Viral fossils in marsupial genomes: secret cellular guardians
Emma F Harding A B , Grace JH Yan A and Peter A White A CA School of Biotechnology and Biomolecular Sciences, University of New South Wales
B Email: e.harding@unsw.edu.au
C Email: p.white@unsw.edu.au
Microbiology Australia 42(3) 134-137 https://doi.org/10.1071/MA21036
Submitted: 16 August 2021 Accepted: 25 August 2021 Published: 6 September 2021
Journal Compilation © The Authors 2021 Open Access CC BY-NC-ND, published (by CSIRO Publishing) on behalf of the ASM
Abstract
Genomic viral integrations, termed endogenous viral elements (EVEs), are fragments of viruses in host chromosomes that provide information about viral evolution and could even help protect the host from infection. In the present study we examined EVEs in thirteen different Australian marsupial species to identify trends in their integration, commonality and to investigate their possible cellular function. We found that marsupial EVEs are commonly derived from viruses of the Bornaviridae, Filoviridae and Parvoviridae families, and circulated up to 160 million years ago. We also show the EVEs are actively transcribed into both long and short RNA molecules in marsupials, and propose they are involved in a cellular defence mechanism to protect the germline from viral genomic invasion.
Every time you get infected with a virus, there is a chance it will leave some of its genetic material behind in your DNA. This happens commonly during infection with retroviruses, like human immunodeficiency virus (HIV), which have a reverse transcriptase and integrase enzymes that allows them to integrate into our chromosomes. Other viruses, including filoviruses like Ebola and bornaviruses like Borna disease virus, can also be integrated into chromosomes by host retrotransposons1. If these integrations occur in germline cells, they are inherited by progeny and can spread through a population. These viral remnants, also known as endogenous viral elements (EVEs), are in every known vertebrate and invertebrate and provide an extensive history of viral evolution2,3.
Few functions have been proposed for EVEs, with the common consensus that they are accidental integrations which degrade through slow mutagenesis over time. EVEs from structural viral genes rapidly lose their coding ability through negative selection to avoid immunological stimulation due to the production of viral capsids or envelopes. However, some EVEs are preserved and exapted for a new cellular function. For example, the syncytin gene, integral to placenta development, is a captured retroviral envelope gene from an ancient integration4. Similarly, a nucleoprotein gene from a bornavirus is translated into a protein in an American squirrel to impede viral infection5.
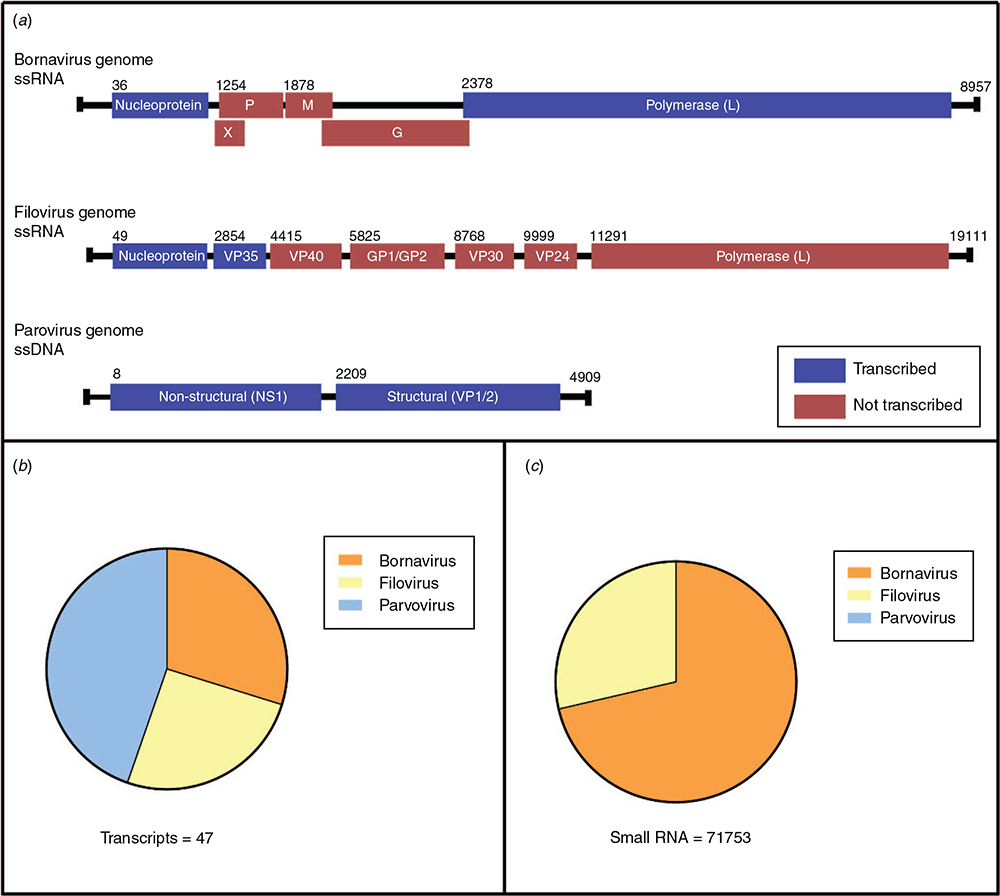
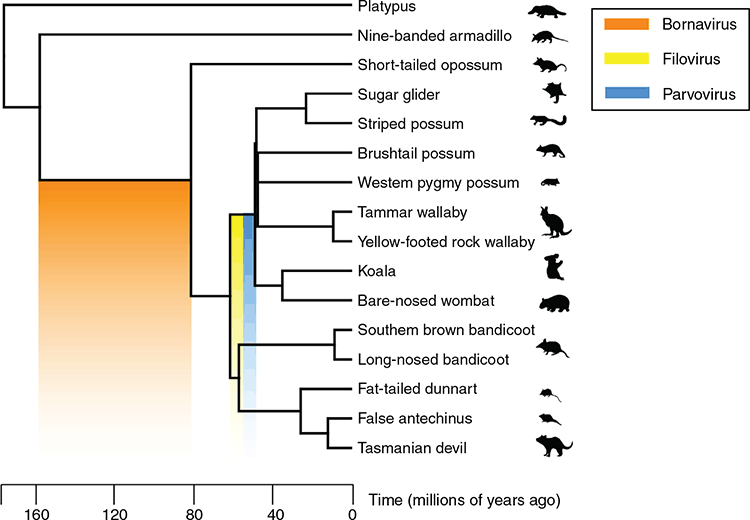
Our study conducted at UNSW Sydney by PhD students Emma Harding, Grace Yan and Alice Russo, and led by Professor Peter White, investigated EVEs in Australian marsupials6. With the exception of rodents, marsupials dominate the mammalian fauna of Australia, in contrast to every other terrestrial ecosystem. We looked at 13 different species – the Tasmanian devil, koala, bare-nosed wombat, sugar glider, long-nosed bandicoot, Southern brown bandicoot, Western pygmy possum, brushtail possum, striped possum, tammar wallaby, fat-tailed dunnart, false antechinus and yellow-footed rock wallaby (Figure 1). The study examined EVEs to trace virus evolution and investigated if EVEs served any functions in marsupial cells.
RNA-Sequencing datasets were screened for the presence of EVE-derived transcripts – RNA transcripts with identity to viral genes which were endogenous in origin. Transcripts were classified as endogenous if they mapped back to the representative marsupial genome (where available), contained premature stop codons or were substantially shorter than the viral counterpart (<50% length).
All viral families were searched for, however only Filoviridae, Bornaviridae, Parvoviridae and Retroviridae EVE transcripts were identified. Whilst many viruses commonly integrate into host DNA during infection, only the small percentage that integrate into germline cells are passed on to progeny. Subsequently, only a small percentage of those become fixated in a population due to selection pressures, leading to the loss of many endogenized sequences in a short space of time. EVEs that successfully ‘survive’ this infancy and become fixated can endure within the genomes of a species for millions of years.
EVEs from Bornaviridae, Filoviridae and Parvoviridae viruses were ubiquitous throughout all of the animals sampled (Figure 2a). These viral families have been shown previously to be endogenised and are seen in almost every lineage of eutherian mammals. Bornaviridae EVEs, in particular, are ubiquitous in vertebrates and are estimated to have been infecting animals for at least 100 million years. These viral families, two of which are RNA and one of which is DNA, have never been sequenced from marsupials, however the widespread presence of EVEs indicates a shared history. The EVEs are extremely divergent from any extant eutherian counterparts – their closest relative viruses are avian and reptilian-infecting rather than those that infect mammals like us.
In addition to their presence in marsupial chromosomes, many EVEs are actively transcribed into RNA (Figure 2b). While transcription is a likely key step for performing a cellular function, the EVEs in marsupials were not being translated into protein, or if they are, the proteins are very short. Each EVE transcript was riddled with stop codons, making the likelihood of its translation into anything functional very low. Instead, we set our sights on their role as non-coding RNA.
Non-coding RNA is present in every cell and contributes to many vital cellular functions, including transcriptional regulation, development and RNA defence. In plants and invertebrates, the primary immune defence against viruses is a system of non-coding small RNA molecules involved in the RNA interference system7–9. We considered, instead of being translated into an antiviral protein, that perhaps the EVE transcripts were being spliced into small RNA to bind and destroy incoming target viral nucleic acid.
To investigate this hypothesis, we looked at small RNA sequencing data from the koala. We found that EVEs were indeed giving rise to small RNA molecules – both small interfering RNA (siRNA) and P-element-induced wimpy testis protein-interacting RNA (piRNA) – associated with antiviral defence in invertebrates (Figure 2c)10. In addition, these molecules were enriched in the testis tissue – a prime location to protect gametes and hence protect offspring from genetic invasion by viruses. This suggests the tantalising possibility of this RNA defence system, previously thought to be abandoned in mammals in favour of the interferon system, still being active and protecting marsupial cells.
Of interest was the common trend that only two types of EVEs were contributing to the small RNA produced in the testis; bornavirus polymerase and filovirus nucleoprotein EVEs. Both Bornaviridae and Filoviridae viruses are single stranded, negative sense RNA viruses with genomes 7–10 kb, encoding 5–8 proteins (Figure 2a). Of these proteins, two were consistently represented in RNA sequencing data – the nucleocapsid, or shell, and the polymerase or replication gene. Bornavirus and filovirus nucleoprotein EVEs are commonly found throughout mammals; however, the bornavirus polymerase is much rarer – bats are the only other mammalian group with this type of EVE3. In contrast to trends in other mammals, marsupials had high amounts of RNA derived from these bornavirus polymerase EVEs, and the small RNA was predominantly from these. One theory suggests the nucleocapsid and polymerase proteins are prime targets for negative sense ssRNA viruses11. In these viruses, the nucleocapsid encapsulates the other viral proteins, and any degradation of this protein will lead to the exposure of pathogen-associated molecular patterns (PAMPs) and inhibition of viral replication12. Following this, it would make sense that these EVEs from these two viral genes are retained in host chromosomes to act as an effective antiviral defence against invasion from similar negative sense RNA viruses.
For additional evidence that this was a conserved protective system, we looked for clues on how old these EVEs were. By looking for orthologous integrations – EVEs in the same genomic loci – in different marsupials, we were able to estimate the age of each EVE that was making RNA transcripts. Most of the transcriptionally active EVEs were estimated to have integrated into marsupial genomes ~40 million years ago, however one bornavirus polymerase EVE was aged between 80 and 160 million years old (Figure 3). This EVE was present in all Australian marsupial genomes, as well as the American opossum genome, indicating it was an ancient virus that circulated during the time of the dinosaurs. We believe the conservation of these EVEs is no accident.
Whilst our study uncovered some very interesting observations, it is far from explaining their occurrence. It has led to many new questions, including why these three viral families (Bornaviridae, Filoviridae, Parvoviridae) are so common in marsupial genomes, why they are still being transcribed and if they do act to protect germline cells. If they protect marsupials, could they hint at a defensive system conserved throughout mammals? Could similar integrations in our DNA be protecting us from infection?
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This research did not receive any specific funding.
References
[1] Belyi, V.A. et al. (2010) Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6, e1001030.| Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes.Crossref | GoogleScholarGoogle Scholar | 20686665PubMed |
[2] Holmes, E.C. (2011) The evolution of endogenous viral elements. Cell Host Microbe 10, 368–377.
| The evolution of endogenous viral elements.Crossref | GoogleScholarGoogle Scholar | 22018237PubMed |
[3] Kawasaki, J. et al. (2021) 100-My history of bornavirus infections hidden in vertebrate genomes. Proc. Natl. Acad. Sci. USA 118, e2026235118.
| 100-My history of bornavirus infections hidden in vertebrate genomes.Crossref | GoogleScholarGoogle Scholar | 33990470PubMed |
[4] Fujino, K. et al. (2014) Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc. Natl. Acad. Sci. USA 111, 13175–13180.
| Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome.Crossref | GoogleScholarGoogle Scholar | 25157155PubMed |
[5] Harding, E. et al. (2021) Ancient viral integrations in marsupials: a potential antiviral defence. Virus Evol. , veab076.
| Ancient viral integrations in marsupials: a potential antiviral defence.Crossref | GoogleScholarGoogle Scholar |
[6] Johnson, W.E. (2019) Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17, 355–370.
| Origins and evolutionary consequences of ancient endogenous retroviruses.Crossref | GoogleScholarGoogle Scholar | 30962577PubMed |
[7] Blair, C.D. (2011) Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6, 265–277.
| Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission.Crossref | GoogleScholarGoogle Scholar | 21449839PubMed |
[8] Schuster, S. et al. (2019) Antiviral RNAi in insects and mammals: parallels and differences. Viruses 11, 448.
| Antiviral RNAi in insects and mammals: parallels and differences.Crossref | GoogleScholarGoogle Scholar |
[9] ter Horst, A.M. et al. (2019) Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWi-interacting RNAs. J. Virol. 93, e02124-18.
| Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWi-interacting RNAs.Crossref | GoogleScholarGoogle Scholar | 30567990PubMed |
[10] Ophinni, Y. et al. (2019) piRNA-guided CRISPR-like immunity in eukaryotes. Trends Immunol. 40, 998–1010.
| piRNA-guided CRISPR-like immunity in eukaryotes.Crossref | GoogleScholarGoogle Scholar | 31679813PubMed |
[11] Teng, D. et al. (2019) A small interfering RNA cocktail targeting the nucleoprotein and large protein genes suppresses borna disease virus infection. Front. Microbiol. 10, 2781.
| A small interfering RNA cocktail targeting the nucleoprotein and large protein genes suppresses borna disease virus infection.Crossref | GoogleScholarGoogle Scholar | 31849913PubMed |
[12] Blanco-Melo D.et al2020 An inability to maintain the ribonucleoprotein genomic structure is responsible for host detection of negative-sense RNA viruses.bioRxiv 2020.2003.2012.989319
Biographies
Emma Harding is a PhD candidate in Professor Peter White’s laboratory in the School of Biotechnology and Biomolecular Sciences at UNSW Sydney. Her research investigates endogenous viruses in mammalian DNA, their evolution and possible cellular functions.
Grace Yan is a PhD candidate under the supervision of Professor Peter White at the School of Biotechnology and Biomolecular Sciences, UNSW Sydney. Her research combines the use of clinical and sewage samples to study the molecular epidemiology of viral gastroenteritis.
Peter White is a Professor in Microbiology at the School of Biotechnology and Biomolecular Sciences at UNSW. His research interests are currently viral gastroenteritis, viral discovery and evolution, and the development of antiviral agents.