Manipulating the soil microbiome for improved nitrogen management
Hang-Wei Hu A and Ji-Zheng He BA School of Agriculture and Food, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, Vic. 3010, Australia. Tel: +61 3 9035 7639, Email: hang-wei.hu@unimelb.edu.au
B School of Agriculture and Food, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, Vic. 3010, Australia. Tel: +61 3 9035 8890, Email: jizheng.he@unimelb.edu.au
Microbiology Australia 39(1) 24-27 https://doi.org/10.1071/MA18007
Published: 21 February 2018
The soil microbiome, including bacteria, archaea, fungi, viruses, and other microbial eukaryotes, has crucial roles in the biogeochemical cycling of nitrogen (N), the maintenance of soil fertility, and the plant N use efficiency (NUE) in agro-ecosystems1. Recent advances in omics-based technologies (e.g. metagenomics, metatranscriptomics, and metaproteomics) have expanded our understanding of the soil microbiome and their controls on specific N-cycling processes1–3. Given the growing N-based fertiliser consumption and continuous land degradation, innovative technologies are needed to manipulate the soil microbiome to improve crop NUE, reduce N losses and increase N reservation in soil. This article discusses the research directions to facilitate the development of microbiome-manipulating technologies for sustainable management of N transformation processes.
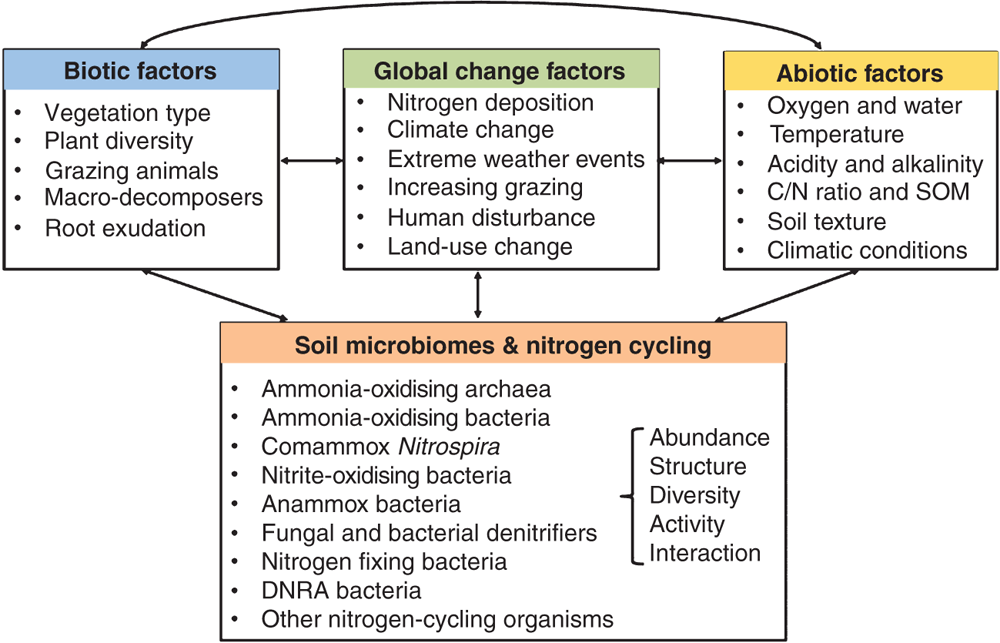
The crop nitrogen use efficiency (NUE) in modern agro-ecosystems is notoriously low, as more than 50% of N fertiliser applied is lost to the environment through ammonia volatilisation, nitrate leaching, and emissions of nitrous oxide (N2O), the third most important greenhouse gas4,5. These losses are mostly driven by a myriad of N-cycling processes (in particular, nitrification and denitrification) that can be modulated by a broad range of soil microorganisms (Figure 1)6,7. Conventional agricultural practices mainly rely on agronomic measures and chemical inputs to improve NUE, which could lead to soil degradation and loss of biodiversity, with detrimental consequences for soil health and ecosystem functioning8. For example, long-term use of synthetic fertilisers, herbicides, and pesticides can negatively influence bacteria and fungi that create organic matter essential to plants. To meet the increasing food demand of a global population of more than 11 billion by 2100, there is an urgent need to discover new intervention points to manage N-cycling microorganisms for improved NUE and sustainable agricultural production.
Propelled by the evidence in manipulating gut microbiomes for improved human health, there are growing interests focused towards the manipulation of the soil microbiome to reduce soil erosion, to enhance plant growth and disease resistance in agro-ecosystems, and to promote the remediation of heavy metal-contaminated soils1,3. In this article, we discuss the currently-used technologies and emerging microbial biotechnologies that can manipulate the soil microbiome in situ to mitigate the processes of agricultural N loss and improve crop NUE, leading to both enhanced crop yield and positive environmental and social outcomes.
Physicochemical approaches to manipulate the soil microbiome
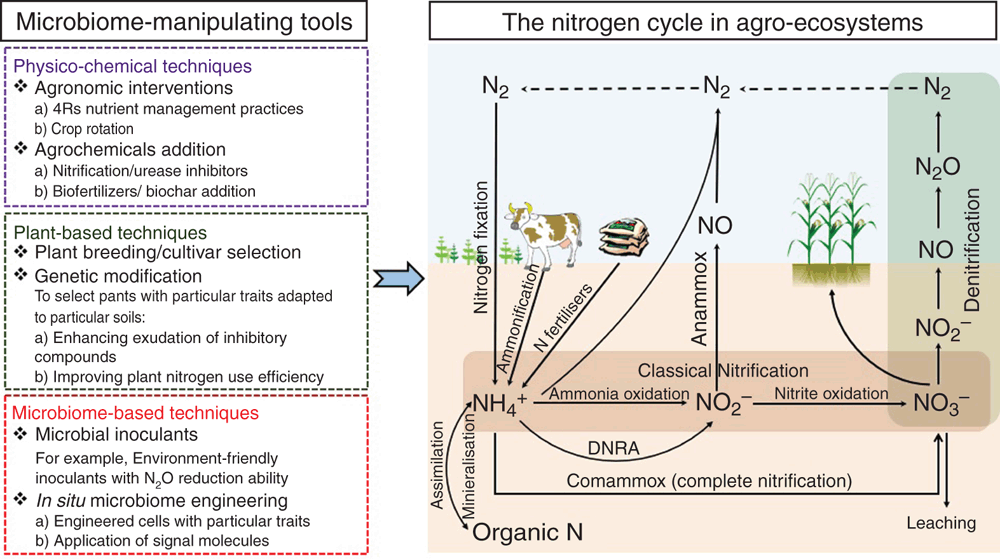
Physicochemical approaches have been put forward to reduce agricultural N losses through manipulating the abundance, structure and activities of soil N-cycling microorganisms or controlling the amount of N resources available to microorganisms (Figure 2). Some practical tools utilised in agro-ecosystems to improve NUE include: (1) use of synthetic nitrification inhibitors (e.g. DMPP and DCD) to inhibit the activity of ammonia oxidisers and reduce the N loss through N2O emission and nitrate leaching9; (2) use of urease inhibitors (e.g. N-(n-butyl) thiophosphoric triamide (NBPT)) to inhibit the expression of genes encoding ureases that catalyse urea hydrolysis10; (3) manipulation of soil properties (e.g. soil pH, C:N ratio, and moisture) by agrochemical amendments and agronomic practices to indirectly reshape the abundance, diversity and structure of soil microbiomes; (4) incorporation of plant residues to enhance microbial N immobilisation and reduce the amount of inorganic N available to soil microbes11; and (5) use of precise nutrient management practices and high-efficiency fertilisers to better synchronise N supply and crop N demand and reduce N available to soil microorganisms. Tools (1) and (2) are direct practices that impact soil microorganisms while the other three tools are indirect practices.
The outcomes of these physicochemical technologies are variable across soils, primarily owing to their largely unknown impacts on soil microorganisms. For example, the nitrification inhibitor DMPP could effectively inhibit nitrification and N2O emissions in alkaline soils through influencing the abundance and metabolic activity of ammonia-oxidising bacteria, but had no significant effects in many acidic soils probably due to the fast degradation of DMPP9,10. Other drawbacks of synthetic inhibitors include difficulties in application, rapid degradation, increased ammonia volatilisation, and migration into the food system5,12. In addition, long-term use of chemicals has detrimental environmental impacts, resulting in the accumulation of residues in fields, loss of beneficial microorganisms, and disruption of the plant and soil microbiota association13. An improved understanding of the key functional genes, enzymes and regulatory mechanisms of N-cycling processes, and their responses to interactions between different climatic, soil and biotic properties (Figure 1), should be essential to improvement of physicochemical strategies.
Plant-based approaches to manipulate the soil microbiome
Plant physiological traits can be selected by plant breeding (cultivar selection) or genetic modification techniques to secrete specific compounds or signalling molecules for the direct manipulation of the soil microbiome in situ8,11. Plants have developed intimate relationships with their interacting soil microbiomes and the environment (termed as the ‘phytobiome’)14. Some plant and crop roots (e.g. Fallopia spp. and Brachiaria humidicola) can exudate organic compounds to inhibit the ammonia monooxygenase (enzyme capable of oxidising NH3 to NH2OH) and hydroxylamine oxidoreductase (enzyme capable of oxidising NH2OH to NO2–) of ammonia oxidisers15, or to inhibit the metabolic activity of denitrifiers16. Screening agricultural crops with similar traits may greatly enhance our ability to improve crop NUE by using them directly for in situ microbiome engineering. A conventional plant breeding programme, however, rarely takes into account the interactions within phytobiome14, which may result in loss of beneficial microbiota, disruption of symbiosis associations, and unknown consequences for other ecosystem processes13. Future plant-based strategies should integrate the knowledge of the phytobiome into the programme, by which specific N-cycling microorganisms are manipulated in situ without compromising beneficial microbiota and other ecosystem functions3.
Emerging microbial biotechnology approaches to manipulate the soil microbiome
Microbial biotechnologies have shown enormous potential in reducing N losses via N2O emissions in soybean root systems where denitrifiers harbouring N2O reductase, enzyme capable of reducing N2O to N2, were amended17. These is evidence that the application of organic fertilisers inoculated with N2O-reducing denitrifiers decreased N2O emissions in agricultural soils at field scales18. However, the persistence and functionality of these inoculated microbiota are uncertain, as most of them are unlikely to persist in soil due to the strong competition from indigenous microbiota. When using specific bacterial or mycorrhizal inocula as a strategy to manipulate the soil microbiome, there is an urgent need to modify the mode of delivery to increase their colonisation potential. Some approaches8 include: (1) use of consortia of multiple compatible microbes, rather than single-strain formulations, to better compete with indigenous microbiota; (2) use of synbiotics to provide support for colonisation of the inoculated strains; (3) use of slow release systems for inocula to provide continual inoculation under field conditions; and (4) using chemical pesticides or predators for the indigenous microbiota to create new niches for the introduced microbiota. The combination of these approaches might help to achieve maximum benefits and improved crop NUE.
Emerging microbial biotechnology tools are proposed to precisely manipulate the soil microbiome in situ, by adding or withdrawing chemicals19, to regulate N transformation processes under various conditions. Multidisciplinary approaches, especially genome engineering and synthetic biology, by fully taking advantages of microbiome knowledge, are needed for maximising the contribution of microbiome-based biotechnologies to sustainable management of the N cycle. Here, we highlight the key opportunities and research priorities to harness the soil microbiome to manage N transformation processes:
-
Exploration of the core soil microbiome components involved in N cycling processes and their signalling compounds (or their inhibitors) for chemical conversations, and how they are impacted by plants, climate, soil properties, and agronomic practices (Figure 1). These efforts will lead to the identification of a set of functional taxa that should be prioritised for further research, and provide new ways through direct manipulation of the microbiome activities or via genetically engineering the native microbiomes in situ. Microbiome-based approaches targeting at reducing rates of nitrification and denitrification (pathways leading to N losses) and increasing rates of dissimilatory nitrate reduction to ammonium (DNRA, the pathway capable of reserving N in soil), would have multiple benefits such as reduced N2O emissions, increased farm productivity, reduced water contamination, and higher farm profitability through reduced use of fertilisers.
-
Technological improvements are needed to decipher the ‘dark matter’ of microbial chemistries, as current metabolomics studies can only match a small fraction of data to known chemical compounds and biochemical pathways20. Quorum sensing signals have been found to regulate the communication between ammonia oxidisers and nitrite oxidisers, and to regulate the production and consumption of N oxide gases in a model nitrite oxidiser21. We are just beginning to recognise the diversity and specificity of signalling molecules, with the advancement of integrated metabolome and proteome technologies14, and thus becoming more reliable to develop microbiome-engineering strategies that could utilise the natural signalling channels of the N-cycling microorganisms.
-
Harnessing the emerging synthetic biology and genome editing tools to directly engineer the genomes and metabolic pathways of indigenous soil microbiome mediating N-cycling processes in situ with high specificity and efficacy19. We need a comprehensive knowledge of the gene regulation frameworks and modelling tools (through integrating various components of microbiome datasets, soil parameters, weather data and new computational methods) to predict the effects of microbiome manipulations in situ and reliably monitor the engineering outcomes. Precision tools such as sequence-specific gene editing using CRISPR/Cas9 delivered by phage or conjugative elements22, and synthetic microbial consortia engineered to disrupt or replace existing communities, are needed for modifying microbiota and their genes in situ.
-
The emerging in situ microbiome-manipulation tools (in particular, use of genetically modified organisms) in the natural environment are subject to regulatory requirements and societal concerns13. Coordinated efforts and multidisciplinary networks of policy makers, industry stakeholders, engineers, public and private partners, and agricultural communities will consolidate and translate new microbiome-related innovations into practical solutions for farmers and ensure that risks associated with microbiome research are properly addressed. In addition to traditional agency-specific requests for proposals, strategic funding investments by national-level interdisciplinary initiatives (e.g. USA National Microbiome Initiative) could ensure availability of sufficient resources for developing broadly applicable microbiome-based tools2,19,23.
Concluding remarks and future perspectives
Although there are a range of ways in which crop NUE and agricultural productivity could be improved by the management of the soil microbiome, this is an area of great challenge which requires advances in multi-omics technologies, systems biology, synthetic biology, data analytics, standardised protocols, and modelling, as well as new collaborative efforts among scientists, engineers, agribusiness professionals and agricultural communities. Therefore, utilisation of existing physicochemical technologies will be the major approaches to manipulate the soil microbiome in short or medium terms. Over a longer term, we envision the innovation in in situ genome engineering technology will offer precise microbiome management approaches to sustainably increase agriculture productivity. These technologies will show enormous potential in managing N transformation processes and can be integrated into next-generation precision agriculture for site-specific management. Under a context of global change and a growing human population, harnessing the capabilities of Earth’s microbiomes will potentially lead to reduced chemical inputs, improved soil and water health, and increased productivity and sustainability of global agro-ecosystems.
Acknowledgements
This work was financially supported by Australian Research Council (DE150100870, DP160101028 and LP160101134).
References
[1] Fierer, N. (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590.| Embracing the unknown: disentangling the complexities of the soil microbiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhtlGktbfN&md5=b79aeb90c64d09f110fd78ab6a3d9586CAS |
[2] Thompson, L.R. et al. (2017) A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463.
| A communal catalogue reveals Earth’s multiscale microbial diversity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhslehurzL&md5=3c9acc3e93eae5c4dd3365fb6c68e566CAS |
[3] Trivedi, P. et al. (2017) Tiny microbes, big yields: enhancing food crop production with biological solutions. Microb. Biotechnol. 10, 999–1003.
| Tiny microbes, big yields: enhancing food crop production with biological solutions.Crossref | GoogleScholarGoogle Scholar |
[4] Hu, H.W. et al. (2015) Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749.
| Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhs1ektrnO&md5=943ad334f9e26eb47172bac459efceacCAS |
[5] Coskun, D. et al. (2017) Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 3, 17074.
| Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXpt1yjs7k%3D&md5=bd98e8cb593e7a1fe0a3351d8cf30f60CAS |
[6] Zhang, L.M. et al. (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045.
| Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlvVGktrw%3D&md5=5cea00b9ab880ef964fd12c7ed406817CAS |
[7] Hu, H.W. and He, J.Z. (2017) Comammox – a newly discovered nitrification process in the terrestrial nitrogen cycle. J. Soils Sediments 17, 2709–2717.
| Comammox – a newly discovered nitrification process in the terrestrial nitrogen cycle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhslGhtrrP&md5=3647a6bbb6d31d5d0dd27dd7582d4d00CAS |
[8] Hu, H.W. et al. (2017) Harnessing microbiome-based biotechnologies for sustainable mitigation of nitrous oxide emissions. Microb. Biotechnol. 10, 1226–1231.
| Harnessing microbiome-based biotechnologies for sustainable mitigation of nitrous oxide emissions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhsFOntLvF&md5=a842bcd5688ac66dc2b3cc3a6238f928CAS |
[9] Shi, X. et al. (2016) Effects of the nitrification inhibitor 3,4-dimethylpyrazole phosphate on nitrification and nitrifiers in two contrasting agricultural soils. Appl. Environ. Microbiol. 82, 5236–5248.
| Effects of the nitrification inhibitor 3,4-dimethylpyrazole phosphate on nitrification and nitrifiers in two contrasting agricultural soils.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XitVahurvN&md5=6a15180298b7f54c308a0cbbd0d3ff79CAS |
[10] Shi, X.Z. et al. (2017) Response of ammonia oxidizers and denitrifiers to repeated applications of a nitrification inhibitor and a urease inhibitor in two pasture soils. J. Soils Sediments 17, 974–984.
| Response of ammonia oxidizers and denitrifiers to repeated applications of a nitrification inhibitor and a urease inhibitor in two pasture soils.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhvVyns73I&md5=f8a1a162cda396dbcf35bb1a58acee0aCAS |
[11] Fisk, L.M. et al. (2015) Root exudate carbon mitigates nitrogen loss in a semi-arid soil. Soil Biol. Biochem. 88, 380–389.
| Root exudate carbon mitigates nitrogen loss in a semi-arid soil.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtVyiurnN&md5=8b629fa03e023cf18df6153533bd0f5bCAS |
[12] Lam, S.K. et al. (2017) Using nitrification inhibitors to mitigate agricultural N2O emission: a double-edged sword? Glob. Chang. Biol. 23, 485–489.
| Using nitrification inhibitors to mitigate agricultural N2O emission: a double-edged sword?Crossref | GoogleScholarGoogle Scholar |
[13] Singh, B.K. and Trivedi, P. (2017) Microbiome and the future for food and nutrient security. Microb. Biotechnol. 10, 50–53.
| Microbiome and the future for food and nutrient security.Crossref | GoogleScholarGoogle Scholar |
[14] Leach, J.E. et al. (2017) Communication in the phytobiome. Cell 169, 587–596.
| Communication in the phytobiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXnt1Cmsb8%3D&md5=e1afc132f5dfe69ee7c9fcffbcfd71aaCAS |
[15] Subbarao, G.V. et al. (2009) Evidence for biological nitrification inhibition in Brachiaria pastures. Proc. Natl. Acad. Sci. USA 106, 17302–17307.
| Evidence for biological nitrification inhibition in Brachiaria pastures.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhs1WktrjE&md5=ba98009760b893f13c7f5f972201acf3CAS |
[16] Bardon, C. et al. (2014) Evidence for biological denitrification inhibition (BDI) by plant secondary metabolites. New Phytol. 204, 620–630.
| Evidence for biological denitrification inhibition (BDI) by plant secondary metabolites.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhslOjs7jO&md5=a2f2a68a49d255a2282a1c312c640735CAS |
[17] Itakura, M. et al. (2013) Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation. Nat. Clim. Change 3, 208–212.
| Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjtVajsr8%3D&md5=b2b5887b1438e68023f15f5101f69c49CAS |
[18] Gao, N. et al. (2017) Nitrous oxide (N2O)-reducing denitrifier-inoculated organic fertilizer mitigates N2O emissions from agricultural soils. Biol. Fertil. Soils 53, 885–898.
| Nitrous oxide (N2O)-reducing denitrifier-inoculated organic fertilizer mitigates N2O emissions from agricultural soils.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXht1yltL%2FM&md5=373202f519ac722361df221995203767CAS |
[19] Sheth, R.U. et al. (2016) Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200.
| Manipulating bacterial communities by in situ microbiome engineering.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XitlSgtbw%3D&md5=5c627b40e940984b0ae0fd748c736b4dCAS |
[20] Alivisatos, A.P. et al. (2015) A unified initiative to harness Earth’s microbiomes. Science 350, 507–508.
| A unified initiative to harness Earth’s microbiomes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhsl2nsL%2FM&md5=017c0a759b18ee933d6dcb83da560ddbCAS |
[21] Mellbye, B.L. et al. (2016) Quorum quenching of Nitrobacter winogradskyi suggests that quorum sensing regulates fluxes of nitrogen oxide(s) during nitrification. MBio 7, e01753–16.
| Quorum quenching of Nitrobacter winogradskyi suggests that quorum sensing regulates fluxes of nitrogen oxide(s) during nitrification.Crossref | GoogleScholarGoogle Scholar |
[22] Citorik, R.J. et al. (2014) Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145.
| Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsFyhsrjP&md5=0f1eb87f8b3f5893d9f0675053886990CAS |
[23] Mueller, U.G. and Sachs, J.L. (2015) Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617.
| Engineering microbiomes to improve plant and animal health.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhsFKhtL7L&md5=85771a94a2e5267c5c2b377ec5c9d1a5CAS |
Biographies
Dr Hangwei Hu is an ARC DECRA Fellow in the School of Agriculture and Food, Faculty of Veterinary and Agricultural Sciences at The University of Melbourne. His research is focused on soil nitrogen cycling processes, environmental microbiomes, and soil antibiotic resistance genes.
Dr Jizheng He is Professor of Soil Ecology at the University of Melbourne. His research interests focus on the soil microbial biogeography, microbial processes of carbon and nitrogen cycles, soil fertility and nutrients management in agricultural ecosystems.