Revisiting biodiscovery from microbial sources in the light of molecular advances
İpek KurtbökeGeneCology Research Centre and Faculty of Science, Health, Education and Engineering, University of the Sunshine Coast, Maroochydore DC, Qld 4558, Australia
Email: ikurtbok@usc.edu.au
Microbiology Australia 38(2) 58-61 https://doi.org/10.1071/MA17028
Published: 26 April 2017
Since the discovery of penicillin microorganisms have been an unexhausted source of novel bioactive compounds that served as scaffolds for potential drug candidates as well for the development of new antibiotics via fermentative processes. However, after 30 glorious years of biodiscovery begun in the 1940s, discovery of new antibiotic or therapeutic compounds with medicinal value entered a decline phase from the late 1970s onwards. At the same time, significant increases in the numbers of antibiotic or multi-drug resistant bacteria resulting in serious infections were reported. Although natural product discovery research was encouraged to continue due to the need to treat these infections only a few discoveries of potent antibiotics were made in the years of decline such as the discovery of Nikkomycin and Spinosyn. However, at the dawn of the 21st century advances in molecular biology such as genome mining and metabolic engineering changed the scene providing new avenues to the field of drug discovery. This article will highlight some of these advances.
With the technical superiority provided by whole-genome sequencing and mining, identification and cloning of secondary metabolite biosynthetic gene clusters, and subsequently their expression in heterologous hosts has become possible1,2. These technological advances have also enabled metabolic engineering approaches for the optimisation of production yields as well as to direct manipulation of responsible synthetic pathways for metabolite secretion to generate modified products3. Moreover, combined with emerging strain prioritisation, bioinformatics, and analytical technologies, natural product dereplication was also revolutionised, in turn enabling structural prediction, rapid detection, and isolation of the most promising natural products at a rapid pace. Thus, a new golden age of natural product discovery has commenced4.
Genome mining
In bacteria, the biosynthesis of natural products (e.g. antibiotics, enzyme inhibitors, signalling molecules) is directed by groups of genes called Biosynthetic Gene Clusters (BGCs)5,6. The target-directed approach that traces such groups of genes to metabolite production revealed the presence of putative BGCs involved in the synthesis of diverse metabolites6–9. Knowledge of the biosynthetic gene cluster organisation then facilitated the cloning of complete pathways and combined with the current bioinformatics approaches the genome mining concept was born6,7, which has allowed the interrogation of microbial genomes to determine their biosynthetic abilities. Moreover, in-depth understanding of natural products biosynthesis and the assembly of gene clusters (e.g. polyketides and peptides) was generated via the application of the genomic methods10. The combination of genetics-based approaches and mass spectrometry (MS)-based analytical methods has also provided a platform for development of reliable dereplication strategies for natural products discovery and identification11. Many cryptic gene clusters that encode secondary metabolism in the most bioactive bacterial members were located through examination of the genomic data12. In addition, a significant degree of chemical diversity encoded in silent/cryptic biosynthetic gene clusters were detected in the genomes of such bacteria. Examples include detection of genes coding for synthesis of stambomycin in Streptomyces ambofaciens13. Successful implementation of genome-guided discovery of natural products and biosynthetic pathways from Australia’s untapped microbial megadiversity has also recently been revealed10.
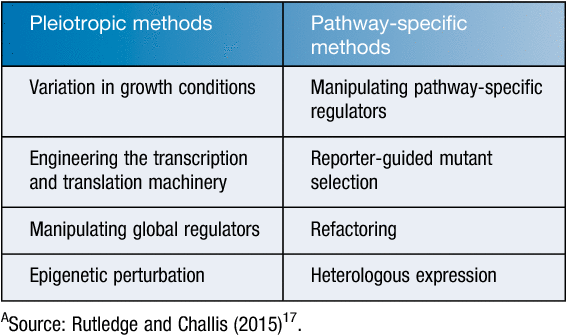
Moreover, for ‘poorly or not at all’ expressed gene clusters under laboratory conditions novel approaches have been trialled such as the control of regulator genes, ribosome engineering, and fermentation co-culture14. Effect of rare earth elements (REEs) were tested for the activation of silent or poorly expressed genes in bacteria. Addition of low concentrations of scandium into the cultures of Streptomyces coelicolor (an actinohordin producer), S. antibioticus (an actinomycin producer), and S. griseus (a streptomycin producer) were found to enhance antibiotic production by 2–25-fold14,15. Moreover, HPLC analysis of several compounds indicated that they could only be detected in the extracts obtained from REE-treated cultures14. The REE treatment, especially with scandium (Sc), was also found to stimulate enzyme production in Bacillus subtilis for both α-amylase and bacilysin at the transcriptional level14,16. Other approaches used to activate silent genes have included Pleiotropic and Pathway-specific methods17 (Table 1).
|
Proteomining
Another widely applicable strategy called natural product proteomining has been developed18. Differential biosynthesis of the bioactivity of interest was achieved with this method using fluctuating growth conditions that ensure differential biosynthesis of the bioactivity of interest. Subsequent combination of metabolomics and quantitative proteomics facilitated establishment of correlations between abundance of natural products and concomitant changes in the protein pool, which allows identification of the relevant biosynthetic gene cluster. Gubbens et al. (2014)18 used this approach to elucidate gene clusters for different natural products in Bacillus and Streptomyces, and revealed the presence of the biosynthetic cluster coding for the synthesis of a novel juglomycin-type antibiotic. One of the major advantages of the natural product proteomining is claimed to be the lack of requirement of prior knowledge of the gene cluster or secondary metabolite: therefore, this represents a general strategy for identification of all types of gene clusters18.
Metabolomic profiling
Metabolomic profiling, using liquid chromatography coupled to high-resolution mass spectrometry has also been used to enable the optimisation of media ingredients with regards to antibiotic production. Examples include the optimisation of the salt media concentration to promote rifamycin production by an obligate marine actinobacterium Salinispora arenicola. Results obtained from these metabolomic analysis and chemical profiling studies then enabled the determination of the link between the days of incubation and different salt concentrations19. Metabolomic profiling thus forms the foundation for the future development of optimum growth media resulting in the target-directed production of the bioactive compounds19.
Advances in natural product chemistry
Improved strategy to frontload NP extracts with lead- and drug-like molecules to facilitate the NP-based drug discovery process has also been reported20. Their approach consisted of the generation of lead-like enhanced (LLE) fractions that contained components with desirable physicochemical properties. Subsequently, a 1H NMR metabolic fingerprinting approach was developed to uncover and reveal unique spectral patterns of the drug-like natural product metabolome of an Australian marine sponge allowing the identification of a novel compound, iotrochotazine A20.
Another successful application of the above-mentioned NMR-based method was its use to access the unique components of the drug-like natural product metabolome of a termite-gut associated bioactive Streptomyces species21. This approach again accelerated the identification of lead-like enhanced fractions containing small molecules with unique spectral patterns and resulted in the isolation and identification of six new natural products (actinoglycosidines A (1) and B (2), actinopolymorphol D (3), niveamycin A (7), B (8) and C (9))21.
Imaging mass spectrometry of natural products
Another fascinating development has been the use of imaging mass spectrometry (IMS) in the investigation of natural products22. IMS was applied to metabolites that possess biological functions (e.g. polyketides, nonribosomal peptides, alkaloids, lantibiotics, microcins, flavonoids, terpenes etc.) to address the functional roles of natural products in metabolic exchange, communication and defence. IMS also revealed the roles of such metabolites as entities that control morphological changes in organisms, thus subsequently aiding in the spatial analysis of natural products from heterogeneous samples22.
Moreover, the use of matrix-assisted laser desorption ionisation-time of flight (MALDI-TOF) IMS applied directly to microorganisms on agar-based medium enabled the capture of the global information about microbial molecules that in turn allowed direct correlation of chemotypes to phenotypes23. Investigations related to metabolic exchange factors of intraspecies, interspecies, and polymicrobial interactions thus became possible23.
One strain-many compounds (OSMAC) approach
Bode et al. (2002)24 investigated the systematic alteration of easily accessible cultivation parameters such as medium composition, aeration, culture vessel and addition of enzyme inhibitors into media to increase the number of secondary metabolites available from one microbial source. They termed their approach the OSMAC: One Strain-Many Compounds. With this approach, they could isolate up to 20 different metabolites in yields up to 2.6 gL–1 from a single organism. The compounds detected covered nearly all major natural product families, and in some cases the increased titres opened new possibilities for semisynthetic methods to enhance even more of the chemical diversity of selected compounds. The OSMAC approach has been a superior alternative to industrial high-throughput screening that focuses on the active principle in a distinct bioassay. English et al. (2016)25 using the same approach detected the induction of antibacterial metabolites when different growth parameters were used.
Single-cell- and metagenomics-based approaches
Metagenomics utilises a range of genomic technologies and bioinformatics tools to directly access the genetic content of entire communities of organisms26,27. The functional gene composition of microbial communities can be accessed, thus resulting in a much broader description than phylogenetic surveys that are often based only on the diversity of one gene (e.g. 16S rRNA gene). Metagenomics provides genetic information on potentially novel biocatalysts or enzymes, genomic linkages between function and phylogeny for uncultured organisms, and evolutionary profiles of community function and structure. It can also be complemented with meta-transcriptomic or meta-proteomic approaches to describe expressed activities26,28,29.
Single-cell genomics (SCG) that enable the sequencing of any genome region in an uncultured cell, played a key role in identifying metabolic features, evolutionary histories and inter-organismal interactions of the uncultured microbial groups that dominate many environments and biogeochemical cycles30. SCG provides information on the cell’s phylogenetic (e.g. 16S rRNA genes) and metabolic markers to generate in-depth understanding of the roles and metabolic functions of each organism. Thus, the SCG approach is a powerful complement to cultivation-based and microbial community-focused research approaches30. Examples include detection of two bioactive phylotypes of the candidate genus ‘Entotheonella’ with multiple, distinct biosynthetic gene clusters and with genomes of greater than 9 megabases, co-inhabiting within the chemically and microbially rich marine sponge Theonella swinhoei, via the use of a single-cell- and metagenomics-based approach31.
The continued importance of culturing
Through functional metagenomics insights might be gained into the metabolic potential of an organism and its community. However, prediction on the ecological significance of the annotated genes without first considering their function in the broad context of metabolism might be difficult32. Thus, reconstruction of metabolic pathways and testing predictions by experimentation with axenic cultures might facilitate generation of a broad understanding on functional diversity32. Design of effective selective culturing techniques thus remains vital in the detection and isolation of the reference strains needed for laboratory experimentation to generate information on the metabolic pathways33–36.
Conclusions
Molecular advances such as metabolic engineering of host strains to express cryptic gene clusters, as well as improved understanding of the chemical and biological mechanisms defining the secondary metabolite biosynthesis, and an architectural understanding of the biosynthetic enzymes combined with bioinformatic and synthetic biology approaches will lead to a new renaissance in biodiscovery3.
We are thus at the dawn of a new golden age of novel bioactive natural product discovery4,12,27. Those interested learning more about this topic and the latest advances are referred to the book Microbial resources: from functional existence in nature to applications37, which is also reviewed in this issue.
References
[1] Zhang, H. et al. (2010) Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 17, 1232–1240.| Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsVGntrfK&md5=4ad5d62623aa9b89499899c0e4b01b5eCAS |
[2] Weber, T. et al. (2015)a antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243.
| antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters.Crossref | GoogleScholarGoogle Scholar |
[3] Weber, T. et al. (2015)b Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26.
| Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitFCrtLzM&md5=d2eaa29f4d8e407bae73709d6715af09CAS |
[4] Shen, B. (2015) A new golden age of natural products drug discovery. Cell 163, 1297–1300.
| A new golden age of natural products drug discovery.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhvFKksb%2FM&md5=2216def94e1026100b46122532206bc2CAS |
[5] Traxler, M.F. and Kolter, R. (2015) Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970.
| Natural products in soil microbe interactions and evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXptVKgtL0%3D&md5=187d929457a54817eac9817a36bd1ecbCAS |
[6] Cruz-Morales, P. et al. (2016) Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes. Genome Biol. Evol. 8, 1906–1916.
| Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes.Crossref | GoogleScholarGoogle Scholar |
[7] Medema, M.H. and Fischbach, M.A. (2015) Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648.
| Computational approaches to natural product discovery.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtlKntrjP&md5=25e832085ae4ab214e576a481b00888dCAS |
[8] Hadjithomas, M. et al. (2015) IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio 6, e00932-15.
| IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites.Crossref | GoogleScholarGoogle Scholar |
[9] Medema, M.H. et al. (2015) Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625–631.
| Minimum information about a biosynthetic gene cluster.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtlKntrjN&md5=ca7ed18c4c60ea08a3bb9f8eb0ceb4f1CAS |
[10] Kalaitzis, J.A. et al. (2016) Genome-guided discovery of natural products and biosynthetic pathways from Australia’s untapped microbial megadiversity. Aust. J. Chem. 69, 129–135.
| Genome-guided discovery of natural products and biosynthetic pathways from Australia’s untapped microbial megadiversity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XitF2nu78%3D&md5=cce1d6a7b1109c3935211bb3d773b1eaCAS |
[11] Mohimani, H. et al. (2014) NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909.
| NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtlalu7bO&md5=2eca6a937d8678992d0aa7186aaa7450CAS |
[12] Choi, S.S. et al. (2015) Genome mining of rare actinomycetes and cryptic pathway awakening. Process Biochem. 50, 1184–1193.
| Genome mining of rare actinomycetes and cryptic pathway awakening.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXnvFShsLg%3D&md5=db339d907e1309cbf0b6dfd1a7983014CAS |
[13] Aigle, B. et al. (2014) Stambomycin and derivatives, their production and their uses as drugs. U.S. Patent 8,664,186.
[14] Ochi, K. and Hosaka, T. (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 97, 87–98.
| New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjtlGhtQ%3D%3D&md5=9da4e84fc6409b838e1ad15955ebf4b6CAS |
[15] Kawai, K. et al. (2007) The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 274, 311–315.
| The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVaisLfL&md5=ffa69b60b90d9c7956bdb0d1d29e914eCAS |
[16] Inaoka, T. and Ochi, K. (2011) Scandium stimulates the production of amylase and bacilysin in Bacillus subtilis. Appl. Environ. Microbiol. 77, 8181–8183.
| Scandium stimulates the production of amylase and bacilysin in Bacillus subtilis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1Gqt77L&md5=4279eead594242d4cb82f58cfc884153CAS |
[17] Rutledge, P.J. and Challis, G.L. (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523.
| Discovery of microbial natural products by activation of silent biosynthetic gene clusters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtFaitr3O&md5=b2064896126d3b1be63b3d05b922ec45CAS |
[18] Gubbens, J. et al. (2014) Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products. Chem. Biol. 21, 707–718.
| Natural product proteomining, a quantitative proteomics platform, allows rapid discovery of biosynthetic gene clusters for different classes of natural products.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXnslGntbs%3D&md5=e973bf74fdc51a3bc75846fa96cdb102CAS |
[19] Bose, U. et al. (2017) Production of N-acyl homoserine lactones by the sponge-associated marine actinobacteria Salinispora arenicola and Salinispora pacifica. FEMS Microbiol. Lett. 364, fnx002.
| Production of N-acyl homoserine lactones by the sponge-associated marine actinobacteria Salinispora arenicola and Salinispora pacifica.Crossref | GoogleScholarGoogle Scholar |
[20] Camp, D. et al. (2012) Drug-like properties: guiding principles for the design of natural product libraries. J. Nat. Prod. 75, 72–81.
| Drug-like properties: guiding principles for the design of natural product libraries.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs12hsb7N&md5=9b030f8ea60c85255f19592e39aed69bCAS |
[21] Romero, C.A. et al. (2015) NMR fingerprints, an integrated approach to uncover the unique components of the drug-like natural product metabolome of termite gut-associated Streptomyces species. RSC Advances 5, 104 524–104 534.
| NMR fingerprints, an integrated approach to uncover the unique components of the drug-like natural product metabolome of termite gut-associated Streptomyces species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhvFajs73K&md5=e42791896efa34baafb3c2ca7f2b3466CAS |
[22] Esquenazi, E. et al. (2009) Imaging mass spectrometry of natural products. Nat. Prod. Rep. 26, 1521–1534.
| Imaging mass spectrometry of natural products.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVCjt7bE&md5=cd03898b1ee0b1e48b800513eb19fcbeCAS |
[23] Yang, J.Y. et al. (2012) Primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 194, 6023–6028.
| Primer on agar-based microbial imaging mass spectrometry.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1Cju7zF&md5=b79e011798955d279b61dd2445fb6dddCAS |
[24] Bode, H.B. et al. (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3, 619–627.
| Big effects from small changes: possible ways to explore nature’s chemical diversity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xlt1Kmtb0%3D&md5=1cf8f183454745eb87cefca847fe3041CAS |
[25] English, A.L. et al. (2016) Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species. Synthetic and Systems Biotechnology , .
| Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species.Crossref | GoogleScholarGoogle Scholar |
[26] Thomas, T. et al. (2012) Metagenomics-a guide from sampling to data analysis. Microb. Inform. Exp. 2, 3.
| Metagenomics-a guide from sampling to data analysis.Crossref | GoogleScholarGoogle Scholar |
[27] Zotchev, S.B. et al. (2016) Metagenomics of marine actinomycetes: from functional gene diversity to biodiscovery. In: Marine OMICS: Principles and Applications (Kim, S.-K., Ed.). pp. 165–186. CRC Press.
[28] Wilmes, P. and Bond, P.L. (2006) Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 14, 92–97.
| Metaproteomics: studying functional gene expression in microbial ecosystems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtlGjt7o%3D&md5=bc9b8b8c9d0145bcd8dcd31f56a91448CAS |
[29] Gilbert, J.A. et al. (2008) Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3, e3042.
| Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities.Crossref | GoogleScholarGoogle Scholar |
[30] Stepanauskas, R. (2012) Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 15, 613–620.
| Single cell genomics: an individual look at microbes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVemtLvN&md5=cb12bc4121fbc2b4170d5603ffd37f85CAS |
[31] Wilson, M.C. et al. (2014) An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62.
| An environmental bacterial taxon with a large and distinct metabolic repertoire.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsl2hsL0%3D&md5=ca715a0d21393e841de9f66d450d7adcCAS |
[32] Temperton, B. and Giovannoni, S.J. (2012) Metagenomics: microbial diversity through a scratched lens. Curr. Opin. Microbiol. 15, 605–612.
| Metagenomics: microbial diversity through a scratched lens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtV2ms7%2FK&md5=fab19648441c7935af2233710ce30ff6CAS |
[33] Kurtböke, D.I. (Ed.) (2003) Selective Isolation of Rare Actinomycetes. Queensland Complete Printing Services, Australia.
[34] Kurtböke, D.I. (2011) Exploitation of phage battery in the search for bioactive actinomycetes. Appl. Microbiol. Biotechnol. 89, 931–937.
| Exploitation of phage battery in the search for bioactive actinomycetes.Crossref | GoogleScholarGoogle Scholar |
[35] Kurtböke, D.I. (2012) A phage-guided route to discovery of bioactive rare actinomycetes. in bacteriophages. InTech , 237–256.
[36] Kurtböke, D.I. (2016) Actinomycetes in biodiscovery: genomic advances and new horizons. Chapter 35, pp. 567–590. In: The Handbook of Microbial Resources (Gupta, V.K., et al., Eds). CAB International Publications, Oxfordshire, UK.
[37] Kurtböke, D.I. (Ed.) (2017) Microbial resources: from functional existence in nature to applications. Elsevier, in press.
Biography
Dr Kurtböke has been working in the field of biodiscovery and has been an active member of the international actinomycete research community since 1982. She currently conducts research and teaches in the field of applied microbiology and biotechnology and is senior lecturer at the University of the Sunshine Coast (USC), Queensland. She has been a member of the Biodiscovery Industry Panel established by the AusBiotech and DEHWA, which networks Australian biodiscovery operators. She has also established a bioactive actinomycete library used for research and teaching activities at the USC as well as in partnership with regional, national and international collaborators for discovery of new therapeutic agents, agrobiologicals, enzymes and environmentally friendly biotechnological innovations. She has also been an active member of the World Federation of Culture Collections (WFCC) including serving as the Vice-President of the Federation (2010–2013).


