Microbial strain typing in surveillance and outbreak investigation: past, present and future
Matthew VN O’SullivanCentre for Infectious Diseases and Microbiology
Pathology West: Institute for Clinical Pathology and Medical Research (ICPMR)
Marie Bashir Institute for Infectious Diseases and Biosecurity
Westmead Hospital
Locked Bag 9001
Westmead, NSW 2145, Australia
Tel: +61 2 9845 6255
Fax: +61 2 9893 8659
Email: matthew.osullivan@health.nsw.gov.au
Microbiology Australia 35(1) 52-53 https://doi.org/10.1071/MA14014
Published: 5 February 2014
It is estimated that 180 000 cases of hospital-acquired infections occur in Australian hospitals each year1. Many of these follow colonisation of patients by nosocomial bacterial pathogens. Identifying these acquisition events is necessary to target infection control interventions and to accurately estimate the burden of hospital-acquired infections on our healthcare system. Strain typing is required to reliably monitor these nosocomial acquisition events, particularly for organisms with high background incidence like Staphylococcus aureus. Traditional strain typing methods have been used in a limited, retrospective fashion to confirm suspected outbreaks, but newer technologies allow routine, prospective strain typing to detect transmission soon after it occurs and whole genome sequencing allows the identification of the direction of transmission chains.
The past
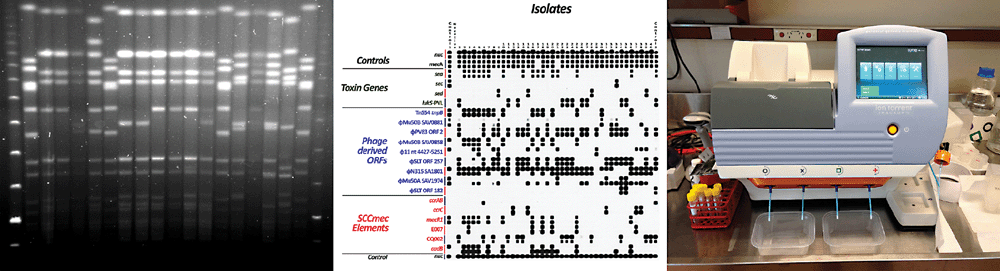
Since its introduction in the 1980s, pulse field gel electrophoresis (PFGE; Figure 1) of restriction enzyme fragmented genomic DNA has been the mainstay of bacterial strain typing for investigation of outbreaks, particularly in the nosocomial setting, but also for other applications, including foodborne illness outbreaks in the community. PFGE has high discriminatory power for most bacteria and guidelines are published for the interpretation of banding patterns to classify isolates as likely to be related or unrelated2. The main disadvantage of PFGE is that it is slow and labour intensive (and therefore expensive). A maximum of ~25 isolates can be examined on a single run, which takes 3–5 days to perform.
Another difficulty with PFGE is interpretation and sharing of results. Interpretation of the fingerprint pattern produced can be somewhat subjective, and inter-run and inter-laboratory comparison of results can be difficult, even with the assistance of software such as BioNumerics (Applied Maths Sint-Martens-Latem, Belgium).
As a result, PFGE-based analysis of nosocomial outbreaks tends to occur in a retrospective fashion where infection control surveillance has identified a cluster of infections (or colonisations) in a discrete geographic area and time period. Strain typing of the isolates is then used to determine whether or not the cluster represents a true clonal outbreak.
While this retrospective approach is useful for surveillance of relatively infrequently occurring pathogens, for more commonly occurring bacteria (such as methicillin-resistant Staphylococcus aureus [MRSA] in settings of high endemicity) this approach is more difficult as clusters representing outbreaks may not be discernible among the background ‘noise’ of endemic cases.
The present
The development of polymerase chain reaction has led to a proliferation of new molecular strain typing methods, which are low cost, rapid and high throughput. Many of these also have the advantage of producing results that can easily be expressed in a digital format, allowing easy inter-run and inter-laboratory comparison. One such method is multi-locus variable number of tandem repeats analysis (MLVA). Here, several loci, each of variable length between isolates (due to a structure composed of varying numbers of repeat units) are amplified, the length of the PCR product precisely determined (ideally by capillary electrophoresis) and the number of repeat units at each locus determined. This series of numbers represent the MLVA type for a given isolate. In Australia, MLVA has become the standard method for strain typing of Salmonella in the surveillance of foodborne outbreaks3. A similar method (mycobacterial interspersed repetitive units, or MIRU, typing) is used for Mycobacterium tuberculosis.
Sequencing of various loci within the bacterial chromosome has also emerged as an important method of bacterial strain typing. In Staphylococcus aureus, sequencing of the staphylococcal protein A (spa) gene has been shown to be a useful typing method and is used routinely for surveillance of nosocomial transmission in some European centres4. Multilocus sequence typing, where several (typically seven) housekeeping genes are examined, is useful for studying overall population genetics of a species but (with some exceptions) lacks the discriminatory power needed for outbreak investigation.
Determining the presence or absence of a panel of genetic targets within the bacterial chromosome (binary typing) can also be a powerful molecular typing method. This approach forms the basis of M. tuberculosis spoligotyping and we have developed a binary typing method for MRSA that is used for surveillance of nosocomial MRSA outbreaks5.
These PCR-based methods are generally inexpensive, high-throughput, highly discriminatory and rapid, allowing them to be used in a different way to the retrospective approach seen with PFGE. These methods can be used prospectively, where all isolates are typed routinely. When a cluster of indistinguishable isolates are seen within a nominated time frame and geographic location, an alert can be conveyed to the public health or infection control unit (in the case of nosocomial surveillance) for further investigation. This prospective approach is essential for pathogens of high endemicity and is used routinely for surveillance of Salmonella, M. tuberculosis and MRSA.
The future
While whole genome sequencing (WGS) currently has many limitations – special bioinformatics skills, software and computing equipment are required to analyse the huge volume of data generated and much work is yet to be done to determine how best to interpret results to define outbreaks – studies are beginning to be published investigating the utility of whole genome sequencing in outbreak investigation, both in hospital and community settings6,7. One major advantage of WGS is the ability to infer the directionality of transmission events and therefore the timeline of an outbreak, by examining the accumulation of single nucleotide polymorphisms in outbreak isolates over time. With the promise of ultimate discriminatory power and with the cost falling dramatically in recent years, WGS is likely to replace existing strain typing methods over the coming decades.
References
[1] Australian Government Productivity Commission (2009) Healthcare associated infections. In Public and Private Hospitals: Productivity Commission Research Report, pp. 123-141, Commonwealth of Australia.[2] Tenover, F.C. et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
| 1:CAS:528:DyaK2MXoslSisLc%3D&md5=543d113b72b07baa2c4c66ab4d010425CAS | 7494007PubMed |
[3] Sintchenko, V. et al. (2012) Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect. Dis. 12, 78.
| Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA).Crossref | GoogleScholarGoogle Scholar | 22462487PubMed |
[4] Harmsen, D. et al. (2003) Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448.
| Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjtFyqtA%3D%3D&md5=41fd7667200e79783b08594f8635a61fCAS | 14662923PubMed |
[5] O’Sullivan, M.V.N. et al. (2012) Prospective genotyping of hospital-acquired methicillin-resistant Staphylococcus aureus isolates by use of a novel, highly discriminatory binary typing system. J. Clin. Microbiol. 50, 3513–3519.
| Prospective genotyping of hospital-acquired methicillin-resistant Staphylococcus aureus isolates by use of a novel, highly discriminatory binary typing system.Crossref | GoogleScholarGoogle Scholar |
[6] Harris, S.R. et al. (2013) Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13, 130–136.
| Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1Gksr0%3D&md5=0da6418c748f7f577b189e67ba22859cCAS | 23158674PubMed |
[7] Walker, T.M. et al. (2013) Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146.
| Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1Git7o%3D&md5=cb1fc27d5e200ea265f8b19fd2ab5454CAS | 23158499PubMed |
Biography
Matthew O’Sullivan is an infectious diseases physician and clinical microbiologist at Westmead Hospital, Sydney, and Senior Lecturer at the Marie Bashir Institute for Infectious Diseases and Biosecurity, University of Sydney. He has a research interest in genotyping of pathogens responsible for hospital-acquired infections, particularly MRSA.