Understanding microbiomes through trait-based ecology
Jennifer L Wood A B and Ashley E Franks A CA Applied Environmental Microbiology, Department of Physiology, La Trobe University, Bundoora, Vic. 3086, Australia
B Email: Jen.Wiltshire@latrobe.edu.au
C Email: a.franks@latrobe.edu.au
Microbiology Australia 39(1) 53-56 https://doi.org/10.1071/MA18014
Published: 16 February 2018
Ecology is the study of the interactions amongst organisms and their environment1. In microbial ecology, a major goal is to understand how environmental microbiomes impact ecosystem health and function. This desire to mechanistically link micro and macro processes is increasingly highlighting the importance of functional ecology, which aims to develop an understanding of relationships using functional traits, as opposed to species names. A functional trait may be any morphological or physiological trait that influences the performance or fitness of an individual in a given environment, such as regeneration time, size, antibiotic production or motility2. Although it is not possible to measure a given trait for each individual within an environmental microbiome, community-level functional traits can be derived from the community metagenome either directly via shotgun sequencing or predictively (for bacteria) from 16S rRNA profiles3. In understanding environmental microbiomes, functional traits have unique properties that can be utilised to (1) compare microbiomes using an ecological framework, (2) understand processes governing community assembly, and (3) build predictive ecological models.
Functional comparisons of environmental microbiomes
Functional traits are not necessarily conserved across phylogenetically closely-related taxa, but rather are conserved amongst organisms with similar life strategies. As such, trait-based comparisons of environmental microbiomes can be used to elucidate repeated ecological patterns across microbiomes even if they are taxonomically distinct. For example, if one was attempting to understand ecological similarities amongst geographically dispersed microbiomes from comparable ecosystems (e.g. wetland microbiomes), one may find that the communities contain vastly different suites of species. In this instance species names alone cannot be used to identify ecological trends uniting these microbiomes. However, because the microbiomes are from similar ecosystems, it is likely that within each microbiome different species will employ similar life strategies to survive and thus exhibit similar functional traits. In this way, functional traits can identify meaningful ecological patterns across taxonomically distinct microbiomes.
Linking micro to macro: understanding of processes governing community assembly
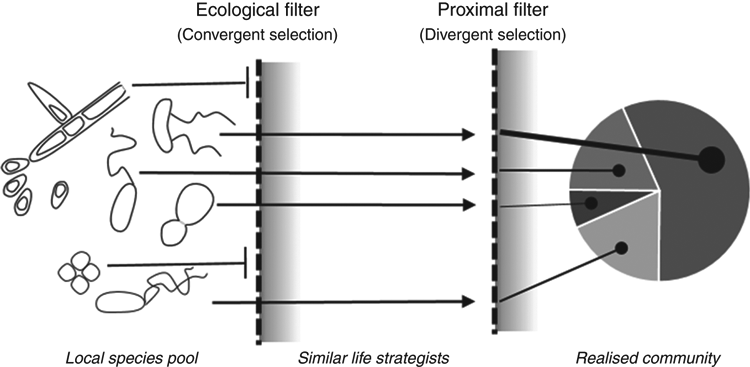
The concept that environmental filters act on traits – not species – can be used to interpret how environmental parameters alter microbiomes in an ecologically meaningful way. The twin-filter hypothesis proposes that a two-step filtering process acts on local species pools: a primary ‘ecological filter’ increases the trait similarity within a community by selecting for similar life strategists (i.e. environments characterised by severe nutrient stress will select for traits that produce stress-tolerant life strategists); secondary ‘proximal filters’ then select against traits which affect survival but are not integral to the broad life strategy (e.g. variation in tolerance to environmental toxins or resistance to local pathogens and predators), creating dissimilarity within the local subset of species and generating the final community structure (Figure 1)4. By examining which traits are enriched by a given environment, or environmental parameter, we can begin to hypothesise how a community is experiencing that environment and why different communities diverge in their ecology. For example, Wood et al.5 demonstrated that the community-level changes induced by the presence of a plant rhizosphere were due to the selection of traits linked to microbial competition for resources (i.e. antibiotic production, siderophore production). Similarly, DeLong et al.6 demonstrated the presence of contrasting ecologies between communities from the phototrophic zone and from near-ocean floor depths, with foraging traits selected for in the phototrophic zone, whilst survival (stress tolerance) traits were characteristic of communities at depth.
|
Building trait-based predictive models
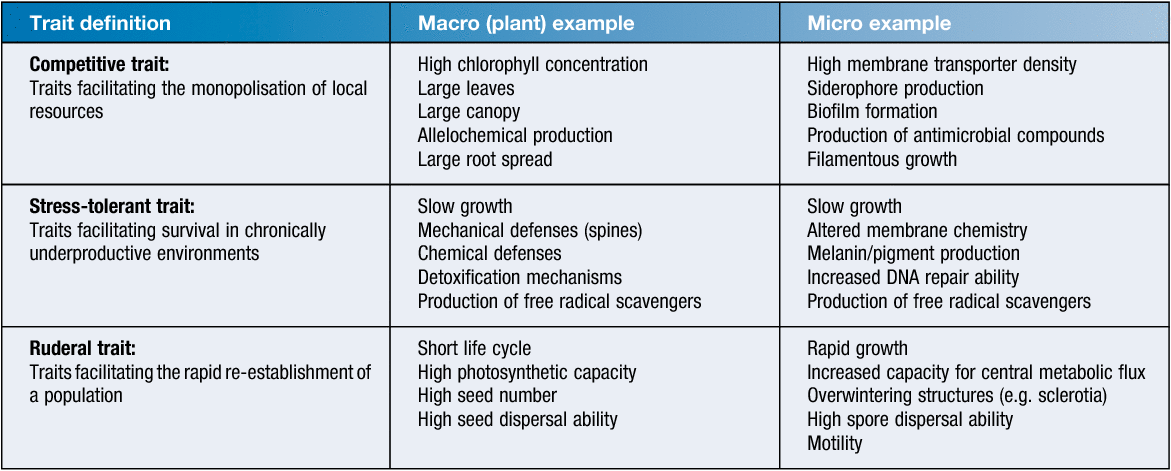
Functional traits can be incorporated into ecological classification frameworks which aim to predict how environmental microbiomes will change over time. Grime’s CSR theory is an ecological classification framework that groups traits in terms of three broad life strategies: competitive, stress tolerance and ruderal (colonisation) life strategies6. Each life strategy group is an umbrella term that encompasses multiple functional traits which achieve the same outcome (Table 1). For example, stress tolerance traits may be defined as any trait that constitutes an investment in the maintenance of organismal biomass. In plants this may be the production of thorns or chemical compounds to deter herbivory. In an environmental microbiome this may manifest as an increase in the prevalence of DNA repair pathways or genes involved in the production of free radical scavengers.
|
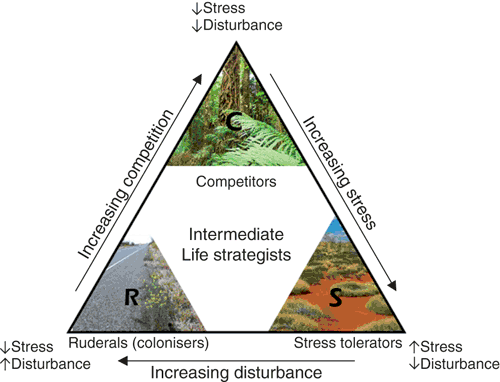
The CSR theory proposes that organisms face a three-way resource trade-off between the investment in C, S or R life strategies, which is governed by the levels of stress (due to resource availability) and disturbance present in an environment4. The theory predicts that when stress and disturbance are minimal, the investment of resources into competitive traits confers a selective advantage that outweighs the loss in fitness due to reduced investment in other adaptive strategies, such as stress-tolerance or colonisation potential (Figure 2). In these communities, competitive interactions – and organisms with traits that contribute to a competitive life strategy – will prevail. Conversely, the theory predicts that stress-tolerance traits and life strategies will prevail when stress is high (i.e. resources are limited) but disturbance is low. Stress tolerant life strategists tend to be slow growing and are adapted to retaining resources. Finally, when stress is low but disturbance is high the theory predicts that ruderal traits, which pertain to re-colonisation potential, will confer a selective advantage and ruderal life strategists will prevail.
|
The use of ecological theories, such as the CSR hypothesis, presents a clear route towards developing predictive models which could be incorporated into ecosystem-level conservation and management practices. Indeed, even though CSR theory has its roots in plant ecology, the core principals are recognised as being applicable to microbial communities7–9. A current barrier to developing predictive trait-based models is that ecological interpretations of microbial traits often rely on prevalent opinions from the literature, rather than on empirical data. For example, the production of antimicrobial metabolites is generally considered to be a competitive trait10. However, compounds recognised for their antibiotic activity in vitro have been shown to influence biofilm formation in Bacillus subtilis suggesting their primary role may be cell–cell communication11.
Future research using controlled microcosms with defined gradients of resource availability (stress) and disturbance are needed to confirm ecological assumptions about functional traits. Trait screening using controlled conditions can also be used to identify core predictor traits that can routinely and robustly discriminate between environmental microbiomes with contrasting ecologies. Ultimately, the development of broad ecological theories that facilitate the classification and comparison of microbiomes from disparate environments will assist in realising the full potential of large-scale collaborative initiatives, such as the TerraGenome project12, the Earth Microbiome project (EMP)13 and Australia’s Biomes of Australian Soil Environments (BASE)14 project.
References
[1] Ganong, W.F. (1904) The cardinal principles of ecology. Science 19, 493–498.| The cardinal principles of ecology.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cvgvVenug%3D%3D&md5=9c1828c2fb92e6df37bcdb74752eb93eCAS |
[2] McGill, B.J. et al. (2006) Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185.
| Rebuilding community ecology from functional traits.Crossref | GoogleScholarGoogle Scholar |
[3] Fierer, N. et al. (2014) Seeing the forest for the genes: using metagenomics to infer the aggregated traits of microbial communities. Front. Microbiol. 5, .
| Seeing the forest for the genes: using metagenomics to infer the aggregated traits of microbial communities.Crossref | GoogleScholarGoogle Scholar |
[4] Grime, J.P. and Pierce, S. (2012) The Evolutionary Strategies that Shape Ecosystems. (Wiley-Blackwell)
[5] Wood, J.L. et al. Competitive traits are more important than stress tolerant traits in a cadmium-contaminated rhizosphere: a role for trait theory in microbial ecology. Front. Microbiol., In press.
[6] DeLong, E.F. et al. (2006) Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503.
| Community genomics among stratified microbial assemblages in the ocean’s interior.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XmvVymuw%3D%3D&md5=62683d260ce0ec9f0cc630c7b5e4401dCAS |
[7] Prosser, J.I. et al. (2007) The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392.
| The role of ecological theory in microbial ecology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkt1Chtr8%3D&md5=00c6af4a0be667db4f5c907d38835f76CAS |
[8] Fierer, N. (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590.
| Embracing the unknown: disentangling the complexities of the soil microbiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhtlGktbfN&md5=b79aeb90c64d09f110fd78ab6a3d9586CAS |
[9] Krause, S. et al. (2014) Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251.
| Trait-based approaches for understanding microbial biodiversity and ecosystem functioning.Crossref | GoogleScholarGoogle Scholar |
[10] Hibbing, M.E. et al. (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25.
| Bacterial competition: surviving and thriving in the microbial jungle.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVylsLvO&md5=d76d25910ce465e70b975a8de3578a60CAS |
[11] López, D. et al. (2009) Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 106, 280–285.
| Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis.Crossref | GoogleScholarGoogle Scholar |
[12] Vogel, T.M. et al. (2009) TerraGenome: a consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 7, 252.
| TerraGenome: a consortium for the sequencing of a soil metagenome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXjtFCisLg%3D&md5=3effd8a568499385db80a488078277bcCAS |
[13] Gilbert, J.A. et al. (2014) The Earth Microbiome project: successes and aspirations. BMC Biol. 12, .
| The Earth Microbiome project: successes and aspirations.Crossref | GoogleScholarGoogle Scholar |
[14] Bissett, A. et al. (2016) Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. Gigascience 5, 21.
| Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database.Crossref | GoogleScholarGoogle Scholar |
Biographies
Dr Jennifer L Wood is post-doctoral fellow in the Applied and Environmental Microbiology Laboratory at La Trobe University. She has recently completed her doctoral research which investigated the utility of functional approaches in understanding soil microbiomes for which she was awarded the ‘Nancy Millis award for theses of exceptional merit’. Her broad research interest is in developing trait-based approaches to interpret and predict how environmental and anthropogenic changes impact microbial community ecology and function. Currently she is working with the Defence Science Technology Group investigating functional changes associated with microbiologically influenced corrosion in marine environments.
Associate Professor Ashley E Franks is head of the Applied and Environmental Microbiology Laboratory at La Trobe University. He conducted his doctorate research as part of the Centre of Marine Biofouling and Bioinnovation at the University of New South Wales by investigating antifungal compounds produced by marine bacteria in biofilms. During his PhD he spent 4 months at the University of Exeter in the UK on an Adrian Lee Fellowship to develop dual bacterial/yeast biofilm systems. On graduating he moved to the Biomerit Research Centre in Cork, Ireland to work on bacterial plant interactions as a Government of Ireland Fellow in Science Technology and Engineering. This research looked at how to use bacteria to help plant growth. He then took a position as a Senior Scientist and Research Professor within the Geobacter Project at the University of Massachusetts Amherst in the USA where he worked on microbes that make electricity. He currently serves as the Chair of the Awards Committee for the International Society of Microbial Electrochemical Technology and was previously the Secretary of Synthetic Biology Australasia.


