washington, DC.)
Giribet, G., and Shear, W. A. (2010). The genus Siro Latreille, 1796 (Opiliones, Cyphophthalmi, Sironidae), in North America with a phylogenetic analysis based on molecular data and the description of four new species. Bulletin of the Museum of Comparative Zoology 160, 1–33.
| The genus Siro Latreille, 1796 (Opiliones, Cyphophthalmi, Sironidae), in North America with a phylogenetic analysis based on molecular data and the description of four new species.Crossref | GoogleScholarGoogle Scholar |
Giribet, G., and Wheeler, W. C. (2002). On bivalve phylogeny: a high-level analysis of the Bivalvia (Mollusca) based on combined morphology and DNA sequence data. Invertebrate Biology 121, 271–324.
| On bivalve phylogeny: a high-level analysis of the Bivalvia (Mollusca) based on combined morphology and DNA sequence data.Crossref | GoogleScholarGoogle Scholar |
Giribet, G., Carranza, S., Baguñà, J., Riutort, M., and Ribera, C. (1996). First molecular evidence for the existence of a Tardigrada + Arthropoda clade. Molecular Biology and Evolution 13, 76–84.
| First molecular evidence for the existence of a Tardigrada + Arthropoda clade.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XhtVylur8%3D&md5=12d70fb007b406aad2b3d1737a48a9adCAS | 8583909PubMed |
Giribet, G., Okusu, A., Lindgren, A. R., Huff, S. W., Schrödl, M., and Nishiguchi, M. K. (2006). Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons. Proceedings of the National Academy of Sciences of the United States of America 103, 7723–7728.
| Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XlsVGksrs%3D&md5=f3c1f9f99e4257d8ba012068df2f5082CAS | 16675549PubMed |
Giribet, G., Sharma, P. P., Benavides, L. R., Boyer, S. L., Clouse, R. M., de Bivort, B. L., Dimitrov, D., Kawauchi, G. Y., Murienne, J. Y., and Schwendinger, P. J. (2012). Evolutionary and biogeographical history of an ancient and global group of arachnids (Arachnida: Opiliones: Cyphophthalmi) with a new taxonomic arrangement. Biological Journal of the Linnean Society 105, 92–130.
| Evolutionary and biogeographical history of an ancient and global group of arachnids (Arachnida: Opiliones: Cyphophthalmi) with a new taxonomic arrangement.Crossref | GoogleScholarGoogle Scholar |
Glover, E. A., and Taylor, J. D. (2010). Needles and pins: acicular crystalline periostracal calcification in venerid bivalves (Bivalvia: Veneridae). The Journal of Molluscan Studies 76, 157–179.
| Needles and pins: acicular crystalline periostracal calcification in venerid bivalves (Bivalvia: Veneridae).Crossref | GoogleScholarGoogle Scholar |
Goto, R., Kawakita, A., Ishikawa, H., Hamamura, Y., and Kato, M. (2012). Molecular phylogeny of the bivalve superfamily Galeommatoidea (Heterodonta, Veneroida) reveals dynamic evolution of symbiotic lifestyle and interphylum host switching. BMC Evolutionary Biology 12, 172.
| Molecular phylogeny of the bivalve superfamily Galeommatoidea (Heterodonta, Veneroida) reveals dynamic evolution of symbiotic lifestyle and interphylum host switching.Crossref | GoogleScholarGoogle Scholar | 22954375PubMed |
Graf, D. L. (2000). The Etherioidea revisited: a phylogenetic analysis of hyriid relationships (Mollusca: Bivalvia: Paleoheterodonta: Unionoida). Occasional Papers of the Museum of Zoology, University of Michigan 729, 1–21.
Graf, D. L., and Cummings, K. S. (2006). Palaeoheterodont diversity (Mollusca: Trigonioida + Unionoida): what we know and what we wish we knew about freshwater mussel evolution. Zoological Society of the Linnean Society 148, 343–394.
| Palaeoheterodont diversity (Mollusca: Trigonioida + Unionoida): what we know and what we wish we knew about freshwater mussel evolution.Crossref | GoogleScholarGoogle Scholar |
Graf, D. L., and Cummings, K. S. (2007). Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida). The Journal of Molluscan Studies 73, 291–314.
| Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida).Crossref | GoogleScholarGoogle Scholar |
Graham, A. (1949). The molluscan stomach. Transactions of the Royal Society of Edinburgh 61, 737–761.
| The molluscan stomach.Crossref | GoogleScholarGoogle Scholar |
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704.
| A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Crossref | GoogleScholarGoogle Scholar | 14530136PubMed |
Gustafson, R. G., and Lutz, R. A. (1992). Larval and early post-larval development of the protobranch bivalve Solemya velum (Mollusca: Bivalvia) . Journal of the Marine Biological Association of the United Kingdom 72, 383–402.
| Larval and early post-larval development of the protobranch bivalve Solemya velum (Mollusca: Bivalvia) .Crossref | GoogleScholarGoogle Scholar |
Gustafson, R. G., and Reid, R. G. B. (1986). Development of the pericalymma larva of Solemya reidi (Bivalvia: Cryptodonta: Solemyidae) as revealed by light and electron microscopy. Marine Biology 93, 411–427.
| Development of the pericalymma larva of Solemya reidi (Bivalvia: Cryptodonta: Solemyidae) as revealed by light and electron microscopy.Crossref | GoogleScholarGoogle Scholar |
Harper, E. M. (1992). Post-larval cementation in the Ostreidae and its implications for other cementing Bivalvia. The Journal of Molluscan Studies 58, 37–47.
| Post-larval cementation in the Ostreidae and its implications for other cementing Bivalvia.Crossref | GoogleScholarGoogle Scholar |
Harper, E. M. (1997). The molluscan periostracum: an important constraint in bivalve evolution. Palaeontology 40, 71–97.
Harper, E. M., Hide, E. A., and Morton, B. (2000). Relationships between the extant Anomalodesmata: a cladistic test. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 129–143. (The Geological Society of London: London.)
Harper, E. M., Dreyer, H., and Steiner, G. (2006). Reconstructing the Anomalodesmata (Mollusca: Bivalvia): morphology and molecules. Zoological Journal of the Linnean Society 148, 395–420.
| Reconstructing the Anomalodesmata (Mollusca: Bivalvia): morphology and molecules.Crossref | GoogleScholarGoogle Scholar |
Harry, H. W. (1985). Synopsis of the supraspecific classification of living oysters (Bivalvia: Gryphaeidae and Ostreidae). The Veliger 28, 121–158.
Haszprunar, G. (1985). The fine structure of the abdominal sense organs of Pteriomorpha (Mollusca, Bivalvia). The Journal of Molluscan Studies 51, 315–319.
Haszprunar, G. (1988). On the origin and evolution of major gastropod groups, with special reference to the Streptoneura (Mollusca). The Journal of Molluscan Studies 54, 367–441.
| On the origin and evolution of major gastropod groups, with special reference to the Streptoneura (Mollusca).Crossref | GoogleScholarGoogle Scholar |
Hautmann, M., and Hagdorn, H. (2013). Oysters and oyster-like bivalves from the Middle Triassic Muschelkalk of the Germanic Basin. Paläontologische Zeitschrift 87, 19–32.
| Oysters and oyster-like bivalves from the Middle Triassic Muschelkalk of the Germanic Basin.Crossref | GoogleScholarGoogle Scholar |
Healy, J. M. (1995). Comparative spermatozoal ultrastructure and its taxonomic and phylogenetic significance in the bivalve order Veneroida. In ‘Advances in Spermatozoal Phylogeny and Taxonomy’. (Eds B. G. Jamieson, J. Ausió and J.-L. Justine.) pp. 155–166. (Éditions du Muséum Paris: Paris.)
Healy, J. M. (1996). Molluscan sperm ultrastructure: correlation with taxonomic units within the Gastropoda, Cephalopoda and Bivalvia. In ‘Origin and Evolutionary Radiation of the Mollusca’. (Ed. J. Taylor.) pp. 99–113 (Oxford University Press: Oxford.)
Healy, J. M. (2000). Mollusca – relict taxa. In ‘Reproductive Biology of Invertebrates’, Vol. IX. Part B. Progress in Male Gamete Biology (Series Eds K. G. Adiyodi and R. G. Adiyodi; Vol. Ed. B. G. M. Jamieson.) pp. 21–79 (Oxford & IBH Publishing Co., New Delhi & Calcutta.)
Healy, J. M., Keys, J. L., and Daddow, L. Y. M. (2000). Comparative sperm ultrastructure in pteriomorphian bivalves with special reference to phylogenetic and taxonomic implications. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 169–190. (The Geological Society of London: London.)
Healy, J. M., Mikkelsen, P. M., and Bieler, R. (2006). Sperm ultrastructure in Glauconome plankta and its relevance to the affinities of the Glauconomidae (Bivalvia: Heterodonta). Invertebrate Reproduction & Development 49, 29–39.
| Sperm ultrastructure in Glauconome plankta and its relevance to the affinities of the Glauconomidae (Bivalvia: Heterodonta).Crossref | GoogleScholarGoogle Scholar |
Healy, J. M., Bieler, R., and Mikkelsen, P. M. (2008a). Spermatozoa of the Anomalodesmata (Bivalvia, Mollusca) with special reference to relationships within the group. Acta Zoologica Stockholm 89, 339–350.
| Spermatozoa of the Anomalodesmata (Bivalvia, Mollusca) with special reference to relationships within the group.Crossref | GoogleScholarGoogle Scholar |
Healy, J. M., Mikkelsen, P. M., and Bieler, R. (2008b). Sperm ultrastructure in Hemidonax pictus (Hemidonacidae, Bivalvia, Mollusca): comparison with other heterodonts, especially Cardiidae, Donacidae and Crassatelloidea. Zoological Journal of the Linnean Society 153, 325–347.
| Sperm ultrastructure in Hemidonax pictus (Hemidonacidae, Bivalvia, Mollusca): comparison with other heterodonts, especially Cardiidae, Donacidae and Crassatelloidea.Crossref | GoogleScholarGoogle Scholar |
Herdman, W. A. (1904). Anatomy of the pearl oyster (Margaritifera vulgaris, Schum.). In ‘Report to the Government of Ceylon on the pearl oyster fisheries of the Gulf of Manaar. Part 2’. (Ed. W. A. Herdman.) pp. 37–69 (The Royal Society: London.)
Hoagland, K. E., and Turner, R. D. (1981). Evolution and adaptive radiation of wood-boring bivalves (Pholadacea). Malacologia 21, 111–148.
| 1:CAS:528:DyaL38Xjt1SjsQ%3D%3D&md5=170fda474c7b6fa8d5b2f3d5756ae31aCAS |
Hodgson, A. N., and Bernard, R. T. F. (1986). Observations on the ultrastructure of the spermatozoon of two mytilids from the south-west coast of England. Journal of the Marine Biological Association of the United Kingdom 66, 385–390.
| Observations on the ultrastructure of the spermatozoon of two mytilids from the south-west coast of England.Crossref | GoogleScholarGoogle Scholar |
Hoeh, W. R., Black, M. B., Gustafson, R. G., Bogan, A. E., Lutz, R. A., and Vrijenhoek, R. C. (1998). Testing alternative hypotheses of Neotrigonia (Bivalvia: Trigonioida) phylogenetic relationships using cytochrome c oxidase subunit I DNA sequences. Malacologia 40, 267–278.
Hoeh, W. R., Bogan, A. E., and Heard, W. H. (2001). A phylogenetic perspective on the evolution of morphological and reproductive characteristics in the Unionoida: In ‘Ecology and Evolution of the Freshwater Mussels Unionoida’. (Eds G. Bauer and K. Wächtler.) pp. 257–280. (Springer-Verlag, Berlin.)
Huelsenbeck, J. P., and Ronquist, F. (2005). Bayesian analysis of molecular evolution using MrBayes. In ‘Statistical Methods in Molecular Evolution’. (Ed. R. Nielsen.) pp. 183–226. (Springer: New York.)
Jablonski, D., and Lutz, R. A. (1980). Molluscan larval shell morphology: ecological and paleontological applications. In ‘Skeletal Growth of Aquatic Organisms.’ (Eds D. C. Rhoads and R. A. Lutz.) pp. 323–377. (Plenum Press: New York.)
Jablonski, D., and Lutz, R. A. (1983). Larval ecology of marine benthic invertebrates: paleobiological implications. Biological Reviews of the Cambridge Philosophical Society 58, 21–89.
| Larval ecology of marine benthic invertebrates: paleobiological implications.Crossref | GoogleScholarGoogle Scholar |
Jablonski, D., Roy, K., and Valentine, J. W. (2006). Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102–106.
| Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVCiurvO&md5=4fbaeb375c53f070ae4060be81f1d2e7CAS | 17023653PubMed |
Jackson, R. T. (1890). Phylogeny of the Pelecypoda: the Aviculidae and their allies. Memoirs of the Boston Society of Natural History 4, 277–400.
Jamieson, B. G. M., and Rouse, G. W. (1989). The spermatozoa of the Polychaeta (Annelida): an ultrastructural review. Biological Reviews of the Cambridge Philosophical Society 64, 93–157.
| The spermatozoa of the Polychaeta (Annelida): an ultrastructural review.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3c%2FgtFequw%3D%3D&md5=993d730433a091807812eb28d9eb2f95CAS |
Janssen, R., and Krylova, E. M. (2012). Bivalves of the family Vesicomyidae from the Neogene Mediterranean basin (Bivalvia: Vesicomyidae). Archiv fuer Molluskenkunde 141, 87–113.
| Bivalves of the family Vesicomyidae from the Neogene Mediterranean basin (Bivalvia: Vesicomyidae).Crossref | GoogleScholarGoogle Scholar |
Jespersen, Å., Lützen, J., and Nielsen, C. (2004). On three species and two new genera (Montacutella and Brachiomya) of galeommatoid bivalves from the irregular sea urchin Brissus latecarinatus with emphasis on their reproduction. Zoologischer Anzeiger 243, 3–19.
| On three species and two new genera (Montacutella and Brachiomya) of galeommatoid bivalves from the irregular sea urchin Brissus latecarinatus with emphasis on their reproduction.Crossref | GoogleScholarGoogle Scholar |
Kappner, I., and Bieler, R. (2006). Phylogeny of venus clams (Bivalvia: Venerinae) as inferred from nuclear and mitochondrial gene sequences. Molecular Phylogenetics and Evolution 40, 317–331.
| Phylogeny of venus clams (Bivalvia: Venerinae) as inferred from nuclear and mitochondrial gene sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XntFWqs70%3D&md5=93360411a67e926dd6857c8ff3e18be8CAS | 16621616PubMed |
Kat, P. W. (1983). Conchiolin layers among the Unionidae and Margaritiferidae (Bivalvia): microstructural characteristics and taxonomic implications. Malacologia 24, 298–311.
Kawaguti, S. (1950). Observations of the heart shell, Corculum cardissa (L.) and its associated zooxanthellae. Pacific Science 4, 43–49.
Kawauchi, G. Y., Sharma, P. P., and Giribet, G. (2012). Sipunculan phylogeny based on six genes, with a new classification and the descriptions of two new families. Zoologica Scripta 41, 186–210.
| Sipunculan phylogeny based on six genes, with a new classification and the descriptions of two new families.Crossref | GoogleScholarGoogle Scholar |
Kennedy, W. J., Morris, N. J., and Taylor, J. D. (1970). The shell structure, mineralogy and relationships of the Chamacea (Bivalvia). Palaeontology 13, 379–413.
Kirkendale, L. (2009). Their day in the sun: molecular phylogenetics and origin of photosymbioisis in the ‘other’ group of photosymbiotic marine bivalves (Cardiidae: Fraginae). Biological Journal of the Linnean Society 97, 448–465.
| Their day in the sun: molecular phylogenetics and origin of photosymbioisis in the ‘other’ group of photosymbiotic marine bivalves (Cardiidae: Fraginae).Crossref | GoogleScholarGoogle Scholar |
Kocot, K. M. (2013). Recent advances and unanswered questions in deep molluscan phylogenetics. American Malacological Bulletin 31, 195–208.
| Recent advances and unanswered questions in deep molluscan phylogenetics.Crossref | GoogleScholarGoogle Scholar |
Kocot, K. M., Cannon, J. T., Todt, C., Citarella, M. R., Kohn, A. B., Meyer, A., Santos, S. R., Schander, C., Moroz, L. L., Lieb, B., and Halanych, K. M. (2011). Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456.
| Phylogenomics reveals deep molluscan relationships.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFers7fE&md5=ed483a49693e0582006f0b202c9109c0CAS | 21892190PubMed |
Korniushin, A. V., and Glaubrecht, M. (2002). Phylogenetic analysis based on the morphology of viviparous freshwater clams of the family Sphaeriidae (Mollusca, Bivalvia, Veneroida). Zoologica Scripta 31, 415–459.
| Phylogenetic analysis based on the morphology of viviparous freshwater clams of the family Sphaeriidae (Mollusca, Bivalvia, Veneroida).Crossref | GoogleScholarGoogle Scholar |
Korschelt, E., and Heider, K. (1858). ‘Text-book of the Embryology of Invertebrates.’ (S. Sonnenschein and Co., Ltd.: London.)
Krug, A. Z., Jablonski, D., Valentine, J. W., and Roy, K. (2009). Generation of Earth’s first-order biodiversity pattern. Astrobiology 9, 113–124.
| Generation of Earth’s first-order biodiversity pattern.Crossref | GoogleScholarGoogle Scholar | 19215204PubMed |
Krylova, E. M., and Sahling, H. (2010). Vesicomyidae (Bivalvia): current taxonomy and distribution. PLoS ONE 5, e9957.
| Vesicomyidae (Bivalvia): current taxonomy and distribution.Crossref | GoogleScholarGoogle Scholar | 20376362PubMed |
Le Pennec, M. (1980). The larval and post-larval hinge of some families of bivalve molluscs. Journal of the Marine Biological Association of the United Kingdom 60, 601–617.
| The larval and post-larval hinge of some families of bivalve molluscs.Crossref | GoogleScholarGoogle Scholar |
Lee, T. (2004). Morphology and phylogenetic relationships of genera of North American Sphaeriidae (Bivalvia, Veneroida). American Malacological Bulletin 19, 1–13.
Lee, S. W., Jang, Y. N., Ryu, K. W., Chae, S. C., Lee, Y. H., and Jeon, C. W. (2011). Mechanical characteristics and morphological effect of complex crossed structure in biomaterials. Fracture mechanics and microstructure of chalky layer in oyster shell. Micron Oxford, England 42, 60–70.
| Mechanical characteristics and morphological effect of complex crossed structure in biomaterials. Fracture mechanics and microstructure of chalky layer in oyster shell.Crossref | GoogleScholarGoogle Scholar |
Lewis, P. O. (2001). A likelihood approach to estimating phylogeny from discrete morphological characters data. Systematic Biology 50, 913–925.
| A likelihood approach to estimating phylogeny from discrete morphological characters data.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38zntVKlsQ%3D%3D&md5=202212a243f461d202522e8b1b6b5353CAS | 12116640PubMed |
Lewy, Z., and Samtleben, C. (1979). Functional morphology and paleontological significance of the conchiolin layers in corbulid pelecypods. Lethaia 12, 341–351.
| Functional morphology and paleontological significance of the conchiolin layers in corbulid pelecypods.Crossref | GoogleScholarGoogle Scholar |
Lindgren, A. R., and Daly, M. (2007). The impact of length-variable data and alignment criterion on the phylogeny of Decapodiformes (Mollusca: Cephalopoda). Cladistics 23, 464–476.
| The impact of length-variable data and alignment criterion on the phylogeny of Decapodiformes (Mollusca: Cephalopoda).Crossref | GoogleScholarGoogle Scholar |
Linton, E. W. (2005). MacGDE: Genetic Data Environment for MacOS X. Software available at http://www.msu.edu/~lintone/macgde/
Lutz, R. A. (1985). Identification of bivalve larvae and postlarvae: a review of recent advances. American Malacological Bulletin, Special Edition 1, 59–78.
Lutz, R. A., Goodsell, J. G., Castagna, M., Chapman, S., Newell, C., Hidu, H., Mann, R., Jablonski, D., Kennedy, V., Siddall, S., Goldberg, R., Beattie, H., Falmagne, C., Chestnut, A., Partridge, A., and Cobleigh, U. T. (1982). Preliminary observations on the usefulness of hinge structures for identification of bivalve larvae. Journal of Shellfish Research 2, 65–70.
MacClintock, C. (1967). Shell structure of patelloid and bellerophontoid gastropods (Mollusca). Bulletin of the Peabody Museum of Natural History 22, 1–140.
Maddison, W. P., and Maddison, D. R. (2011). Mesquite: a modular system for evolutionary analysis. Version 2.75. Software available at http://mesquiteproject.org.
Malchus, N. (2004). Constraints in the ligament ontogeny and evolution of pteriomorphian Bivalvia. Palaeontology 47, 1539–1574.
| Constraints in the ligament ontogeny and evolution of pteriomorphian Bivalvia.Crossref | GoogleScholarGoogle Scholar |
Marshall, B. A. (2002). Some recent Thraciidae, Periplomatidae, Myochamidae, Cuspidariidae and Spheniopsidae (Anomalodesmata) from the New Zealand region and referral of Thracia reinga Crozier, 1966 and Scintillona benthicola Dell, 1956 to Tellimya Brown, 1827 (Montacutidae) (Mollusca: Bivalvia). Molluscan Research 22, 221–288.
| Some recent Thraciidae, Periplomatidae, Myochamidae, Cuspidariidae and Spheniopsidae (Anomalodesmata) from the New Zealand region and referral of Thracia reinga Crozier, 1966 and Scintillona benthicola Dell, 1956 to Tellimya Brown, 1827 (Montacutidae) (Mollusca: Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Matsumoto, M. (2003). Phylogenetic analysis of the subclass Pteriomorphia (Bivalvia) from mtDNA COI sequences. Molecular Phylogenetics and Evolution 27, 429–440.
| Phylogenetic analysis of the subclass Pteriomorphia (Bivalvia) from mtDNA COI sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjs1Wmsr4%3D&md5=f732e16fad35ed6799550a2aa7bf2676CAS | 12742748PubMed |
Mikkelsen, P. M., and Bieler, R. (2007). ‘Seashells of Southern Florida: Living Marine Mollusks of the Florida Keys and Adjacent Regions. Bivalves.’ (Princeton University Press: Princeton, NJ.) [Copyright date given as ‘2008’”]
Mikkelsen, P. M., Bieler, R., Kappner, I., and Rawlings, T. A. (2006). Phylogeny of Veneroidea (Mollusca: Bivalvia) based on morphology and molecules. Zoological Journal of the Linnean Society 148, 439–521.
| Phylogeny of Veneroidea (Mollusca: Bivalvia) based on morphology and molecules.Crossref | GoogleScholarGoogle Scholar |
Milan, M., Coppe, A., Reinhardt, R., Cancela, L. M., Leite, R. B., Saavedra, C., Ciofi, C., Chelazzi, G., Patarnello, T., Bortoluzzi, S., and Bargelloni, L. (2011). Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: genomic tools for environmental monitoring. BMC Genomics 12, 234.
| Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: genomic tools for environmental monitoring.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXmtlKlsLk%3D&md5=f68e521714ebeffa5f44121e67f3021dCAS | 21569398PubMed |
Mittelbach, G. G., Schemske, D. W., Cornell, H. V., Allen, A. P., Brown, J. M., Bush, M. B., Harrison, S. P., Hurlbert, A. H., Knowlton, N., Lessios, H. A., McCain, C. M., McCune, A. R., McDade, L. A., McPeek, M. A., Near, T. J., Price, T. D., Ricklefs, R. E., Roy, K., Sax, D. F., Schluter, D., Sobel, J. M., and Turelli, M. (2007). Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecology Letters 10, 315–331.
| Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography.Crossref | GoogleScholarGoogle Scholar | 17355570PubMed |
Miyazaki, I. (1962). On the identification of lamellibranch larvae. Bulletin of the Japanese Society of Scientific Fisheries 28, 955–966.
| On the identification of lamellibranch larvae.Crossref | GoogleScholarGoogle Scholar |
Monari, S. (2009). Phylogeny and biogeography of pholadid bivalve Barnea (Anchomasa) with considerations on the phylogeny of Pholadoidea. Acta Palaeontologica Polonica 54, 315–335.
| Phylogeny and biogeography of pholadid bivalve Barnea (Anchomasa) with considerations on the phylogeny of Pholadoidea.Crossref | GoogleScholarGoogle Scholar |
Morse, M. P., and Zardus, J. D. (1997). Bivalvia. In ‘Microscopic anatomy of invertebrates, Volume 6A: Mollusca II’. (Eds F. W. Harrison and A. J. Kohn.) pp. 7–11. (Wiley-Liss: New York.)
Morton, B. (1981). The Anomalodesmata. Malacologia 21, 35–60.
Morton, B. (1982). The biology, functional morphology and taxonomic status of Fluviolanatus subtorta (Bivalvia: Trapeziidae), a heteromyarian bivalve possessing ‘zooxanthellae’. Journal of the Malacological Society of Australia 5, 113–140.
Morton, B. (1987). The mantle margin and radial mantle glands of Entodesma saxicola and E. inflata (Bivalvia: Anomalodesmata: Lyonsiidae). The Journal of Molluscan Studies 53, 139–151.
| The mantle margin and radial mantle glands of Entodesma saxicola and E. inflata (Bivalvia: Anomalodesmata: Lyonsiidae).Crossref | GoogleScholarGoogle Scholar |
Morton, B. (2000). The biology and functional morphology of Fragum erugatum (Bivalvia: Cardiidae) from Shark Bay, Western Australia: the significance of its relationship with entrained zooxanthellae. Journal of Zoology 251, 39–52.
| The biology and functional morphology of Fragum erugatum (Bivalvia: Cardiidae) from Shark Bay, Western Australia: the significance of its relationship with entrained zooxanthellae.Crossref | GoogleScholarGoogle Scholar |
Morton, B. (2012). The functional morphology and inferred biology of the enigmatic South African ‘quadrivalve’ bivalve Clistoconcha insignis Smith, 1910 (Thracioidea: Clistoconchidae fam. nov.): another anomalodesmatan evolutionary eccentric. Transactions of the Royal Society of South Africa 67, 59–89.
| The functional morphology and inferred biology of the enigmatic South African ‘quadrivalve’ bivalve Clistoconcha insignis Smith, 1910 (Thracioidea: Clistoconchidae fam. nov.): another anomalodesmatan evolutionary eccentric.Crossref | GoogleScholarGoogle Scholar |
Morton, B. S., Prezant, R. S., and Wilson, B. (1998). Class Bivalvia. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5’. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 195–234. (CSIRO Publishing: Melbourne.)
Nakazima, M. (1958). On the differentiation of stomach of Pelecypoda (I). Venus Fukuyama-Shi, Japan 20, 197–207.
Nelson, T. C. (1918). On the origin, nature, and the function of the crystalline style of lamellibranchs. Journal of Morphology 31, 53–111.
| On the origin, nature, and the function of the crystalline style of lamellibranchs.Crossref | GoogleScholarGoogle Scholar |
Nevesskaja L. A. Skarlato O. A. Starobogatov Y. I. Eberzin A. G. (1971 ). Палеонтологический журнал 2, 3–20 [A new concept of the bivalve system; in Russian].
Newell, N. D. (1965). Classification of the Bivalvia. American Museum Novitates 2206, 1–25.
Ockelmann, K. W. (1965). Developmental types in marine bivalves and their distribution along the Atlantic coast of Europe. In ‘Proceedings of the First European Malacological Congress’. (Eds L. R. Cox and J. F. Peake.) pp. 25–35. (Conchological Society of Great Britain and Ireland and the Malacological Society of London: London.)
Ockelmann, K. W. (1983). Descriptions of mytilid species and definition of the Dacrydiinae, n. subfam. (Mytilacea—Bivalvia). Ophelia 22, 81–123.
| Descriptions of mytilid species and definition of the Dacrydiinae, n. subfam. (Mytilacea—Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Ohno, T., Katoh, T., and Yamasu, T. (1995). The origin of algal-bivalve photo-symbiosis. Palaeontology 38, 1–21.
Oliver, P. G., and Holmes, A. M. (2006). The Arcoidea (Mollusca: Bivalvia): a review of the current phenetic-based systematics. Zoological Journal of the Linnean Society 148, 237–251.
| The Arcoidea (Mollusca: Bivalvia): a review of the current phenetic-based systematics.Crossref | GoogleScholarGoogle Scholar |
Oliver, P. G., and Taylor, J. D. (2012). Bacterial symbiosis in the Nucinellidae (Bivalvia: Solemyida) with descriptions of two new species. The Journal of Molluscan Studies 78, 81–91.
| Bacterial symbiosis in the Nucinellidae (Bivalvia: Solemyida) with descriptions of two new species.Crossref | GoogleScholarGoogle Scholar |
Oliver, G., Rodrigues, C. F., and Cunha, M. R. (2011). Chemosymbiotic bivalves from the mud volcanoes of the Gulf of Cadiz, NE Atlantic, with descriptions of new species of Solemyidae, Lucinidae and Vesicomyidae. ZooKeys 113, 1–38.
| Chemosymbiotic bivalves from the mud volcanoes of the Gulf of Cadiz, NE Atlantic, with descriptions of new species of Solemyidae, Lucinidae and Vesicomyidae.Crossref | GoogleScholarGoogle Scholar |
Oliver, P. G., Southward, E. C., and Dando, P. R. (2013). Bacterial symbiosis in Syssitomya pourtalesiana Oliver, 2012 (Galeommatoidea: Montacutidae), a bivalve commensal with the deep-sea echinoid Pourtalesia. The Journal of Molluscan Studies 79, 30–41.
| Bacterial symbiosis in Syssitomya pourtalesiana Oliver, 2012 (Galeommatoidea: Montacutidae), a bivalve commensal with the deep-sea echinoid Pourtalesia.Crossref | GoogleScholarGoogle Scholar |
Owen, G. (1953). On the biology of Glossus humanus (L.) (Isocardia cor Lam.). Journal of the Marine Biological Association of the United Kingdom 32, 85–106.
Owen, G. (1955). Observations on the stomach and digestive diverticula of the Lamellibranchia. I. The Anisomyaria and Eulamellibranchia. The Quarterly Journal of Microscopical Science 96, 517–537.
Owen, G. (1956). Observations on the stomach and digestive diverticula of the Lamellibranchia. II. The Nuculidae. The Quarterly Journal of Microscopical Science 97, 541–567.
Owen, G. (1978). Classification and the bivalve gill. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 284, 377–385.
| Classification and the bivalve gill.Crossref | GoogleScholarGoogle Scholar |
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290.
| APE: analyses of phylogenetics and evolution in R language.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXms1eitg%3D%3D&md5=4346e988d5184eb70da871d50537f8b8CAS | 14734327PubMed |
Park, J. K., and Ó Foighil, D. (2000). Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Molecular Phylogenetics and Evolution 14, 75–88.
| Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXisFemtQ%3D%3D&md5=544edae28f9acf849b6699a77fdf441bCAS | 10631043PubMed |
Parkhaev, P. Y. (2008). The Early Cambrian radiation of Mollusca. In ‘Phylogeny and Evolution of the Mollusca’. (Eds W. F. Ponder and D. R. Lindberg.) pp. 33–69. (University of California Press: Berkeley, CA.)
Pelseneer, P. (1891). Contribution a l’étude des lamellibranches. Archives de Biologie 9, 147–312.
Pelseneer, P. (1911). ‘Les lamellibranches de l’expédition du Siboga. Partie anatomique’. (E. J. Brill: Leiden.)
Plazzi, F., and Passamonti, M. (2010). Towards a molecular phylogeny of mollusks: bivalves’ early evolution as revealed by mitochondrial genes. Molecular Phylogenetics and Evolution 57, 641–657.
| Towards a molecular phylogeny of mollusks: bivalves’ early evolution as revealed by mitochondrial genes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtlOhtrnO&md5=b31944646a34888f980f25d8a3b712eeCAS | 20817110PubMed |
Plazzi, F., Ceregato, A., Taviani, M., and Passamonti, M. (2011). A molecular phylogeny of bivalve mollusks: ancient radiations and divergences as revealed by mitochondrial genes. PLoS ONE 6, e27147.
| A molecular phylogeny of bivalve mollusks: ancient radiations and divergences as revealed by mitochondrial genes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsV2lu77P&md5=52347cfc7617f5ba49fc3fde780147a1CAS | 22069499PubMed |
Plazzi, F., Ribani, A., and Passamonti, M. (2013). The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genomics 14, 409.
| The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVKrtrrK&md5=2678369e9ec163bc83bf17a8e9835fc2CAS | 23777315PubMed |
Pojeta, J., Jr. (1987). Class Pelecypoda. In ‘Fossil Invertebrates’. (Eds R. S. Boardman, A. H. Cheetham and A. J. Rowell.) pp. 386–435. (Blackwell: Palo Alto, CA.)
Pojeta, J. (1988). The origin and Paleozoic diversification of solemyid pelecypods. New Mexico Bureau of Mines and Mineral Resources Memoir 44, 201–222.
Pojeta, J., and Runnegar, B. (1974). Fordilla troyensis and the early history of pelecypod mollusks. American Scientist 62, 706–711.
Pojeta, J., Runnegar, B., and Kriz, J. (1973). Fordilla troyensis Barrande: the oldest known pelecypod. Science 180, 866–868.
| Fordilla troyensis Barrande: the oldest known pelecypod.Crossref | GoogleScholarGoogle Scholar | 17789257PubMed |
Ponder, W. F., and Lindberg, D. R. (1997). Towards a phylogeny of gastropod molluscs: an analysis using morphological characters. Zoological Journal of the Linnean Society 119, 83–265.
| Towards a phylogeny of gastropod molluscs: an analysis using morphological characters.Crossref | GoogleScholarGoogle Scholar |
Popham, J. D., and Dickson, M. R. (1973). Bacterial associations in the teredo Bankia australis (Lamellibranchia: Mollusca). Marine Biology 19, 338–340.
| Bacterial associations in the teredo Bankia australis (Lamellibranchia: Mollusca).Crossref | GoogleScholarGoogle Scholar |
Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25, 1253–1256.
| jModelTest: phylogenetic model averaging.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXotlKgsb4%3D&md5=a227fa0ae4120a3a8c06cab96986f12aCAS | 18397919PubMed |
Prendini, L. (2001). Species or supraspecific taxa as terminals in cladistic analysis? Groundplans versus exemplars revisited. Systematic Biology 50, 290–300.
| Species or supraspecific taxa as terminals in cladistic analysis? Groundplans versus exemplars revisited.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38zntVOgtA%3D%3D&md5=606a00fdb3e16e481e97a1622815b2a6CAS | 12116933PubMed |
Prendini, L., Weygoldt, P., and Wheeler, W. C. (2005). Systematics of the Damon variegatus group of African whip spiders (Chelicerata: Amblypygi): evidence from behaviour, morphology and DNA. Organisms, Diversity & Evolution 5, 203–236.
| Systematics of the Damon variegatus group of African whip spiders (Chelicerata: Amblypygi): evidence from behaviour, morphology and DNA.Crossref | GoogleScholarGoogle Scholar |
Prezant, R. S. (1981). The arenophilic radial mantle glands of the Lyonsiidae (Bivalvia: Anomalodesmata) with notes on lyonsiid evolution. Malacologia 20, 267–289.
Purchon, R. D. (1954). A note on the biology of the lamellibranch Rocellaria (Gastrochaena) cuneiformis Spengler. Proceedings of the Zoological Society of London 124, 17–33.
| A note on the biology of the lamellibranch Rocellaria (Gastrochaena) cuneiformis Spengler.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1955). The structure and function of the British Pholadidae (rock-boring Lamellibranchia). Proceedings of the Zoological Society of London 124, 859–911.
| The structure and function of the British Pholadidae (rock-boring Lamellibranchia).Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1956). The stomach in the Protobranchia and Septibranchia (Lamellibranchia). Proceedings of the Zoological Society of London 127, 511–525.
| The stomach in the Protobranchia and Septibranchia (Lamellibranchia).Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1957). The stomach in the Filibranchia and Pseudolamellibranchia. Proceedings of the Zoological Society of London 129, 27–60.
| The stomach in the Filibranchia and Pseudolamellibranchia.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1958). The stomach in the Eulamellibranchia; stomach type IV. Proceedings of the Zoological Society of London 131, 487–525.
| The stomach in the Eulamellibranchia; stomach type IV.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1959). Phylogenetic classification of the Lamellibranchia, with special reference to the Protobranchia. Proceedings of the Malacological Society of London 33, 224–230.
Purchon, R. D. (1960). The stomach in the Eulamellibranchia; stomach types IV and V. Proceedings of the Zoological Society of London 135, 431–489.
| The stomach in the Eulamellibranchia; stomach types IV and V.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1978). An analytical approach to a classification of the Bivalvia. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 284, 425–436.
| An analytical approach to a classification of the Bivalvia.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1985). Studies on the internal structure and function of the stomachs of bivalve molluscs: stomach types III, IV and V. In ‘The Malacofauna of Hong Kong and Southern China. II Proceedings of the Second International Workshop on the Malacofauna of Hong Kong and Southern China, Hong Kong, 6–24 April 1983. Vol. 1’. (Eds B. Morton and D. Dudgeon.) pp. 337–361. (Hong Kong University Press: Hong Kong.)
Purchon, R. D. (1987a). The stomach in the Bivalvia. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 316, 183–276.
| The stomach in the Bivalvia.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1987b). Classification and evolution of the Bivalvia: an analytical study. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 316, 277–302.
| Classification and evolution of the Bivalvia: an analytical study.Crossref | GoogleScholarGoogle Scholar |
Purchon, R. D. (1990). Stomach structure, classification and evolution of the Bivalvia. In ‘The Bivalvia–Proceedings of a Memorial Symposium in Honour of Sir Charles Maurice Yonge, 1986.’ (Ed. B. Morton) pp. 73–95. (Hong Kong University Press: Hong Kong.)
Rambaut, A., and Drummond, A. J. (2009). Tracer v. 1.5. Program and Documentation. Software available at http://beast.bio.ed.ac.uk/Tracer/
Ranson, G. (1961). Les espèces d’huîtres perlières du genre Pinctada (Biologie de quelques-unes d’entre elles). Mémoires Institut Royal des Sciences Naturelles de Belgique 67, 3–95.
Rees, C. B. (1950). The identification and classification of lamellibranch larvae. Hull Bulletins of Marine Ecology 3, 73–104.
Reid, R. G. B. (1965). The structure and function of the stomach in bivalve molluscs. Journal of Zoology 147, 156–184.
| The structure and function of the stomach in bivalve molluscs.Crossref | GoogleScholarGoogle Scholar |
Reid, R. G. B. (1980). Aspects of the biology of a gutless species of Solemya (Bivalvia: Protobranchia). Canadian Journal of Zoology 58, 386–393.
| Aspects of the biology of a gutless species of Solemya (Bivalvia: Protobranchia).Crossref | GoogleScholarGoogle Scholar |
Reid, R. G. B. (1998). Order Solemyoidea. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5′. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 241–247.. (CSIRO Publishing: Melbourne.)
Reid, R. G. B., and Bernard, F. R. (1980). Gutless bivalves. Science 208, 609–610.
| Gutless bivalves.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cvhsl2nuw%3D%3D&md5=1c7d3555c3ba628aa66e9cce16f547b1CAS |
Rex, M. A., McClain, C. R., Johnson, N. A., Etter, R. J., Allen, J. A., Bouchet, P., and Warén, A. (2005). A source-sink hypothesis for abyssal biodiversity. American Naturalist 165, 163–178.
| A source-sink hypothesis for abyssal biodiversity.Crossref | GoogleScholarGoogle Scholar | 15729648PubMed |
Reynolds, P. D., and Okusu, A. (1999). Phylogenetic relationships among families of the Scaphopoda (Mollusca). Zoological Journal of the Linnean Society 126, 131–154.
| Phylogenetic relationships among families of the Scaphopoda (Mollusca).Crossref | GoogleScholarGoogle Scholar |
Ridewood, W. G. (1903). On the structure of the gills of lamellibranchs. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 195, 147–284.
| On the structure of the gills of lamellibranchs.Crossref | GoogleScholarGoogle Scholar |
Roe, K. J., and Hoeh, W. R. (2003). Systematics of freshwater mussels (Bivalvia: Unionoida). In ‘Molecular Systematics and Phylogeography of Mollusks’. (Eds C. Lydeard and D.R. Lindberg.) pp. 91–122. (Smithsonian Books: Washington, DC.)
Rouse, G. W., Jermiin, L. S., Wilson, N. G., Eeckhaut, I., Lanterbecq, D., Oji, T., Young, C. M., Browning, T., Cisternas, P., Helgen, L. E., Stuckey, M., and Messing, C. G. (2013). Fixed, free, and fixed: The fickle phylogeny of extant Crinoidea (Echinodermata) and their Permian-Triassic origin. Molecular Phylogenetics and Evolution 66, 161–181.
| Fixed, free, and fixed: The fickle phylogeny of extant Crinoidea (Echinodermata) and their Permian-Triassic origin.Crossref | GoogleScholarGoogle Scholar | 23063883PubMed |
Roy, K., Hunt, G., Jablonski, D., Krug, A. Z., and Valentine, J. W. (2009). A macroevolutionary perspective on species range limits. Proceedings. Biological Sciences 276, 1485–1493.
| A macroevolutionary perspective on species range limits.Crossref | GoogleScholarGoogle Scholar |
Sabatier, A. (1877). ‘Études sur la moule commune (Mytilus edulis)’. (Coulet: Montpelier.)
Salvini-Plawen, L. v. (1988). The structure and function of molluscan digestive systems. In ‘The Mollusca, v. 11, Form and Function’. (Eds E. R. Trueman and M. R. Clarke.) pp. 301–379. . (Academic Press: New York.)
Salvini-Plawen, L. v., and Steiner, G. (1996). Synapomorphies and plesiomorphies in higher classification of Mollusca. In ‘Origin and Evolutionary Radiation of the Mollusca. Centenary Symposium of the Malacological Society of London’. (Ed. J. D. Taylor.) pp. 29–51. (Oxford University Press: Oxford.)
Sanders, H. L., and Allen, J. A. (1973). Studies on deep-sea Protobranchia (Bivalvia); prologue and the Pristiglomidae. Bulletin of the Museum of Comparative Zoology 145, 237–262.
Sartori, A. F., Passos, F. D., and Domaneschi, O. (2006). Arenophilic mantle glands in the Laternulidae (Bivalvia: Anomalodesmata) and their evolutionary significance. Acta Zoologica Stockholm 87, 265–272.
| Arenophilic mantle glands in the Laternulidae (Bivalvia: Anomalodesmata) and their evolutionary significance.Crossref | GoogleScholarGoogle Scholar |
Saul, L. R. (1973). Evidence for the origin of the Mactridae (Bivalvia) in the Cretaceous. University of California Publications in Geological Sciences 97, 1–51.
Scarlato, O. A., and Starobogatov, Y. I. (1979). Osnovye cherty evoliutsii i sistema klassa Bivalvia [General evolutionary patterns and the system of the Class Bivalvia; in Russian]. In ‘Morfologiia, Sistematika i Filogeniia Molliuskov’. [Morphology, Systematics and Phylogeny of Mollusks] (Ed. Y. I. Starobogatov.) Akademiia Nauuk SSSR, Trudy Zoologicheskogo Instituta 80, 5–38. [For English translation, see K. J. Boss and M. K. Jacobson (1985), Museum of Comparative Zoology, Harvard University, Department of Mollusks, Special Occasional Publication 5, 1–67.]
Schneider, J. A. (1992). Preliminary cladistic analysis of the bivalve family Cardiidae. American Malacological Bulletin 9, 145–155.
Schneider, J. A. (1995). Phylogeny of the Cardiidae (Mollusca, Bivalvia): Protocardiinae, Laevicardiinae, Lahilliinae, Tulongocardiinae subfam. n. and Pleuriocardiinae subfam. n. Zoologica Scripta 24, 321–346.
| Phylogeny of the Cardiidae (Mollusca, Bivalvia): Protocardiinae, Laevicardiinae, Lahilliinae, Tulongocardiinae subfam. n. and Pleuriocardiinae subfam. n.Crossref | GoogleScholarGoogle Scholar |
Schneider, J. A. (1998). Phylogeny of the Cardiidae (Bivalvia): phylogenetic relationships and morphological evolution within the subfamilies Clinocardiinae, Lymnocardiinae, Fraginae and Tridacninae. Malacologia 40, 321–373.
| 1:CAS:528:DyaK1cXhsVOmtrg%3D&md5=a3785f4ab8d4047bb64f5ceed7a74bacCAS |
Schneider, J. A. (2001). Bivalve systematics during the 20th century. Journal of Paleontology 75, 1119–1127.
| Bivalve systematics during the 20th century.Crossref | GoogleScholarGoogle Scholar |
Schwendinger, P. J., and Giribet, G. (2005). The systematics of the south-east Asian genus Fangensis Rambla (Opiliones: Cyphophthalmi: Stylocellidae). Invertebrate Systematics 19, 297–323.
| The systematics of the south-east Asian genus Fangensis Rambla (Opiliones: Cyphophthalmi: Stylocellidae).Crossref | GoogleScholarGoogle Scholar |
Seydel, E. (1909). Untersuchen über den Byssusapparat der Lamellibranchiaten. Zoologische Jahrbucher. Abteilung fur Anatomie und Ontogenie der Tiere 27, 465–582.
Sharma, P. P., and Wheeler, W. C. (2013). Revenant clades in historical biogeography: the geology of New Zealand predisposes endemic clades to root age shifts. Journal of Biogeography 40, 1609–1618.
| Revenant clades in historical biogeography: the geology of New Zealand predisposes endemic clades to root age shifts.Crossref | GoogleScholarGoogle Scholar |
Sharma, P. P., Vahtera, V., Kawauchi, G. Y., and Giribet, G. (2011). Running WILD: The case for exploring mixed parameter sets in sensitivity analysis. Cladistics 27, 538–549.
| Running WILD: The case for exploring mixed parameter sets in sensitivity analysis.Crossref | GoogleScholarGoogle Scholar |
Sharma, P. P., González, V. L., Kawauchi, G. Y., Andrade, S. C. S., Guzmán, A., Collins, T. M., Glover, E. A., Harper, E. M., Healy, J. M., Mikkelsen, P. M., Taylor, J. D., Bieler, R., and Giribet, G. (2012). Phylogenetic analysis of four protein-encoding genes largely corroborates the traditional classification of Bivalvia (Mollusca). Molecular Phylogenetics and Evolution 65, 64–74.
| Phylogenetic analysis of four protein-encoding genes largely corroborates the traditional classification of Bivalvia (Mollusca).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFKntrrK&md5=1fc2e24a26eab00dedb57f631bbdaa3eCAS | 22659514PubMed |
Sharma, P. P., Zardus, J. D., Boyle, E. E., González, V. L., Jennings, R. M., McIntyre, E., Wheeler, W. C., Etter, R. J., and Giribet, G. (2013). Into the deep: A phylogenetic approach to the bivalve subclass Protobranchia. Molecular Phylogenetics and Evolution 69, 188–204.
| Into the deep: A phylogenetic approach to the bivalve subclass Protobranchia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtVOjtLvE&md5=6e3aade72d44b7f22afb718203f7b737CAS | 23742885PubMed |
Simone, L. R. L. (2009). Comparative morphology among representatives of main taxa of Scaphopoda and basal protobranch Bivalvia (Mollusca). Papéis Avulsos de Zoologia 49, 405–457.
Simone, L. R. L., and Chichvarkhin, A. (2004). Comparative morphological study of four species of Barbatia occurring on the southern Florida coast (Arcoidea, Arcidae). Malacologia 46(2, Bivalve Studies in the Florida Keys, R. Bieler and P. M. Mikkelsen, eds.), 355–379.
Simone, L. R. L., and Wilkinson, S. (2008). Comparative morphological study of some Tellinidae from Thailand (Bivalvia: Tellinoidea). The Raffles Bulletin of Zoology 18, 151–190.
Simone, L. R. L., Mikkelsen, P. M., and Bieler, R. (). Comparative anatomy of selected marine bivalves from Florida and a comparison with Brazilian taxa (Mollusca: Bivalvia). Malacologia XX, xx–yy.
Sipe, A. R., Wilbur, A. E., and Cary, S. C. (2000). Bacterial symbiont transmission in the wood-boring shipworm Bankia setacea (Bivalvia: Teredinidae). Applied and Environmental Microbiology 66, 1685–1691.
| Bacterial symbiont transmission in the wood-boring shipworm Bankia setacea (Bivalvia: Teredinidae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXisVWls78%3D&md5=dcafb0034be6745cf6e8c21082bc43caCAS | 10742261PubMed |
Slack-Smith, S. M. (1998a). Order Ostreoida. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5’. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 268–282. (CSIRO Publishing: Melbourne.)
Slack-Smith, S. M. (1998b). Superfamily Carditoidea. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5’. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 322–325.. (CSIRO Publishing: Melbourne.)
Slack-Smith, S. M. (1998c). Superfamily Crassatelloidea. In ‘Mollusca: The Southern Synthesis. Fauna of Australia. Vol. 5’. (Eds P. L. Beesley, G. J. B. Ross and A. Wells.) pp. 325–328.. (CSIRO Publishing: Melbourne.)
Smith, S., Wilson, N. G., Goetz, F., Feehery, C., Andrade, S. C. S., Rouse, G. W., Giribet, G., and Dunn, C. W. (2011). Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367.
| Resolving the evolutionary relationships of molluscs with phylogenomic tools.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlOmsrnJ&md5=9d9c59e21a03fce7c25f1cc4da0e541bCAS | 22031330PubMed |
Spagna, J. C., and Álvarez-Padilla, F. (2008). Finding an upper limit for gap costs in direct optimization parsimony. Cladistics 24, 787–801.
| Finding an upper limit for gap costs in direct optimization parsimony.Crossref | GoogleScholarGoogle Scholar |
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
| RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFKlsbfI&md5=b0df768ad6679df0c2d42f77d14048ddCAS | 16928733PubMed |
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57, 758–771.
| A rapid bootstrap algorithm for the RAxML Web servers.Crossref | GoogleScholarGoogle Scholar | 18853362PubMed |
Stasek, C. R. (1963). Synopsis and discussion of the association of ctenidia and labial palps in the bivalved Mollusca. The Veliger 6, 91–97.
Steiner, G. (1999). What can 18S rDNA do for bivalve phylogeny? Response. Journal of Molecular Evolution 48, 258–261.
| What can 18S rDNA do for bivalve phylogeny? Response.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXhs1yms7o%3D&md5=bde14f23e5262f58debce1b81eb2d5c7CAS |
Steiner, G., and Hammer, S. (2000). Molecular phylogeny of the Bivalvia inferred from 18S rDNA sequences with particular reference to the Pteriomorphia. In ‘The Evolutionary Biology of the Bivalvia’. (Eds E. M. Harper, J. D. Taylor and J. A. Crame.) pp. 11–29.. (The Geological Society of London: London.)
Steiner, G., and Müller, M. (1996). What can 18S rDNA do for bivalve phylogeny? Journal of Molecular Evolution 43, 58–70.
| What can 18S rDNA do for bivalve phylogeny?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xks1Grtbg%3D&md5=8d44cdcba7e8d0bae1f63475d67de14aCAS | 8660430PubMed |
Stempell, W. (1898). Beiträge zur Kenntniss der Nuculiden. Zoologische Jahrbücher. Supplementheft 4, 339–430.
Stuardo, J. R. (1968). On the phylogeny, taxonomy and distribution of the Limidae (Mollusca: Bivalvia). Ph.D. thesis, Harvard University.
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Tanaka, M., Ikuta, T., Shoguchi, E., Fujiwara, M., Shinzato, C., Hisata, K., Fujie, M., Usami, T., Nagai, K., Maeyama, K., Okamoto, K., Aoki, H., Ishikawa, T., Masaoka, T., Fujiwara, A., Endo, K., Endo, H., Nagasawa, H., Kinoshita, S., Asakawa, S., Watabe, S., and Satoh, N. (2012). Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Research 19, 117–130.
| Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlsFektr8%3D&md5=b8495543a4906a6fac43282af175558fCAS | 22315334PubMed |
Taylor, J. D. (1990). Field observations of prey selection by the muricid gastropods Thais clavigera and Morula musiva feeding on the intertidal oyster Saccostrea cucullata. In ‘Proceedings of the Second International Marine Biological Workshop: the Marine Flora and Fauna of Hong Kong and Southern China, Hong Kong’. (Ed. B. Morton.) pp. 837–855. (Hong Kong University Press: Hong Kong.)
Taylor, J. D., and Glover, E. A. (2000). Functional anatomy, chemosymbiosis and evolution of the Lucinidae. Geological Society of London Special Publications 177, 207–225.
| Functional anatomy, chemosymbiosis and evolution of the Lucinidae.Crossref | GoogleScholarGoogle Scholar |
Taylor, J. D., and Glover, E. A. (2006). Lucinidae (Bivalvia) – the most diverse group of chemosymbiotic molluscs. Zoological Journal of the Linnean Society 148, 421–438.
Taylor, J. D., and Glover, E. A. (2010). Chemosymbiotic bivalves. Topics in Geobiology 33, 107–135.
| Chemosymbiotic bivalves.Crossref | GoogleScholarGoogle Scholar |
Taylor, J. D., Kennedy, W. J., and Hall, A. (1969). The shell structure and mineralogy of the Bivalvia. Introduction. Nuculacea-Trigonacea. Bulletin of the British Museum Natural History, Zoology Suppl. 3, 1–125.
Taylor, J. D., Kennedy, W. J., and Hall, A. (1973). The shell structure and mineralogy of the Bivalvia II. Lucinacea–Clavagellacea, Conclusions. Bulletin of the British Museum Natural History, Zoology 22, 255–294.
Taylor, J. D., Glover, E. A., and Williams, S. T. (2005). Another bloody bivalve: anatomy and relationships of Eucrassatella donacina from south Western Australia (Mollusca: Bivalvia: Crassatellidae). In ‘The Marine Flora and Fauna of Esperance, Western Australia’. (Eds F. E. Wells, D. I. Walker and G. A. Kendrick.) pp. 261–288. (Western Australian Museum: Perth.)
Taylor, J. D., Williams, S. T., and Glover, E. A. (2007a). Evolutionary relationships of the bivalve family Thyasiridae (Mollusca: Bivalvia), monophyly and superfamily status. Journal of the Marine Biological Association of the United Kingdom 87, 565–574.
| Evolutionary relationships of the bivalve family Thyasiridae (Mollusca: Bivalvia), monophyly and superfamily status.Crossref | GoogleScholarGoogle Scholar |
Taylor, J. D., Williams, S. T., Glover, E. A., and Dyal, P. (2007b). A molecular phylogeny of heterodont bivalves (Mollusca: Bivalvia: Heterodonta): new analyses of 18S and 28S rRNA genes. Zoologica Scripta 36, 587–606.
| A molecular phylogeny of heterodont bivalves (Mollusca: Bivalvia: Heterodonta): new analyses of 18S and 28S rRNA genes.Crossref | GoogleScholarGoogle Scholar |
Taylor, J. D., Glover, E. A., and Williams, S. T. (2008). Ancient chemosynthetic bivalves: systematics of Solemyidae from eastern and southern Australia (Mollusca: Bivalvia). Memoirs of the Queensland Museum 54, 75–104.
Taylor, J. D., Glover, E. A., and Williams, S. T. (2009). Phylogenetic position of the bivalve family Cyrenoididae—removal from (and further dismantling of) the superfamily Lucinoidea. The Nautilus 123, 9–13.
Taylor, J. D., Glover, E. A., Smith, L., Dyal, P., and Williams, S. T. (2011). Molecular phylogeny and classification of the chemosymbiotic bivalve family Lucinidae (Mollusca: Bivalvia). Zoological Journal of the Linnean Society 163, 15–49.
Tëmkin, I. (2006). Morphological perspective on classification and evolution of Recent Pterioidea (Mollusca: Bivalvia). Zoological Journal of the Linnean Society 148, 253–312.
| Morphological perspective on classification and evolution of Recent Pterioidea (Mollusca: Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Tëmkin, I. (2010). Molecular phylogeny of pearl oysters and their relatives (Mollusca, Bivalvia, Pterioidea). BMC Evolutionary Biology 10, 342.
| Molecular phylogeny of pearl oysters and their relatives (Mollusca, Bivalvia, Pterioidea).Crossref | GoogleScholarGoogle Scholar | 21059254PubMed |
Tëmkin, I., and Strong, E. E. (2013). New insights on stomach anatomy of carnivorous bivalves. Journal of Molluscan Studies 79, 332–339.
Terwilliger, R. C., Terwilliger, N. B., and Arp, A. (1983). Thermal vent clam (Calyptogena magnifica) hemoglobin. Science 219, 981–983.
| Thermal vent clam (Calyptogena magnifica) hemoglobin.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXht1eisb4%3D&md5=cd43b040d86c158784981af4527db1daCAS | 17817936PubMed |
Thiele, J. (1886). Die Mundlappen der Lamellibranchiaten. Zeitschrift fur Wissenschartliche Zoologie 44, 239–272.
Thiele, J. (1934). Handbuch der systematischen Weichtierkunde. Vol. 2(3) [Scaphopoda; Bivalvia; Cephalopoda; additions and corrections for parts I and 2; index for part 3], 779–1022 (Gustav Fischer: Jena). Reprinted as ‘Handbook of Systematic Malacology’ by A. Asher & Co., Amsterdam, 1963; English language edition (Eds R. Bieler and P. M. Mikkelsen) by Smithsonian Institution Libraries/Amerind Publishing Co., 1998.
Thorson, G. (1950). Reproduction and larval ecology of marine bottom invertebrates. Biological Reviews of the Cambridge Philosophical Society 25, 1–45.
| Reproduction and larval ecology of marine bottom invertebrates.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaG3c%2FlsVKitg%3D%3D&md5=a20609ebdbe23a339e00ad14fec0a840CAS | 24537188PubMed |
Trench, R. K., Wethey, D. S., and Porter, J. W. (1981). Observations on the symbiosis with zooxanthellae among the Tridacninae. The Biological Bulletin 161, 180–198.
| Observations on the symbiosis with zooxanthellae among the Tridacninae.Crossref | GoogleScholarGoogle Scholar |
Ungvari, Z., Ridgway, I., Philipp, E. E. R., Campbell, C. M., McQuary, P., Chow, T., Coelho, M., Didier, E. S., Gelino, S., Holmbeck, M. A., Kim, I., Levy, E., Sosnowska, D., Sonntag, W. E., Austad, S. N., and Csiszar, A. (2011). Extreme longevity is associated with increased resistance to oxidative stress in Arctica islandica, the longest-living non-colonial animal. The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences 66, 741–750.
| Extreme longevity is associated with increased resistance to oxidative stress in Arctica islandica, the longest-living non-colonial animal.Crossref | GoogleScholarGoogle Scholar | 21486920PubMed |
Varón, A., Sy Vinh, L., and Wheeler, W. C. (2010). POY version 4: phylogenetic analysis using dynamic homologies. Cladistics 26, 72–85.
| POY version 4: phylogenetic analysis using dynamic homologies.Crossref | GoogleScholarGoogle Scholar |
Veniot, A., Bricelj, V. M., and Beninger, P. G. (2003). Ontogenetic changes in gill morphology and potential significance for food acquisition in the scallop Placopecten magellanicus. Marine Biology 142, 123–131.
von Ihering, H. (1891). Anodonta und Glabaris. Zoologischer Anzeiger 14, 474–484.
Waller, T. R. (1978). Morphology, morphoclines and a new classification of the Pteriomorphia (Mollusca: Bivalvia). Philosophical Transactions of the Royal Society of London, Series B 284, 345–365.
| Morphology, morphoclines and a new classification of the Pteriomorphia (Mollusca: Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Waller, T. R. (1990). The evolution of ligament systems in the Bivalvia. In ‘The Bivalvia—Proceedings of a Memorial Symposium in Honour of Sir Charles Maurice Yonge, Edinburgh, 1986’. (Ed. B. Morton.) pp. 49–71. (Hong Kong University Press: Hong Kong.)
Waller, T. R. (1998). Origin of the molluscan class Bivalvia and a phylogeny of major groups. In ‘Bivalves: An Eon of evolution—Palaeobiological studies honoring Norman D. Newell’. (Eds P. A. Johnston and J. W. Haggart.) pp. 1–45. (University of Calgary Press: Calgary.)
Waterbury, J. B., Calloway, C. B., and Turner, R. D. (1983). A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221, 1401–1403.
| A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXlsFCqu7o%3D&md5=e1d962d6ed56860814257bc3285d62fbCAS | 17759016PubMed |
Wheeler, W. C. (1995). Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Systematic Biology 44, 321–331.
Wheeler, W. (1996). Optimization alignment: the end of multiple sequence alignment in phylogenetics? Cladistics 12, 1–9.
| Optimization alignment: the end of multiple sequence alignment in phylogenetics?Crossref | GoogleScholarGoogle Scholar |
Wheeler, W. C., Aagesen, L., Arango, C. P., Faivovich, J., Grant, T., D’Haese, C., Janies, D., Smith, W. L., Varón, A., and Giribet, G. (2005). ‘Dynamic Homology and Phylogenetic Systematics: a Unified Approach using POY.’ (American Museum of Natural History: New York.)
Whelan, N. V., Geneva, A. J., and Graf, D. L. (2011). Molecular phylogenetic analysis of tropical freshwater mussels (Mollusca: Bivalvia: Unionoida) resolves the position of Coelatura and supports a monophyletic Unionidae. Molecular Phylogenetics and Evolution 61, 504–514.
| Molecular phylogenetic analysis of tropical freshwater mussels (Mollusca: Bivalvia: Unionoida) resolves the position of Coelatura and supports a monophyletic Unionidae.Crossref | GoogleScholarGoogle Scholar | 21827862PubMed |
White, K. M. (1937). ‘Mytilus.’ (University Press of Liverpool: Liverpool.)
Whitfield, R. P. (1891). Observations on some Cretaceous fossils from the Beyrut district of Syria, in the collection of the American Museum of Natural History, with descriptions of some new species. Bulletin of the American Museum of Natural History 3, 381–441.
Whiting, M. F., Carpenter, J. M., Wheeler, Q. D., and Wheeler, W. C. (1997). The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Systematic Biology 46, 1–68.
| 1:STN:280:DC%2BD383js1yqtQ%3D%3D&md5=86df74a7ce3b490b4da8ad4ca25d6b71CAS | 11975347PubMed |
Williams, S. T., Taylor, J. D., and Glover, E. A. (2004). Molecular phylogeny of the Lucinoidea (Bivalvia): non-monophyly and separate acquisition of bacterial chemosymbiosis. The Journal of Molluscan Studies 70, 187–202.
| Molecular phylogeny of the Lucinoidea (Bivalvia): non-monophyly and separate acquisition of bacterial chemosymbiosis.Crossref | GoogleScholarGoogle Scholar |
Wilson, N. G., Huang, D., Goldstein, M. C., Cha, H., Giribet, G., and Rouse, G. W. (2009). Field collection of Laevipilina hyalina McLean, 1979 from southern California, the most accessible living monoplacophoran. The Journal of Molluscan Studies 75, 195–197.
| Field collection of Laevipilina hyalina McLean, 1979 from southern California, the most accessible living monoplacophoran.Crossref | GoogleScholarGoogle Scholar |
Wilson, N. G., Rouse, G. W., and Giribet, G. (2010). Assessing the molluscan hypothesis Serialia (Monoplacophora + Polyplacophora) using novel molecular data. Molecular Phylogenetics and Evolution 54, 187–193.
| Assessing the molluscan hypothesis Serialia (Monoplacophora + Polyplacophora) using novel molecular data.Crossref | GoogleScholarGoogle Scholar | 19647088PubMed |
Xiong, B., and Kocher, T. D. (1991). Comparison of mitochondrial DNA sequences of seven morphospecies of black flies (Diptera: Simuliidae). Genome 34, 306–311.
| Comparison of mitochondrial DNA sequences of seven morphospecies of black flies (Diptera: Simuliidae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXkvVKnu7o%3D&md5=9432fb0c4b25a3f374f7f88a17a3a9efCAS | 2055453PubMed |
Xue, D.-X., Wang, H.-Y., Zhang, T., Zhang, S.-P., and Xu, F.-S. (2012). 基于 28S rRNA 基因片段的翼形亚纲 (Bivalvia: Pteriomorphia) 系统发育的初步研究 Oceanologia et Limnologia Sinica 43, 348–356.
| 1:CAS:528:DC%2BC38XhtFSqsLzE&md5=d3f78ed6b7263b9b70756ea515ecba73CAS |
Yonge, C. M. (1928). Structure and function of the organs of feeding and digestion in the septibranchs, Cuspidaria and Poromya. Philosophical Transactions of the Royal Society of London, Series B 216, 221–263.
| Structure and function of the organs of feeding and digestion in the septibranchs, Cuspidaria and Poromya.Crossref | GoogleScholarGoogle Scholar |
Yonge, C. M. (1939). The protobranchiate Mollusca: a functional interpretation of their structure and evolution. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences 230, 79–147, pl. 15.
Yonge, C. M. (1949). On the structure and adaptations of the Tellinacea, deposit-feeding Eulamellibranchia. Philosophical Transactions of the Royal Society 234, 29–76.
| On the structure and adaptations of the Tellinacea, deposit-feeding Eulamellibranchia.Crossref | GoogleScholarGoogle Scholar |
Yonge, C. M. (1962). On the primitive significance of the byssus in the Bivalvia and its effects in evolution. Journal of the Marine Biological Association of the United Kingdom 42, 113–125.
| On the primitive significance of the byssus in the Bivalvia and its effects in evolution.Crossref | GoogleScholarGoogle Scholar |
Yonge, C. M. (1978). Significance of the ligament in the classification of the Bivalvia. Proceedings of the Royal Society of London. Series B, Biological Sciences 202, 231–248.
| Significance of the ligament in the classification of the Bivalvia.Crossref | GoogleScholarGoogle Scholar |
Yonge, C. M. (1979). Cementation in bivalves. In ‘Pathways in Malacology’. (Eds S. van der Spoel, A. C. van Bruggen and J. Lever.) pp. 83–106. (Bohn, Scheltema and Holkema: Utrecht.)
Yonge, C. M. (1981). Functional morphology and evolution in the Tridacnidae (Mollusca: Bivalvia: Cardiacea). Records of the Australian Museum 33, 735–777.
| Functional morphology and evolution in the Tridacnidae (Mollusca: Bivalvia: Cardiacea).Crossref | GoogleScholarGoogle Scholar |
Yonge, C. M., and Morton, B. (1980). Ligament and lithodesma in the Pandoracea and the Poromyacea with a discussion on evolutionary history in the Anomalodesmata (Mollusca: Bivalvia). Journal of Zoology 191, 263–292.
| Ligament and lithodesma in the Pandoracea and the Poromyacea with a discussion on evolutionary history in the Anomalodesmata (Mollusca: Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Yuan, Y., Li, Q., Kong, L. F., and Yu, H. (2012a). The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): a novel gene order and unusual non-coding region. Molecular Biology Reports 39, 1287–1292.
| The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): a novel gene order and unusual non-coding region.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVOnsg%3D%3D&md5=6e2d85f113a1af39fe86b89b52ddd7afCAS | 21598108PubMed |
Yuan, Y., Li, Q., Yu, H., and Kong, L. (2012b). The complete mitochondrial genomes of six heterodont bivalves (Tellinoidea and Solenoidea): variable gene arrangements and phylogenetic implications. PLoS ONE 7, e32353.
| The complete mitochondrial genomes of six heterodont bivalves (Tellinoidea and Solenoidea): variable gene arrangements and phylogenetic implications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjsFyitbg%3D&md5=97862a64ac502e9cf171802b3cf7c497CAS | 22384227PubMed |
Zardus, J. D. (2002). Protobranch bivalves. Advances in Marine Biology 42, 1–65.
| Protobranch bivalves.Crossref | GoogleScholarGoogle Scholar | 12094722PubMed |
Zardus, J. D., and Martel, A. L. (2002). Phylum Mollusca: Bivalvia. In ‘Atlas of Marine Invertebrate Larvae’. (Eds C. M. Young, M. A. Sewell and M. E. Rice.) pp. 289–325. (Academic Press: London.)
Zardus, J. D., and Morse, M. P. (1998). Embryogenesis, morphology and ultrastructure of the pericalymma larva of Acila castrensis (Bivalvia: Protobranchia: Nuculoida). Invertebrate Biology 117, 221–244.
| Embryogenesis, morphology and ultrastructure of the pericalymma larva of Acila castrensis (Bivalvia: Protobranchia: Nuculoida).Crossref | GoogleScholarGoogle Scholar |
Zardus, J. D., Etter, R. J., Chase, M. R., Rex, M. A., and Boyle, E. E. (2006). Bathymetric and geographic population structure in the pan-Atlantic deep-sea bivalve Deminucula atacellana (Schenck, 1939). Molecular Ecology 15, 639–651.
| Bathymetric and geographic population structure in the pan-Atlantic deep-sea bivalve Deminucula atacellana (Schenck, 1939).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjsVGnsLg%3D&md5=9a89b7437e4cf212a1e5536c1918ca6bCAS | 16499691PubMed |
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., Yang, P., Zhang, L., Wang, X., Qi, H., Xiong, Z., Que, H., Xie, Y., Holland, P. W. H., Paps, J., Zhu, Y., Wu, F., Chen, Y., Wang, J., Peng, C., Meng, J., Yang, L., Liu, J., Wen, B., Zhang, N., Huang, Z., Zhu, Q., Feng, Y., Mount, A., Hedgecock, D., Xu, Z., Liu, Y., Domazet-Lošo, T., Du, Y., Sun, X., Zhang, S., Liu, B., Cheng, P., Jiang, X., Li, J., Fan, D., Wang, W., Fu, W., Wang, T., Wang, B., Zhang, J., Peng, Z., Li, Y., Li, N., Wang, J., Chen, M., He, Y., Tan, F., Song, X., Zheng, Q., Huang, R., Yang, H., Du, X., Chen, L., Yang, M., Gaffney, P. M., Wang, S., Luo, L., She, Z., Ming, Y., Huang, W., Zhang, S., Huang, B., Zhang, Y., Qu, T., Ni, P., Miao, G., Wang, J., Wang, Q., Steinberg, C. E. W., Wang, H., Li, N., Qian, L., Zhang, G., Li, Y., Yang, H., Liu, X., Wang, J., Yin, Y., and Wang, J. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54.
| The oyster genome reveals stress adaptation and complexity of shell formation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlKrtrvL&md5=4aec1c5294eb05b87ce01df353fabeb8CAS | 22992520PubMed |
Zieritz, A., Checa, A. G., Aldridge, D. C., and Harper, E. M. (2011). Variability, function and phylogenetic significance of periostracal microprojections in unionoid bivalves. Journal of Zoological Systematics and Evolutionary Research 49, 6–15.
| Variability, function and phylogenetic significance of periostracal microprojections in unionoid bivalves.Crossref | GoogleScholarGoogle Scholar |