Multilocus and mitogenomic phylogenetic analyses reveal a new genus and species of freshwater mussel (Bivalvia: Unionidae) from Guangxi, China
Yu-Ting Dai A , Xiao-Chen Huang A , Chen-Hui-Zi Wu A , Zhong-Guang Chen A , Liang Guo B , Feng-Yue Shu C , Shan Ouyang A and Xiao-Ping Wu
A , Chen-Hui-Zi Wu A , Zhong-Guang Chen A , Liang Guo B , Feng-Yue Shu C , Shan Ouyang A and Xiao-Ping Wu  A *
A *
A Jiangxi Province Key Laboratory of Watershed Ecosystem Change and Biodiversity, Center for Watershed Ecology, Institute of Life Science and School of Life Sciences, Nanchang University, Nanchang, 330031, PR China.
B Fuzhou Wilds of Insects Cultural Creativity Co., Ltd, Fuzhou, 350000, PR China.
C College of Life Sciences, Qufu Normal University, Qufu, 273165, PR China.
Invertebrate Systematics 37(2) 152-166 https://doi.org/10.1071/IS22048
Submitted: 14 September 2022 Accepted: 16 February 2023 Published: 14 March 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing.
Abstract
Freshwater mussels are essential for the integrity of freshwater ecosystems but numbers of these organisms are declining rapidly at regional and global scales. The phylogenetic and biogeographic aspects of the rich unionoid fauna of the Indo-Burma region are becoming increasingly well understood. Guangxi is part of the Chinese portion of the Indo-Burma biodiversity hotspot but regional studies of the freshwater mussel diversity are scarce. In this study, we report a new genus and species of freshwater mussel from Guangxi, China. Genetic datasets including three genes (COI, 16S rRNA and 28S rRNA) and complete maternal mitogenomes were compiled to infer the phylogenetic history of the group. Molecular phylogenetic analyses showed that the new species formed a monophyletic group and was closely related to Obovalis and Ptychorhynchus in the tribe Gonideini of the subfamily Gonideinae. Morphological and molecular evidence supported that these specimens represent an undescribed genus and species that we describe as Postolata guangxiensis gen. nov., sp. nov. The discovery of this new taxon adds to the known level of endemism of freshwater mussels in Guangxi and a detailed survey of uncharted areas should reveal new diversity in the future. We also suggest that complete mitogenomes or even genome-scale nuclear data should be used for phylogenetic reconstructions when proposing major taxonomic changes.
ZooBank: urn:lsid:zoobank.org:pub:76FC5A1D-7507-4F26-A12C-EC08AB333274
Keywords: conservation, Gonideinae, Indo-Burma, integrative taxonomy, mitochondrial genome, molecular systematics, morphological characters, Unionidae.
Introduction
Freshwater mussels (Mollusca: Bivalvia: Unionida) that include ~192 genera and 958 species worldwide (Graf and Cummings 2021) are essential for the integrity of freshwater ecosystems but numbers of these organisms are declining rapidly at regional and global scales (Lopes-Lima et al. 2021). China abounds in rivers and lakes, and is one of the major biodiversity hotspots for freshwater mussels (Zieritz et al. 2018). However, field investigations and studies of freshwater bivalves in China are geographically biased and have mainly been concentrated in the middle and lower reaches of the Yangtze River (Liu et al. 2022). Although a new species (Pseudocuneopsis sichuanensis Huang, Dai, Chen & Wu, 2022) from the upper Yangtze River in Sichuan Province and a newly recorded species (Cuneopsis demangei Haas, 1929) from the Pearl River basin in Guangdong Province were discovered during a recent study (Wu et al. 2022), there are still many localities that have been largely ignored and deserve comprehensive investigation.
The Guangxi Zhuang Autonomous Region (henceforth referred to as Guangxi), located in South China and bordering Vietnam, is part of the Chinese portion of the Indo-Burma biodiversity hotspot (Tordoff et al. 2012 and references therein). The Pearl River Basin, the second largest river basin in China, spans the entire territory of Guangxi. In addition, Guangxi has three climatic zones: northern tropics, southern subtropics and central subtropics (Kuang et al. 2007). Guangxi may harbour unique freshwater mussel fauna due to these unique climatic regimes and also karst topography.
According to He and Zhuang (2013), only eight freshwater mussel species were recorded in Guangxi, among which three specimens (i.e. Chamberlainia hainesiana (Lea, 1856), Cristaria plicata (Leach, 1814) and Gibbosula crassa (Wood, 1815)) from the National Zoological Museum of China and two specimens of Lens eximius (Lea, 1856) collected from South Guangxi were displayed. However, due to possible species misidentifications, whether the distribution range of Chamberlainia hainesiana and Lens eximius includes Guangxi remains to be further investigated (Pfeiffer et al. 2021; Goncalves et al. 2022). The flagship species of these Guangxi freshwater mussels, Gibbosula crassa, attracts the most because of its rarity and economic value (Liu et al. 1979a). The species was first recorded as Lamprotula mansuyi (Dautzenberg & Fischer, 1908) in China in the Youjiang River, Longzhou County, Guangxi (Liu et al. 1979b) and had been the only freshwater bivalve species under Grade-II conservation on the List of Wild Animals under State Priority Conservation in China until 2021 when seven other species (i.e. Margaritifera dahurica (Middendorff, 1850), Aculamprotula fibrosa (Heude, 1877), A. polysticta (Heude, 1877), A. scripta (Heude, 1875), Lamprotula leaii (Gray, 1833), Sinosolenaia carinata (Heude, 1877), and Novaculina chinensis Liu & Zhang, 1979) were also added to the Grade-II category. Unfortunately, populations of Gibbosula crassa may have been extirpated in China and can only be found in Vietnam (Lopes-Lima et al. 2018). Apart from these, little is known regarding the diversity and distribution of freshwater mussel species in Guangxi. The classification and identification of Guangxi freshwater mussels mainly refer to the shell morphology of the species in other regions, and may therefore result in an underestimation of the true diversity. Recently, the discovery of many unknown or cryptic species in Southeast Asia has greatly improved our understanding of species-level freshwater mussel diversity in the Indo-Burma region (e.g. Bolotov et al. 2019, 2020; Konopleva et al. 2020; Pfeiffer et al. 2021; Zieritz et al. 2021b), and this has reference significance for the study of freshwater mussel diversity in Guangxi.
During a field trip to Guilin, Guangxi, we found a unique freshwater mussel species from a tributary of the Luoqing River. Morphologically, this species belongs to the subfamily Gonideinae (ascending lamella of the inner demibranch not completely fused to visceral mass; Pfeiffer et al. 2019) but owns some unique conchological characters. After examination of Gonideinae species from China and neighbouring countries and reference to the literature (e.g. Heude 1875, 1877a, 1877b, 1878, 1879, 1880a, 1880b, 1881, 1883, 1885; Haas 1969; Brandt 1974; Liu et al. 1979a; He and Zhuang 2013), we were unable to match this species to any of the recorded species. We conducted phylogenetic analyses based on multi-locus data (i.e. COI, 16S rRNA and 28S rRNA) to reveal the systematic position. To further clarify the relationships with neighbouring genera, the complete maternal mitogenome of this species was determined and used for phylogenetic analyses alongside the other 52 maternal mitogenomes of freshwater mussels. Results permitted us to confirm this as a new genus and species, and we describe Postolata guangxiensis gen. nov., sp. nov. herein. Our findings highlight that Guangxi has a high level of endemism of freshwater mussels and a detailed survey of uncharted areas will help reveal new diversity in future.
Materials and methods
Specimen sampling and morphological observations
In July 2021, 18 specimens were collected from a tributary of the Luoqing River, Yongfu County, Guilin City, Guangxi, China (25°04′42″N, 110°06′39″E) (Fig. 1, Supplementary Fig. S1). A Digital Vernier caliper (±0.01 mm) was used to measure shell length, height and width. Specimens were dissected (Fig. 2, Supplementary Fig. S2) and the gonadal tissue of each individual was examined using a microscope. The adductor muscle was used for subsequent DNA extraction. The remaining soft parts were preserved at −80°C. All voucher specimens were deposited in the Museum of Biology, Nanchang University (NCUMB), China.

|
We examined the following morphological characters: shell shape, shell size, shell thickness, umbo, surface sculpture, teeth and adductor muscle scars. These characters were compared with images from MUSSELp online database (see http://mussel-project.uwsp.edu; Graf and Cummings 2021) and taxonomic literature (He and Zhuang 2013).
DNA extraction, PCR, sequencing and mitogenome assembly
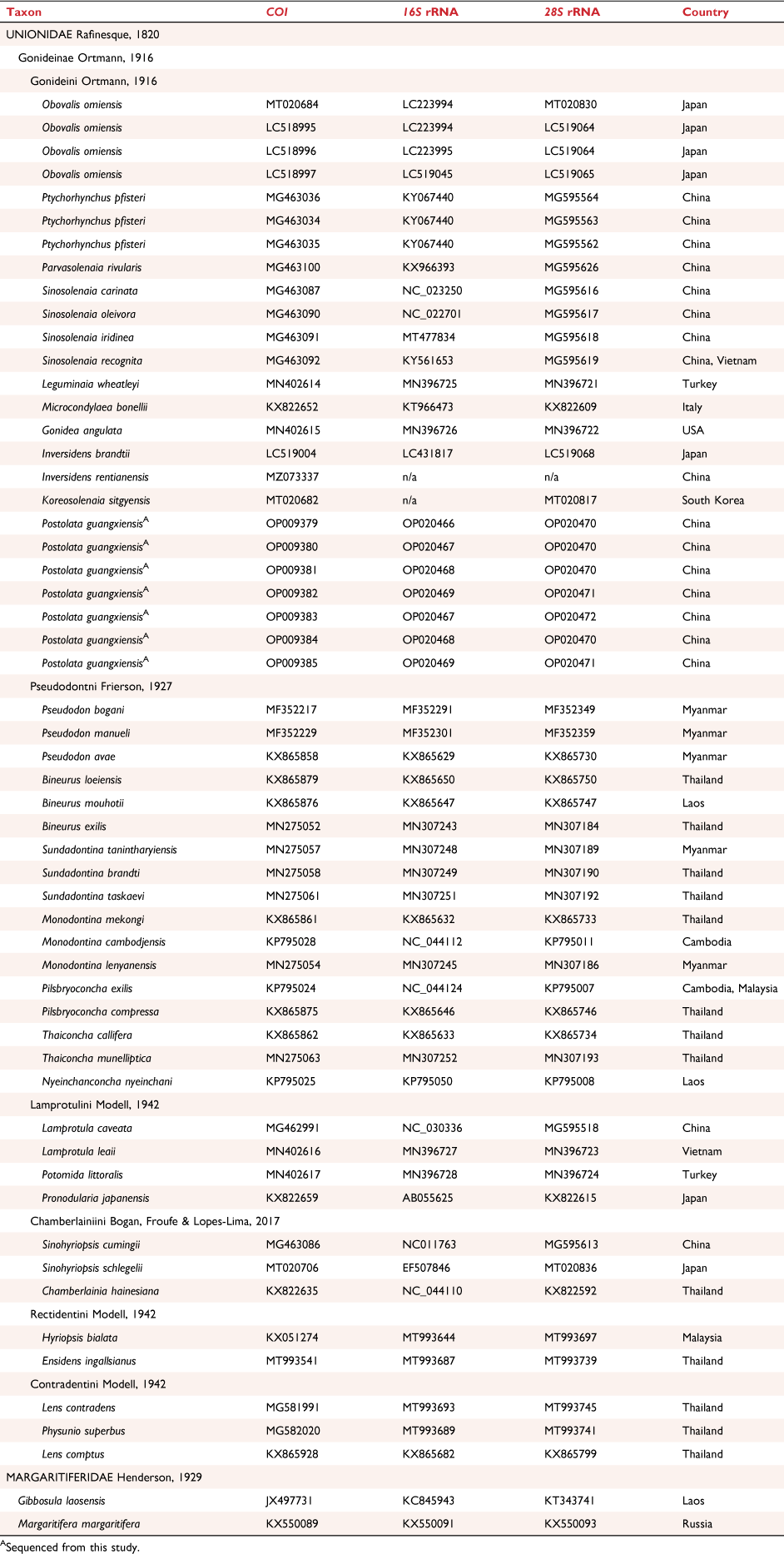
Total genomic DNA was extracted from the dissected tissue using the E.Z.N.A. Mollusc DNA kit (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s protocol. We amplified and sequenced fragments from the mitochondrial cytochrome c oxidase subunit-I gene (COI) (LCO22me2 + HCO700dy2) (Walker et al. 2007), 16S small ribosomal RNA gene (16S) (16sar-L-myt + 16sbr-H-myt) (Bolotov et al. 2018) and nuclear 28S ribosomal RNA gene (28S) (D23F + D4RB) (Park and Ó Foighil 2000). The polymerase chain reaction (PCR) was conducted using a 25-µL mixture of 2× Taq Plus Master MixII (Vazyme, PR China) (12.5 µL), ddH2O (9.5 µL), 10 µM primers (1 µL each) and genomic DNA (1 µL, ~100 ng μL−1). Thermal cycling was initiated at 98°C for 10 s, followed by 35 cycles of 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 1 min and a final extension at 72°C for 7 min. The PCR products were commercially sequenced by Sangon Biotech (Shanghai, PR China). The newly obtained sequences were deposited in GenBank under accession numbers OP009379–OP009385 for COI, OP020466–OP020469 for 16S and OP020470–OP020472 for 28S (Table 1).
|
As for mitogenome sequencing, the qualified genomic DNA sample was sent to Novogene (Beijing, PR China). After quality controls, the library was successfully prepared and sequenced in the Illumina NovaSeq. 6000 platform yielding ~4 Gb of data, with 2 × 150-bp paired-end reads. After discarding low-quality reads, clean reads were obtained and assembled de novo using CLC Genomic Workbench (ver. 12.0, see https://digitalinsights.qiagen.com/). Contigs identified as mitogenome sequences were inspected manually for overlap at the beginning and end, resulting in a circular mitogenome. Geneious (ver. 11.0, see https://www.geneious.com; Kearse et al. 2012) was used to check the complete mitogenome and analyse nucleotide composition. Strand asymmetry was calculated with the following formulae:
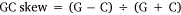
and
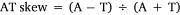
The mitogenome was initially annotated using the MITOS web server (see http://mitos.bioinf.uni-leipzig.de/index.py; Bernt et al. 2013). ARWEN (see http://130.235.244.92/ARWEN/; Laslett and Canbäck 2008) was also used to identify the locations of all tRNA genes. The annotations of two rRNA genes were refined based on the locations of adjacent genes. Protein-coding genes (PCGs) were confirmed based on the NCBI ORF Finder (see http://www.ncbi.nlm.nih.gov/orffinder/) and BLAST searches (see http://blast.ncbi.nlm.nih.gov/). The annotated mitochondrial genome was submitted to GenBank under accession number OP009366 (Table 2).
|
Alignments and partitioning strategies
Three datasets were constructed in this study: (i) COI dataset (25 haplotypes including newly sequenced species and four neighbouring species) (Supplementary Table S1); (ii) three-gene dataset (containing COI, 16S and 28S; 56 sequences) (Table 1); (iii) mitogenome dataset (containing 12 PCGs (except for ATP8) and two rRNA genes; 53 taxa) (Table 2).
All PCGs were codon-aligned by MUSCLE (Edgar 2004) implemented in MEGA X (ver. 10.2, see http://www.megasoftware.net; Kumar et al. 2018), whereas ribosomal genes (12S, 16S and 28S rRNAs) were aligned in MAFFT (ver. 7, see https://mafft.cbrc.jp/alignment/server/; Katoh et al. 2019) using the Q-INS-i algorithm. We used Gblocks (ver. 0.91b, see http://molevol.cmima.csic.es/castresana/Gblocks.html; Castresana 2000) to exclude ambiguous areas of the alignment for each gene. Gap positions were all removed for the mitogenome dataset, whereas half were allowed for the three-gene dataset. DnaSP (ver. 6, see http://www.ub.edu/dnasp/; Rozas et al. 2017) was used to calculate the number of haplotypes.
We recognised five partitions (three codons of COI, 16S and 28S) for the three-gene dataset and used MEGA X to find the best substitution models based on the corrected Akaike Information Criterion (AICc) (Supplementary Table S2). As for the mitogenome dataset, we used PartitionFinder2 (ver. 2.3.4, see http://www.robertlanfear.com/partitionfinder/; Lanfear et al. 2017) to determine the best-fit partitioning schemes and substitution models under greedy search. Predefined data blocks for the partitioning scheme search were designated according to gene region (rRNA gene) or codon positions (PCGs). Branch lengths were allowed to be unlinked, and model selection and partitioning schemes were selected based on the AICc (Supplementary Table S2).
Phylogenetic analyses
Maximum‐likelihood (ML) analyses were performed in raxmlGUI (ver. 2.0, see https://antonellilab.github.io/raxmlGUI/; Edler et al. 2020), with the ML + rapid bootstrap method and 1000 replicates. Bayesian inference (BI) analyses were conducted in MrBayes (ver. 3.2.6, see http://nbisweden.github.io/MrBayes/; Ronquist et al. 2012). Four simultaneous runs with four independent Markov Chain Monte Carlo (MCMC) were implemented for 10 million generations, and trees were sampled every 1000 generations with a burn-in of 25%. The convergence was checked with the average standard deviation of split frequencies <0.01 and the potential scale reduction factor (PSRF) ~1.
Inter and intra-specific distances based on the COI dataset were calculated in MEGA X using the uncorrected p-distance. Standard error estimates were obtained by 1000 bootstrap replicates.
Results
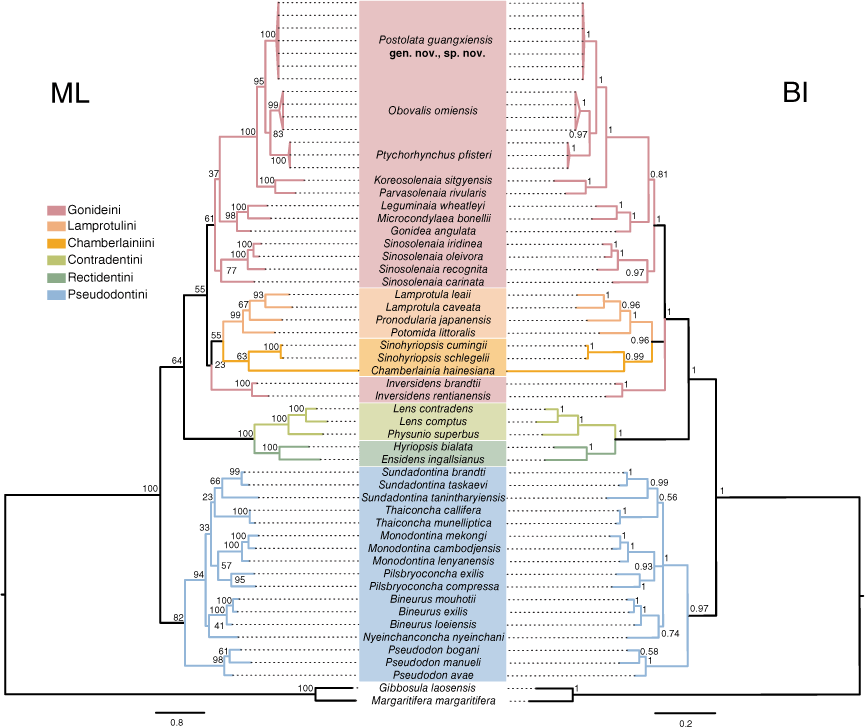
Multilocus phylogenetic analyses
In total, seven COI haplotypes were detected for the 18 sequenced specimens from Guangxi. We also obtained the 16S and 28S sequences corresponding to the individuals of these COI haplotypes. The COI dataset had an aligned length of 603 characters, of which 139 were variable sites, and 13 were parsimony informative sites. The trimmed and concatenated three-gene dataset consisted of 1484 characters, with 581 variable sites and 525 parsimony informative sites.
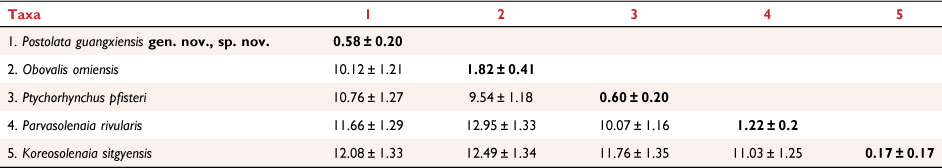
ML and BI trees based on the three-gene dataset resulted in largely congruent topologies, except for two nodes containing polytomies in the BI tree (Fig. 3). These clades were not well-resolved in the ML tree either, with both receiving low nodal support (BS < 50). Specimens from Guangxi formed a robust monophyletic clade (BS/BPP = 100/1.00) in the tribe Gonideini that did not belong to any previously known species or genera in the subfamily Gonideinae. The intraspecific divergence of this new species was 0.58 ± 0.20% (within the mean group uncorrected p-distance). This species was recovered as the sister group of Obovalis + Ptychorhynchus (BS/BPP = 95/1.0). In the focal clade (including Obovalis, Ptychorhynchus, Koreosolenaia and Parvasolenaia) in Gonideini, the pairwise uncorrected COI p-distance ranged from 10.12% (between this species and Obovalis omiensis (Heimburg, 1884)) to 12.08% (between this species and Koreosolenaia sitgyensis Lee, Kim, Lopes-Lima & Bogan, 2020) (Table 3), providing compelling evidence for the founding of the new genus (Jeratthitikul et al. 2021; Wu et al. 2022). Occupying a unique phylogenetic position and displaying distinctive morphological characteristics (Table 4, Fig. 1c), this species is described herein as Postolata guangxiensis gen. nov., sp. nov.
|
|
In our multilocus phylogenetic trees, five tribes of Gonideinae were monophyletic, except for Gonideini (Fig. 3). Inversidens was not nested within Gonideini or Lamprotulini but the placement of this genus lacked statistical support (BS = 23).
Mitochondrial genome structure
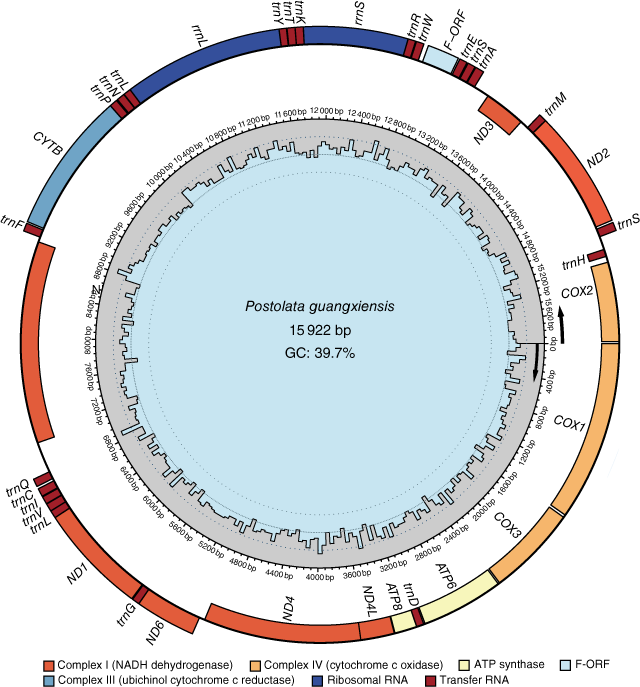
The maternal mitogenome of Postolata guangxiensis was 15 922 bp in length and consisted of 14 PCGs (including one F-orf), two rRNA genes, 22 tRNA genes and one putative control region. The location of the F-orf was between tRNATrp and tRNAGlu and the gene order of this mitogenome was consistent with other Gonideinae species except for Chamberlainia hainesiana (Froufe et al. 2020) (Fig. 4, Supplementary Table S3). COI, COII, COIII, ND3, ND4, ND4L, ND5, ATP6, ATP8, tRNAAsp and tRNAHis were encoded on the heavy strand and the remaining genes were light strand-encoded. Most PCGs used the standard ATN (ATC, ATT and ATG) as start codons, whereas COI and ATP8 started with TTG and GTG respectively. No incomplete stop codons were found in this mitogenome. The most commonly used stop codon was TAA that was found in 8 genes of the 14 PCGS. All 22 tRNA genes were in the range of 61–73 bp, with the classic clover-leaf structure (Supplementary Fig. S3). The rrnS gene was located between tRNAArg and tRNALys, whereas the rrnL gene was located between tRNATyr and tRNALeu. The overall base composition was A (23.0%), T (37.3%), C (12.9%) and G (26.8%), with the A + T content of 60.3%. Nucleotide asymmetry of the mitochondrial strands was assessed by AT skew and GC skew that were −0.24 and 0.35 respectively.
|
Mitochondrial phylogenomic analyses
The fully aligned data matrix used for mitochondrial phylogenomics was 12 071 bp long and included 7152 variable sites, of which 6506 were parsimony-informative. ML and BI trees based on the mitogenome dataset yielded an identical topology and were statistically well-supported by 100% maximum likelihood bootstrap (BS) support values and Bayesian posterior probabilities (BPP) in most nodes (Fig. 5). In both trees, P. guangxiensis was placed in the focal clade of Gonideini with higher support values (BS/BPP = 100/1.0) than those of multilocus phylogenetic trees (BS/BPP = 95/1.0). Our mitochondrial phylogenomic analyses supported Parvasolenaia rivularis (Heude, 1877) and Ptychorhynchus pfisteri (Heude, 1874) as successive sister taxa to P. guangxiensis (BS/BPP = 100/1.0). This relationship is congruent with the multilocus phylogenetic analyses, although mitogenome data of Obovalis omiensis and Koreosolenaia sitgyensis were absent.
Six of the eight tribes from the subfamily Gonideinae were covered in the present study, with exceptions being Schepmaniini and Ctenodesmini (Fig. 5). These tribes formed robust monophyletic clades (BS/BPP = 100/1.0), with Chamberlainiini being resolved as sister to the remainder of the subfamily. The sister-group relationship between Gonideini and Pseudodontini was recovered (BS/BPP = 95/1.0).
Taxonomy
Family UNIONIDAE Rafinesque, 1820
Subfamily GONIDEINAE Ortmann, 1916
Tribe GONIDEINI Ortmann, 1916
Genus Postolata Dai, Huang & Wu, gen. nov.
Etymology
The genus epithet is compiled by two Latin words ‘Postʼ and ‘Lataʼ that mean ‘rear’ and ‘wide’ respectively, attributed to the shell morphological characters. The letter ‘o’ was added in the middle according to the Latin nomenclature.
Diagnosis
Shell medium size, moderately thick; irregularly rectangular, anterior rounded and short, posterior long and wide. Dorsal margin slightly curved downwards; ventral margin nearly straight. Posterior margin nearly perpendicular to ventral margin. Umbo inflated, below hinge line, often eroded. Hinge short. Mantle off-white to light-brownish, aperture margins black or brown, flap margin papillate. Papillae white to brown, short, incurrent aperture bearing larger papillae than excurrent aperture. Gills light brownish.
Differential diagnosis
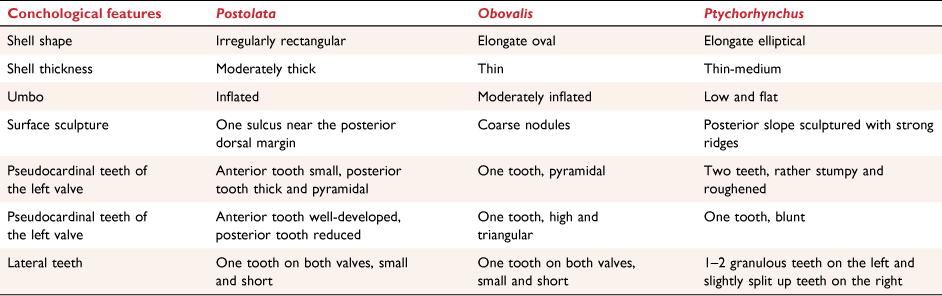
Phylogenetic analyses showed that the new genus was closely related to Obovalis and Ptychorhynchus. This new genus differs from other genera in Gonideini by having an irregularly rectangular shell shape. The shell shape is rounded at the anterior and wide at the posterior, and the posterior margin is almost perpendicular to the ventral margin. However, the shells of Obovalis and Ptychorhynchus are elongated and oval. In addition, compared with these two genera, the new genus has a more inflated umbo and a thicker shell. The new genus also differs from Obovalis and Ptychorhynchus in the shell teeth and surface sculpture. In the new genus, there is one sulcus near the posterior dorsal margin on the surface sculpture, but coarse nodules in Obovalis and strong ridges on the posterior slope in Ptychorhynchus. A comparison of morphological characteristics with related genera is shown in Table 4.
Distribution
The genus is known only from tributaries of the Luoqing River, Guilin City, Guangxi, China.
Remarks
The new genus belongs to the tribe Gonideini of the subfamily Gonideinae that currently consists of only one species. Molecular data and morphological features simultaneously support this notion.
Postolata guangxiensis Dai, Huang, Guo & Wu, gen. nov., sp. nov.
(Fig. 1c, 2, Supplementary Fig. S1, S2.)
Etymology
The name of this species is derived from Guangxi, in which the type locality is located. We recommend ‘Guangxi rear-wide mussel’ (English) and ‘Guang Xi Hou Ju Bang’ (广西后矩蚌) (Chinese) as the common names of Postolata guangxiensis.
Material examined
Holotype: CHINA: ♀; Guangxi, Guilin City, Yongfu County, Luoqing River; 25°04′42″N, 110°06′39″E; 31 December 2020; F. Y. Shu, Z. G. Chen and L. Guo leg.; 22_NCU_XPWU_PGF01.
Paratypes: CHINA: 17 shells; same collection data as for holotype; specimen vouchers are shown in Supplementary Table S4.
Shell description
Shell medium size, moderately thick; length 44.99–60.82 mm, height 31.02–40.33 mm, width 16.72–22.82 mm (Supplementary Table S4). Anterior rounded, short, posterior long and wide. Dorsal margin slightly curved downwards; ventral margin nearly straight. Posterior margin nearly perpendicular to ventral margin. Umbo inflated, below hinge line, located at 1/3 of the dorsal margin and often eroded. Periostracum brownish or blackish, with thin growth lines and one sulcus near the posterior dorsal margin. Lines arranged in irregular concentric circles. Hinge short. Ligament short and strong. Mantle muscle scars obvious. Anterior adductor muscle scars oval, deep and rough; posterior adductor muscle scars long, oval, smooth. Left valve with two pseudocardinal teeth, anterior tooth small, posterior tooth thick, high and pyramidal; anterior pseudocardinal tooth of the right valve well developed, posterior pseudocardinal tooth reduced, connected to lateral teeth. Lateral teeth of both valves short and thick. Nacre white.
Soft anatomy description
Mantle off-white to light-brownish, aperture margins black or brown, flap margin papillate. Excurrent aperture bearing reduced, with two rows of papillae, inner row inflated, off-white; free papillae stalks black to reddish-brown. Incurrent aperture bearing larger papillae than excurrent aperture. Papillae white to brown, with two rows, inner row usually larger, conical and elongated with a simple tip. Gills light-brownish, inner gills slightly longer and wider than outer gills. Labial palps light brownish to brown, distally pointed and irregularly fan-shaped in appearance. Visceral mass creamy white to pale-orange, maybe off-white adjacent to foot; foot off-white or pale orange.
Distribution
The species is known only from the Luoqing River, Guilin City, Guangxi, China.
Habitat
P. guangxiensis occurs in a small tributary of the Luoqing River, Guangxi, China. The species lives in shallow rivers with slow flow, clear water and silt substrate (Fig. 1a).
Discussion
Phylogenetic implications from multilocus and mitogenomic data
Based on morphological and molecular data, we herein describe a new genus and species of freshwater mussel endemic to Guangxi, China. Though our multilocus data strongly support the validity of Postolata guangxiensis gen. nov, sp. nov. and resolve the phylogenetic relationships with closely related genera in Gonideini, higher-level evolutionary relationships in the subfamily Gonideinae are still elusive. The reason for low nodal support values and the presence of polytomy in the BI tree is that the three-gene data are insufficient in informative sites to resolve deep divergences such as tribal level relationships, as has been discussed in the utility of nuclear genome data (i.e. the Unioverse probe set) (Pfeiffer et al. 2019). In addition, previous phylogenetic investigations showed that Inversidens belonged to Gonideini (Lopes-Lima et al. 2020; Sano et al. 2020) but this classification is unsupported in our phylogenetic trees based on the three-gene dataset. Inversidens is not within any tribe of Gonideinae, although further investigations using mitochondrial or nuclear genome data are needed.
Compared with a single gene or limited number of molecular markers, complete mitochondrial genomes are rich in informative loci and have been increasingly applied to clarify disputes regarding the phylogenetic relationships among freshwater mussels (Huang et al. 2018, 2019; Zieritz et al. 2021a; Wu et al. 2022). Therefore, to corroborate our multilocus phylogenetic results, the maternal mitochondrial genome of Postolata guangxiensis was used to reconstruct the phylogenetic trees combining the other 52 maternal mitogenomes. Our mitochondrial phylogenomic results provide a well-supported phylogeny for the placement of P. guangxiensis. Additionally, our mitogenomic results support six tribes of Gonideinae as monophyletic, including Rectidentini, Contradentini and Pseudodontini, and this is fully congruent with recent phylogenomic analyses using anchored hybrid enrichment data (Pfeiffer et al. 2019) or mitogenomic data (Zieritz et al. 2021a). This indicates that the higher-level relationships in freshwater mussels are converging significantly from both nuclear and mitochondrial genome data. However, previous phylogenetic studies based on a handful of genes assigned Contradentini and Rectidentini to Rectidentinae (Lopes-Lima et al. 2017; Bolotov et al. 2017a), and Pseudodontini and Pilsbryoconchini to Pseudodontinae (Bolotov et al. 2017b). Therefore, we suggest that deep genealogical relationships in freshwater mussels should be reconstructed and verified by genome-scale data (at least the maternal/F-type mitogenome), especially when proposing new taxa.
Morphological characteristics and species distribution ranges provide additional support for the classification of this new species. On the one hand, the species has a similar shell shape to the other Gonideini species, with trapezoidal to rectangular shells. Another distinct morphological characteristic of the tribe is having no or only vestigial hinge teeth and tetragenous brooding type (Lopes-Lima et al. 2017; Froufe et al. 2020). Nevertheless, P. guangxiensis has strong pseudocardinal teeth and vestigial lateral teeth that can be a new diagnostic feature in this tribe. Regrettably, since no brooding individuals were collected in this study, we could not examine the glochidia of this new species and this will require longer sampling times and more in-depth testing in future studies. On the other hand, Postolata, Obovalis and Ptychorhynchus are allopatrically distributed. Postolata has only been found in a tributary of the Luoqing River in the Pearl River Basin but we speculate that this species may have a wider distribution range. Ptychorhynchus species occur in the Hainan Island, Yangtze River Basin and Yellow River Basin in China, whereas the monotypic genus Obovalis only occurs in Japan (MUSSELp Database, see http://mussel-project.uwsp.edu).
Endangered status and conservation implications
Freshwater mussels are valued for providing important ecosystem functions and services (Vaughn 2018). Assessment and monitoring of species diversity are critical during times of ongoing biodiversity loss and new species discovery is a fundamental first step. Species identification and description are traditionally performed using morphological characters. However, sometimes speciation does not necessarily translate into clear morphological differentiation (Daïnou et al. 2016), that, coupled with the plasticity of shell morphology (Inoue et al. 2013) in freshwater bivalves, makes species identification difficult. We integrate comprehensive molecular evidence into the identification and classification of the new taxon from Guangxi. This contributes to the further expansion of Unionidae in China and suggests that more undescribed species may exist in understudied areas with highly endemic freshwater habitats.
Stream habitats harbouring endemic mussel species are vulnerable to threats due to the impact of urbanisation and human activities, and need urgent attention and protection (e.g. Zieritz et al. 2021b; Wu et al. 2022). The type locality of Postolata guangxiensis is a small tributary of the Luoqing River that has high water quality and silt substrate and there are many homes nearby. The discharge of domestic sewage and agricultural runoff can affect the water quality in the habitat. In addition, freshwater mussels are highly sensitive to direct drying and drought-induced secondary effects in situ (Haag and Warren 2008). Consequently, habitat degradation and loss are major threats to this newly discovered species based on available distribution information.
In recent decades, hydraulic projects, urban development, environmental pollution and other issues have led to the change or even loss of habitat function of freshwater fauna in Guangxi. We speculate that some freshwater mussels in this area may have been seriously threatened. Nonetheless, Guangxi, of which the southern part is situated in the Indo-Burma hotspot, has received extensive attention and protection. The conservation of unionoid fauna in this region is severely hampered by a lack of knowledge of freshwater mussel distribution, population trends, threats and accurate taxonomic information (Liu et al. 2022). This is also the case in other areas where freshwater mussel research is lacking. Along with the increasing number of large rivers where water projects have been established or are being planned, small rivers and tributaries have become the refuges for aquatic organisms (Jiang et al. 2011; Sabo et al. 2012). Nevertheless, comprehensive biodiversity surveys for tributary basins are lacking, rendering aquatic biodiversity and threatening factors unclear (Yin et al. 2022). In China, only half of the freshwater mussel species have been assessed for conservation status and approximately half the assessed species are threatened with extinction (International Union for Conservation of Nature 2021; Liu et al. 2022). Although in situ conservation of specific endangered freshwater mussel species can be realised by establishing nature reserves, such as the Lamprotula mansuyi nature reserve in the Zuojiang River of Guangxi, most of the species live outside protected areas and are facing the impacts of anthropogenic disturbances without any specific restoration actions being undertaken. Fortunately, the Chinese government has realised the seriousness of this problem and systematically updated the List of Wild Animals under State Priority Conservation in 2021. Seven freshwater bivalve species were newly added as wild animals in the Grade-II conservation rating. This new list will increase protection awareness of this long-neglected group of aquatic wildlife in China for the general public and fishery bureaux. Meanwhile, in addition to traditional pearl-farmed mussels (e.g. Sinohyriopsis cumingii), more than 10 unionid species, including Aculamprotula tortuosa, Lamprotula leaii and Sinosolenaia oleivora (Feng-Yue Shu unpubl. data) are undergoing mass propagation and captive breeding in China. These mussels are either being sold for consumption or used in the restoration of freshwater ecosystems in environmental projects. Therefore, we hope that more researchers and relevant companies will be involved in the study and application of artificial propagation of freshwater mussels that is critical for preventing overexploitation of wild populations and restoring endangered species.
Supplementary material
Supplementary material is available online.
Data availability
The data that support this study are available in the article and accompanying online Supplementary material.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This study was funded by the National Natural Science Foundation of China (number 32100354), the National Key R&D Program of China (2018YFD0900801) and the Innovation Fund Designated for Graduate Students of Jiangxi Province (YC2022-B026).
References
Bernt, M, Donath, A, Jühling, F, Externbrink, F, Florentz, C, Fritzsch, G, Pütz, J, Middendorf, M, and Stadler, PF (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution 69, 313–319.| MITOS: improved de novo metazoan mitochondrial genome annotation.Crossref | GoogleScholarGoogle Scholar |
Bolotov, IN, Kondakov, AV, Vikhrev, IV, Aksenova, OV, Bespalaya, YV, Gofarov, MY, Kolosova, YS, Konopleva, ES, Spitsyn, VM, Tanmuangpak, K, and Tumpeesuwan, S (2017a). Ancient river inference explains exceptional oriental freshwater mussel radiations. Scientific Reports 7, 2135.
| Ancient river inference explains exceptional oriental freshwater mussel radiations.Crossref | GoogleScholarGoogle Scholar |
Bolotov, IN, Vikhrev, IV, Kondakov, AV, Konopleva, ES, Gofarov, MY, Aksenova, OV, and Tumpeesuwan, S (2017b). New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Scientific Reports 7, 11573.
| New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia.Crossref | GoogleScholarGoogle Scholar |
Bolotov, IN, Aksenova, OV, Bakken, T, Glasby, CJ, Gofarov, MY, Kondakov, AV, Konopleva, ES, Lopes-Lima, M, Lyubas, AA, Wang, Y, Bychkov, AY, Sokolova, AM, Tanmuangpak, K, Tumpeesuwan, S, Vikhrev, IV, Shyu, JBH, Win, T, and Pokrovsky, OS (2018). Discovery of a silicate rock-boring organism and macrobioerosion in fresh water. Nature Communications 9, 2882.
| Discovery of a silicate rock-boring organism and macrobioerosion in fresh water.Crossref | GoogleScholarGoogle Scholar |
Bolotov, IN, Konopleva, ES, Vikhrev, IV, Lopes-Lima, M, Bogan, AE, Lunn, Z, Chan, N, Win, T, Aksenova, OV, Gofarov, MY, Tomilova, AA, and Kondakov, AV (2019). Eight new freshwater mussels (Unionidae) from tropical Asia. Scientific Reports 9, 12053.
| Eight new freshwater mussels (Unionidae) from tropical Asia.Crossref | GoogleScholarGoogle Scholar |
Bolotov, IN, Konopleva, ES, Vikhrev, IV, Gofarov, MY, Lopes-Lima, M, Bogan, AE, Lunn, Z, Chan, N, Win, T, Aksenova, OV, Tomilova, AA, Tanmuangpak, K, Tumpeesuwan, S, and Kondakov, AV (2020). New freshwater mussel taxa discoveries clarify biogeographic division of Southeast Asia. Scientific Reports 10, 6616.
| New freshwater mussel taxa discoveries clarify biogeographic division of Southeast Asia.Crossref | GoogleScholarGoogle Scholar |
Brandt, RAM (1974). The non-marine aquatic Mollusca of Thailand. Archiv für Molluskenkunde 105, 1–423.
Castresana, J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552.
| Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis.Crossref | GoogleScholarGoogle Scholar |
Daïnou, K, Blanc-Jolivet, C, Degen, B, Kimani, P, Ndiade-Bourobou, D, Donkpegan, ASL, Tosso, F, Kaymak, E, Bourland, N, Doucet, JL, and Hardy, OJ (2016). Revealing hidden species diversity in closely related species using nuclear SNPs, SSRs and DNA sequences – a case study in the tree genus Milicia. BMC Evolutionary Biology 16, 259.
| Revealing hidden species diversity in closely related species using nuclear SNPs, SSRs and DNA sequences – a case study in the tree genus Milicia.Crossref | GoogleScholarGoogle Scholar |
Edgar, RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797.
| MUSCLE: multiple sequence alignment with high accuracy and high throughput.Crossref | GoogleScholarGoogle Scholar |
Edler, D, Klein, J, Antonelli, A, and Silvestro, D (2020). raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12, 373–377.
| raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML.Crossref | GoogleScholarGoogle Scholar |
Froufe, E, Bolotov, I, Aldridge, DC, Bogan, AE, Breton, S, Gan, HM, Kovitvadhi, U, Kovitvadhi, S, Riccardi, N, Secci-Petretto, G, Sousa, R, Teixeira, A, Varandas, S, Zanatta, D, Zieritz, A, Fonseca, MM, and Lopes-Lima, M (2020). Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Heredity 124, 182–196.
| Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution.Crossref | GoogleScholarGoogle Scholar |
Goncalves, A, Zieritz, A, Lopes-Lima, M, Deein, G, and Pfeiffer, J (2022). Taxonomic revision and conservation assessment of the Southeast Asian freshwater mussel genus Chamberlainia Simpson, 1900. Journal of Molluscan Studies 88, eyac008.
| Taxonomic revision and conservation assessment of the Southeast Asian freshwater mussel genus Chamberlainia Simpson, 1900.Crossref | GoogleScholarGoogle Scholar |
Graf, DL, and Cummings, KS (2021). A ‘big data’ approach to global freshwater mussel diversity (Bivalvia: Unionoida), with an updated checklist of genera and species. Journal of Molluscan Studies 87, eyaa034.
| A ‘big data’ approach to global freshwater mussel diversity (Bivalvia: Unionoida), with an updated checklist of genera and species.Crossref | GoogleScholarGoogle Scholar |
Haag, WR, and Warren, ML (2008). Effects of severe drought on freshwater mussel assemblages. Transactions of the American Fisheries Society 137, 1165–1178.
| Effects of severe drought on freshwater mussel assemblages.Crossref | GoogleScholarGoogle Scholar |
Haas F (1969) ‘Superfamilia Unionacea.’ [‘Superfamily Unionacea.’] (Walter de Gruyter: Berlin, Germany) [In German]
He J, Zhuang Z (2013) ‘The Freshwater Bivalves of China.’ (ConchBooks: Harxheim, Germany)
Heude PM (1875) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Premier Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 1.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1877a) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Deuxième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 2.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1877b) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Troisième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 3.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1878) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Quatrième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 4.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1879) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Cinquième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 5.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1880a) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Sixième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 6.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1880b) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Dixième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 10.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1881) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Septième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 7.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1883) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Huitième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 8.’] (Librairie F. Savy: Paris, France) [In French]
Heude PM (1885) ‘Conchyliologie fluviatile de la province de Nanking et de la Chine centrale. Neuvième Fascicule.’ [‘River conchyliology of Nanking province and central China. Issue 9.’] (Librairie F. Savy: Paris, France) [In French]
Huang, XC, Wu, RW, An, CT, Xie, GL, Su, JH, Ouyang, S, Zhou, CH, and Wu, XP (2018). Reclassification of Lamprotula rochechouartii as Margaritifera rochechouartii comb. nov. (Bivalvia: Margaritiferidae) revealed by time-calibrated multi-locus phylogenetic analyses and mitochondrial phylogenomics of Unionoida. Molecular Phylogenetics and Evolution 120, 297–306.
| Reclassification of Lamprotula rochechouartii as Margaritifera rochechouartii comb. nov. (Bivalvia: Margaritiferidae) revealed by time-calibrated multi-locus phylogenetic analyses and mitochondrial phylogenomics of Unionoida.Crossref | GoogleScholarGoogle Scholar |
Huang, XC, Su, JH, Ouyang, JX, Ouyang, S, Zhou, CH, and Wu, XP (2019). Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): Species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Molecular Phylogenetics and Evolution 130, 45–59.
| Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): Species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns.Crossref | GoogleScholarGoogle Scholar |
Inoue, K, Hayes, DM, Harris, JL, and Christian, AD (2013). Phylogenetic and morphometric analyses reveal ecophenotypic plasticity in freshwater mussels Obovaria jacksoniana and Villosa arkansasensis (Bivalvia: Unionidae). Ecology and Evolution 3, 2670–2683.
| Phylogenetic and morphometric analyses reveal ecophenotypic plasticity in freshwater mussels Obovaria jacksoniana and Villosa arkansasensis (Bivalvia: Unionidae).Crossref | GoogleScholarGoogle Scholar |
International Union for Conservation of Nature (2021) The IUCN Red List of Threatened Species. V.2018-3. Available at https://www.iucnredlist.org [Verified 10 September 2022]
Jeratthitikul, E, Sutcharit, C, Ngor, PB, and Prasankok, P (2021). Molecular phylogeny reveals a new genus of freshwater mussels from the Mekong River Basin (Bivalvia: Unionidae). European Journal of Taxonomy 775, 119–142.
| Molecular phylogeny reveals a new genus of freshwater mussels from the Mekong River Basin (Bivalvia: Unionidae).Crossref | GoogleScholarGoogle Scholar |
Jiang, X, Xiong, J, Xie, Z, and Chen, Y (2011). Longitudinal patterns of macroinvertebrate functional feeding groups in a Chinese river system: a test for river continuum concept (RCC). Quaternary International 244, 289–295.
| Longitudinal patterns of macroinvertebrate functional feeding groups in a Chinese river system: a test for river continuum concept (RCC).Crossref | GoogleScholarGoogle Scholar |
Katoh, K, Rozewicki, J, and Yamada, KD (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20, 1160–1166.
| MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization.Crossref | GoogleScholarGoogle Scholar |
Kearse, M, Moir, R, Wilson, A, Stones-Havas, S, Cheung, M, Sturrock, S, Buxton, S, Cooper, A, Markowitz, S, Duran, C, Thierer, T, Ashton, B, Meintjes, P, and Drummond, A (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Konopleva, ES, Bolotov, IN, Kondakov, AV, Kononov, OD, Gofarov, MY, Tomilova, AA, Lunn, Z, Chan, N, Win, T, and Vikhrev, IV (2020). A taxonomic review of Trapezidens (Bivalvia: Unionidae: Lamellidentini), a freshwater mussel genus endemic to Myanmar, with a description of a new species. Ecologica Montenegrina 27, 45–57.
| A taxonomic review of Trapezidens (Bivalvia: Unionidae: Lamellidentini), a freshwater mussel genus endemic to Myanmar, with a description of a new species.Crossref | GoogleScholarGoogle Scholar |
Kuang, XY, Su, Z, and Tu, FX (2007). 广西气候区划 [Climate regionalisation of Guangxi.] Guangxi Sciences 14, 278–283.
Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549.
| MEGA X: molecular evolutionary genetics analysis across computing platforms.Crossref | GoogleScholarGoogle Scholar |
Lanfear, R, Frandsen, PB, Wright, AM, Senfeld, T, and Calcott, B (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34, 772–773.
| PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |
Laslett, D, and Canbäck, B (2008). ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175.
| ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences.Crossref | GoogleScholarGoogle Scholar |
Liu YY, Zhang WZ, Wang YX, Wang EY (1979a) 中国经济动物志: 淡水软体动物 [‘Economic fauna of China: Freshwater mollusc.’] (Science Press: Beijing, PR China) [In Chinese]
Liu, YY, Zhang, WZ, Wang, YX, and Wang, EY (1979b). 我国蚌类新纪录 [New records of Chinese freshwater clams.] Zoological Systematics 4, 188.
Liu, XJ, Liu, YY, Wu, RW, Zanatta, DT, Lopes-Lima, M, Gonçalves, DV, Bogan, AE, Ouyang, S, and Wu, XP (2022). Systematics, distribution, biology, and conservation of freshwater mussels (Bivalvia: Unionida) in China. Aquatic Conservation: Marine and Freshwater Ecosystems 32, 859–895.
| Systematics, distribution, biology, and conservation of freshwater mussels (Bivalvia: Unionida) in China.Crossref | GoogleScholarGoogle Scholar |
Lopes-Lima, M, Froufe, E, Do, VT, Ghamizi, M, Mock, KE, Kebapçı, Ü, Klishko, O, Kovitvadhi, S, Kovitvadhi, U, Paulo, OS, Pfeiffer JM, III, Raley, M, Riccardi, N, Şereflişan, H, Sousa, R, Teixeira, A, Varandas, S, Wu, X, Zanatta, DT, Zieritz, A, and Bogan, AE (2017). Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution 106, 174–191.
| Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes.Crossref | GoogleScholarGoogle Scholar |
Lopes-Lima, M, Bolotov, IN, Do, VT, Aldridge, DC, Fonseca, MM, Gan, HM, Gofarov, MY, Kondakov, AV, Prié, V, Sousa, R, Varandas, S, Vikhrev, IV, Teixeira, A, Wu, RW, Wu, X, Zieritz, A, Froufe, E, and Bogan, AE (2018). Expansion and systematics redefinition of the most threatened freshwater mussel family, the Margaritiferidae. Molecular Phylogenetics and Evolution 127, 98–118.
| Expansion and systematics redefinition of the most threatened freshwater mussel family, the Margaritiferidae.Crossref | GoogleScholarGoogle Scholar |
Lopes-Lima, M, Hattori, A, Kondo, T, Hee Lee, J, Ki Kim, S, Shirai, A, Hayashi, H, Usui, T, Sakuma, K, Toriya, T, Sunamura, Y, Ishikawa, H, Hoshino, N, Kusano, Y, Kumaki, H, Utsugi, Y, Yabe, S, Yoshinari, Y, Hiruma, H, Tanaka, A, Sao, K, Ueda, T, Sano, I, Miyazaki, J-I, Gonçalves, DV, Klishko, OK, Konopleva, ES, Vikhrev, IV, Kondakov, AV, Yu. Gofarov, M, Bolotov, IN, Sayenko, EM, Soroka, M, Zieritz, A, Bogan, AE, and Froufe, E (2020). Freshwater mussels (Bivalvia: Unionidae) from the rising sun (Far East Asia): phylogeny, systematics, and distribution. Molecular Phylogenetics and Evolution 146, 106755.
| Freshwater mussels (Bivalvia: Unionidae) from the rising sun (Far East Asia): phylogeny, systematics, and distribution.Crossref | GoogleScholarGoogle Scholar |
Lopes-Lima, M, Riccardi, N, Urbanska, M, Köhler, F, Vinarski, M, Bogan, AE, and Sousa, R (2021). Major shortfalls impairing knowledge and conservation of freshwater molluscs. Hydrobiologia 848, 2831–2867.
| Major shortfalls impairing knowledge and conservation of freshwater molluscs.Crossref | GoogleScholarGoogle Scholar |
Park, JK, and Ó Foighil, D (2000). Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Molecular Phylogenetics and Evolution 14, 75–88.
| Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments.Crossref | GoogleScholarGoogle Scholar |
Pfeiffer, JM, Breinholt, JW, and Page, LM (2019). Unioverse: a phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida). Molecular Phylogenetics and Evolution 137, 114–126.
| Unioverse: a phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida).Crossref | GoogleScholarGoogle Scholar |
Pfeiffer, JM, Graf, DL, Cummings, KS, and Page, LM (2021). Taxonomic revision of a radiation of South-east Asian freshwater mussels (Unionidae: Gonideinae: Contradentini+Rectidentini). Invertebrate Systematics 35, 394–470.
| Taxonomic revision of a radiation of South-east Asian freshwater mussels (Unionidae: Gonideinae: Contradentini+Rectidentini).Crossref | GoogleScholarGoogle Scholar |
Ronquist, F, Teslenko, M, van der Mark, P, Ayres, DL, Darling, A, Höhna, S, Larget, B, Liu, L, Suchard, MA, and Huelsenbeck, JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar |
Rozas, J, Ferrer-Mata, A, Sánchez-Delbarrio, JC, Guirao-Rico, S, Librado, P, Ramos-Onsins, SE, and Sánchez-Gracia, A (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34, 3299–3302.
| DnaSP 6: DNA sequence polymorphism analysis of large data sets.Crossref | GoogleScholarGoogle Scholar |
Sabo, JL, Bestgen, K, Graf, W, Sinha, T, and Wohl, EE (2012). Dams in the Cadillac Desert: downstream effects in a geomorphic context. Annals of the New York Academy of Sciences 1249, 227–246.
| Dams in the Cadillac Desert: downstream effects in a geomorphic context.Crossref | GoogleScholarGoogle Scholar |
Sano, I, Saito, T, Miyazaki, J-I, Shirai, A, Uechi, T, Kondo, T, and Chiba, S (2020). Evolutionary history and diversity of unionoid mussels (Mollusca: Bivalvia) in the Japanese archipelago. Plankton & Benthos Research 15, 97–111.
| Evolutionary history and diversity of unionoid mussels (Mollusca: Bivalvia) in the Japanese archipelago.Crossref | GoogleScholarGoogle Scholar |
Tordoff, AW, Baltzer, MC, Fellowes, JR, Pilgrim, JD, and Langhammer, PF (2012). Key biodiversity areas in the Indo-Burma hotspot: process, progress and future directions. Journal of Threatened Taxa 4, 2779–2787.
| Key biodiversity areas in the Indo-Burma hotspot: process, progress and future directions.Crossref | GoogleScholarGoogle Scholar |
Vaughn, CC (2018). Ecosystem services provided by freshwater mussels. Hydrobiologia 810, 15–27.
| Ecosystem services provided by freshwater mussels.Crossref | GoogleScholarGoogle Scholar |
Walker, JM, Bogan, AE, Bonfiglio, EA, Campbell, DC, Christian, AD, Curole, JP, Harris, JL, Wojtecki, RJ, and Hoeh, WR (2007). Primers for amplifying the hypervariable, male-transmitted COII–COI junction region in amblemine freshwater mussels (Bivalvia: Unionoidea: Ambleminae). Molecular Ecology Notes 7, 489–491.
| Primers for amplifying the hypervariable, male-transmitted COII–COI junction region in amblemine freshwater mussels (Bivalvia: Unionoidea: Ambleminae).Crossref | GoogleScholarGoogle Scholar |
Wu, XP, Dai, YT, Yin, N, Shu, FY, Chen, ZG, Guo, L, Zhou, CH, Ouyang, S, and Huang, XC (2022). Mitogenomic phylogeny resolves Cuneopsis (Bivalvia: Unionidae) as polyphyletic: the description of two new genera and a new species. Zoologica Scripta 51, 173–184.
| Mitogenomic phylogeny resolves Cuneopsis (Bivalvia: Unionidae) as polyphyletic: the description of two new genera and a new species.Crossref | GoogleScholarGoogle Scholar |
Yin, S, Yi, Y, Liu, Q, Luo, Q, and Chen, K (2022). A review on effects of human activities on aquatic organisms in the Yangtze River Basin since the 1950s. River 1, 104–119.
| A review on effects of human activities on aquatic organisms in the Yangtze River Basin since the 1950s.Crossref | GoogleScholarGoogle Scholar |
Zieritz, A, Bogan, AE, Froufe, E, Klishko, O, Kondo, T, Kovitvadhi, U, Kovitvadhi, S, Lee, JH, Lopes-Lima, M, Pfeiffer, JM, Sousa, R, Van Do, T, Vikhrev, I, and Zanatta, DT (2018). Diversity, biogeography and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia. Hydrobiologia 810, 29–44.
| Diversity, biogeography and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia.Crossref | GoogleScholarGoogle Scholar |
Zieritz, A, Froufe, E, Bolotov, I, Gonçalves, DV, Aldridge, DC, Bogan, AE, Gan, HM, Gomes-Dos-Santos, A, Sousa, R, Teixeira, A, Varandas, S, Zanatta, D, and Lopes-Lima, M (2021a). Mitogenomic phylogeny and fossil-calibrated mutation rates for all F- and M-type mtDNA genes of the largest freshwater mussel family, the Unionidae (Bivalvia). Zoological Journal of the Linnean Society 193, 1088–1107.
| Mitogenomic phylogeny and fossil-calibrated mutation rates for all F- and M-type mtDNA genes of the largest freshwater mussel family, the Unionidae (Bivalvia).Crossref | GoogleScholarGoogle Scholar |
Zieritz, A, Jainih, L, Pfeiffer, J, Rahim, KAA, Prayogo, H, Anwari, MS, Fikri, AH, Diba, F, Taha, H, Sulaiman, Z, Froufe, E, and Lopes-Lima, M (2021b). A new genus and two new, rare freshwater mussel (Bivalvia: Unionidae) species endemic to Borneo are threatened by ongoing habitat destruction. Aquatic Conservation: Marine and Freshwater Ecosystems 31, 3169–3183.
| A new genus and two new, rare freshwater mussel (Bivalvia: Unionidae) species endemic to Borneo are threatened by ongoing habitat destruction.Crossref | GoogleScholarGoogle Scholar |