Laboratory automation: efficiency and turnaround times
Patrick R MurrayBD Diagnostic Systems
7 Loveton Circle
Sparks, Maryland 21152, USA
Tel: 410 316 4477
Fax: 410 316 4041
Email: patrick_murray@bd.com
Microbiology Australia 35(1) 49-51 https://doi.org/10.1071/MA14013
Published: 4 February 2014
Although automation is widely used in clinical chemistry, hematology and immunology laboratories, the microbiology laboratory has been slow to adopt automation. Some may criticise microbiologists as being overly conservative and this may seem justified when we recognise that many of the fundamental technologies used in today’s laboratories have existed for more than 100 years (e.g. petri dishes of culture media, biochemical tests for organism identification, microscope for observing organisms on glass slides). Some testing in microbiology has been automated, most notably blood cultures, biochemical identification and antibiotic susceptibility testing, but more comprehensive laboratory automation has been challenged by the variety of specimens submitted for microbiology evaluation, complexity of protocols for processing the specimens, and the technical skills required to evaluate and interpret cultured specimens. Despite these challenges and the apparent reluctance for many microbiologists to forsake tradition, we are at the threshold of major changes and opportunities in the microbiology laboratory with the introduction of laboratory automation. In this review I examine the existing automation platforms, how clinically significant value can be realised from the systems and the path forward for additional opportunities. For the sake of clarity, I will restrict my comments to automated platforms currently in routine clinical use and not discuss systems under development or promised.
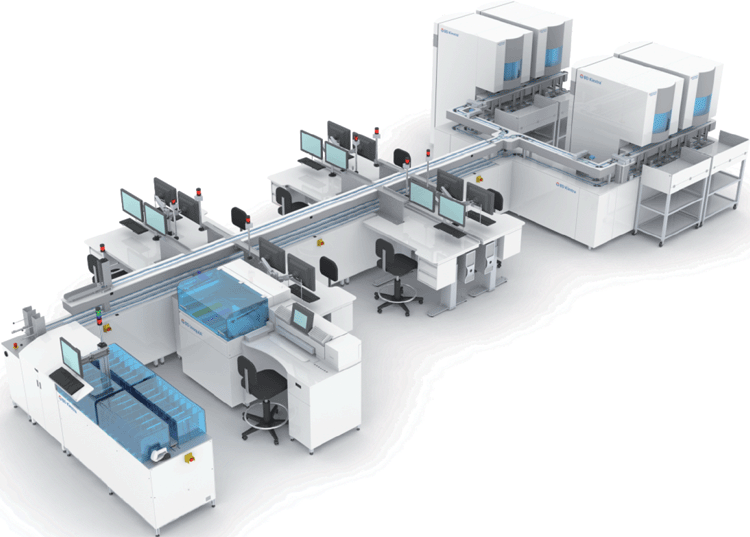
Automation of the clinical microbiology laboratory is a multifunctional process that includes preanalytical processing of specimens; preparation and examination of microscopy slides; incubation and imaging of inoculated cultures; identification and antimicrobial susceptibility testing of isolated organisms; automated movement of plates between specimen processing systems, incubators and work stations; and software to unify the system components and provide laboratory management tools to increase laboratory efficiency and manage microbiology data for analysis and reporting. Three companies are most active in microbiology automation: Becton Dickinson Diagnostic Systems (BDDS; Sparks, MD, USA), bioMerieux (Marcy L’Etoile, France) and Copan Diagnostics (Brescia, Italy). In 2010 BDDS acquired the Innova automated specimen processor from Dynacon and in 2012 BDDS acquired KIESTRA Lab Automation, a company in The Netherlands with extensive experience in microbiology automation with an initial entry into industrial microbiology almost 20 years ago, installation of the first automated clinical laboratory in 2006 and currently with more than 40 clinical systems (Figure 1) in Europe, the Middle East, Australia, Canada and the United States. Copan Diagnostics has recently introduced their automated WASPLab, a track-based system with automated specimen processing and incubation in Canada and Europe, and bioMerieux is currently evaluating their standalone specimen processors and automated incubators in Europe.
|
The most extensive work is with automated specimen processors, with four in current use: Innova (BDDS), InoqulA (BDDS), PREVI Isola (bioMerieux) and WASP (Walkaway Specimen Processing; Copan Diagnostics). Comprehensive summaries of these systems have been previously reported1–3 so my comments here will be limited. Evaluation of the systems should be based on throughput (number of specimens that can be processed); ability to inoculate plates, broths and glass slides; ability to obtain isolated colonies on inoculated plates; cost of consumables (pipettes, loops or other inoculation devices); and safety (ability to uncap and recap specimen containers, use of HEPA filtered air). Limited studies have demonstrated improved recovery of isolated colonies4,5, but systematic comparisons of these instruments with different specimen types have not been reported and, although improved isolated growth should reduce the delays and consumable costs associated with the need for colony subcultures, the clinical value remains to be demonstrated.
Wescor (Logan, UT, USA), bioMerieux, and GG&B Co (Wichita Falls, TX, USA) currently market instruments that automate staining slides inoculated with clinical specimens; however, a systematic evaluation of these instruments with different specimen types is not available. Additionally, no company offers technology to interpret Gram-stained slides, a labour intensive and technically demanding task that would clearly be improved with automated standardisation. Although such technology exists in haematology and cytology laboratories, the requirement to detect and discriminate variably sized bacteria in complex specimen matrices is a technical challenge.
I believe the most innovative component of the automated microbiology laboratory is the incubator-imaging systems that are connected to the specimen processors. Inoculated plates are transferred into and out of the incubator by a moving track system, plates are moved into and retrieved from pre-assigned incubation slots by robotic arms, and automated camera systems are programmed to capture images of the surfaces of inoculated plates at predetermined intervals of time. This level of automation eliminates delays in placing plates into an incubator; allows growth to be monitored over time (such as what we can currently do with our automated blood culture and identification/susceptibility testing platforms); and avoids the inefficiencies of searching for plates in the incubator, premature examination of plates before growth is present and placement of plates at the workbench for hours while the technologist performs his or her work. The advantages of this automation are enormous but the technical complexities should not be underestimated. The speed required to capture images, as well as move more than 1000 plates in the incubators, impact significantly on the utility of this dynamic digital imaging. The clinical value of the incubator-imaging systems will not be realised unless the robotic movements can be done randomly as required by technologists processing the cultured specimens.
Traditionally, culture plates are examined once a day when staff is ready to read the plates rather than when the culture plates are ready to be read. Additionally, inefficiencies and processing delays are introduced because multiple technologists may work with individual specimens and there is no visual record of the growth observed previously. In 2006 the introduction of the KIESTRA automated imaging systems in clinical laboratories allowed sequential images of plates to be examined simultaneously, the magnification of images could be increased to more closely examine colonies and a permanent record of the images was created for quality control purposes. Because the plates remained in the incubator until additional work was required (e.g. subculture, identification tests, antimicrobial susceptibility tests), the bacterial colonies continued to grow under optimal incubation conditions while the images were examined remotely in distraction-free reading rooms.
Currently lacking in the automated laboratories is integration of blood culture instruments, identification platforms such as MALDI mass spectrometry, antimicrobial disk and MIC susceptibility testing, and molecular diagnostic systems. True total laboratory automation will not be realised until this integration occurs. The most critical system is blood culture instruments. Although current instruments can monitor bottles and indicate when microbial growth is detected, a medical technologist must manually perform subcultures, Gram stains, and identification and susceptibility tests. This introduces a significant processing delay during periods of high activity in the laboratory or when staffing is not available such as when the microbiology laboratory is closed. Integration of the workup of positive blood cultures will significantly improve the clinical value of automation.
One aspect of microbiology automation that has not been given sufficient attention is the ability to modify traditional work habits to increase laboratory efficiency and decrease turnaround time for clinically meaningful test results. It is said that automation of pre-analytical specimen processing will decrease the labour needs resulting in personnel savings. That is true but will only be realised if work habits are altered to take full advantage of the automation. For example, most laboratories will prioritise specimen processing throughout the day (such as culturing all urines or all sputa together) or limit the types of specimens processed in evening or night shifts when less experienced technologists are in the laboratory. This is believed to improve efficiency in manual processing; however, this also results in handling specimens multiple times, delays processing specimens and creates additional paperwork. This traditional approach to specimen processing is unnecessary with automated platforms.
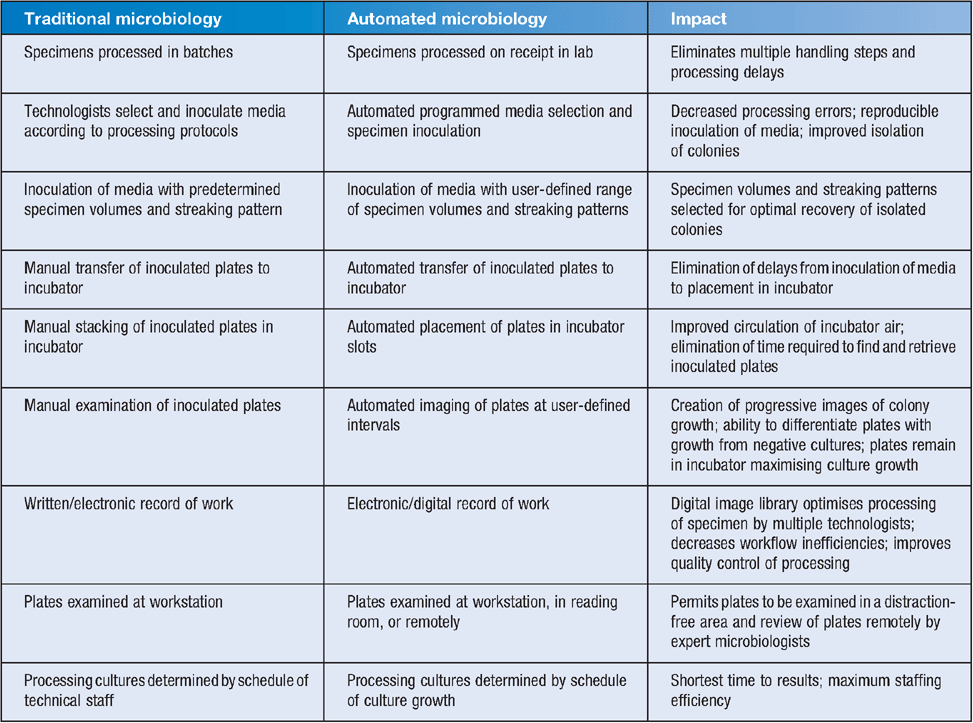
I believe an even more important workflow change can occur using the imaging systems and lab management software. For example, sequential imaging can determine when a specimen has sufficient growth for further processing. This can either be done for individual cultures or, more appropriately, used to define when specific specimen types (e.g. urines, sputa, wound specimens) should be available for processing. Images of specimens with no growth can be reviewed quickly in batch mode and attention then focused on specimens with growth. Software can monitor the number and types of positive cultures in the incubator and determine when the cultures will be ready for further processing. This allows the laboratory manager to predict when work will be available throughout the day and to match technical staffing with the flow of work for each specimen type resulting in increased efficiency and decreased time to results. Using these laboratory management tools significantly decreased staffing needs and increased productivity in one user of the KIESTRA automated platform in England (personal communication, Nicola Newman). The advantages of laboratory automation with a system such as KIESTRA is summarised in Table 1.
|
This is an exciting time for microbiologists. We have witnessed the recent introduction of mass spectrometry for rapid, highly accurate organism identification, molecular platforms for performing highly multiplex assays and now laboratory automation with the promise of improved laboratory efficiency and decreased time to results. I believe our only limitation is the imagination for using these tools.
References
[1] Novak, S.M. and Marlowe, E.M. (2013) Automation in the clinical microbiology laboratory Clin. Lab. Med. 33, 567–588.| Automation in the clinical microbiology laboratoryCrossref | GoogleScholarGoogle Scholar | 23931839PubMed |
[2] Greub, G. and Prod’hom, G. (2011) Automation in clinical bacteriology: what system to choose? Clin. Microbiol. Infect. 17, 655–660.
| Automation in clinical bacteriology: what system to choose?Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MvntVaqtQ%3D%3D&md5=c36743e73b2a8698ca9b4a7c40e72b3fCAS | 21521409PubMed |
[3] Bourbeau, P.P. and Ledeboer, N.A. (2013) Automation in clinical microbiology. J. Clin. Microbiol. 51, 1658–1665.
| Automation in clinical microbiology.Crossref | GoogleScholarGoogle Scholar | 23515547PubMed |
[4] Bourbeau, P.P. and Swartz, B.L. (2009) First evaluation of the WASP, a new automated microbiology plating instruments. J. Clin. Microbiol. 47, 1101–1106.
| First evaluation of the WASP, a new automated microbiology plating instruments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXlvVamt78%3D&md5=c057959d87086719fd158f198ea4ad01CAS | 19158259PubMed |
[5] Mischnik, A. et al. (2012) First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J. Clin. Microbiol. 50, 2732–2736.
| First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach.Crossref | GoogleScholarGoogle Scholar | 22692745PubMed |
Biography
Patrick Murray, PhD, is the Worldwide Director of Scientific Affairs for BD Diagnostic Systems and the former Chief of Microbiology at the NIH Clinical Center in Bethesda, Maryland.


