Aquatic animals; endemic and exotic bacterial pathogens
Nicky BullerAnimal Health Laboratories
Department of Agriculture and Food Western Australia
3 Baron-Hay Court, South Perth
WA 6151, Australia
Tel: +61 8 9368 3425
Fax: +61 8 9474 1881
Email:
Nicky.Buller@agric.wa.gov.au
Microbiology Australia 34(1) 22-24 https://doi.org/10.1071/MA13007
Published: 20 March 2013
The detection and identification of bacterial pathogens from fish and other aquatic animals presents particular challenges with regards to transportation of samples to the laboratory, knowledge of media and growth requirements, and detection of endemic or exotic pathogens. This article highlights the challenges in this specialised area of microbiology with a focus on an endemic and an exotic disease.
Bacteria from marine samples such as kingfish, prawn or seahorse, require media containing NaCl, but in addition to this some bacteria require magnesium and potassium ions. Bacteria from marine mammals such as whales and dolphins may need to be grown on media with and without salt as some organisms that are salt-requiring will adapt to normal physiological conditions in the marine mammal. In these cases initial growth may occur on media without salt, but for subsequent identification tests, NaCl must be added to the inoculum, as some reactions will give a false-negative result without NaCl despite the bacterium adapting to host conditions1.
An incubation temperature of 25°C is generally used for aquatic samples, however some bacteria isolated from cold-water fish may have an optimal growth temperature of 18°C, whereas samples from marine mammals may grow best at 37°C. The best option is to incubate duplicate plates at both 25 and 37°C.
An endemic disease: the challenges of Tenacibaculummaritimum
Often fish farms are located huge distances from a laboratory and samples may not arrive in optimal condition to ensure isolation of the pathogen. An example is a fish farm in the far north of WA; a six hour boat trip to the nearest town with flights to Perth, means samples may take three days to reach the laboratory. When trying to isolate a slow-growing bacterium such as Tenacibaculummaritimum, sometimes the best option is inoculate culture plates at the farm (Figs 1, 2).

|

|
Tenacibaculum maritimum is the causative agent of tenacibaculosis, an ulcerative disease that affects a variety of marine fish species. Clinical signs include ulcers and necrosis on the body surface, eroded mouth, frayed fins and tail rot (Fig. 3)2.

|
Tenacibaculum maritimum is an aerobic, oxidase positive, Gram-negative, slender, flexible rod 0.5 × 2 µm to 30 µm with an occasional cell of 100 µm in length. Cells have gliding motility on solid media3. Growth occurs on a low nutrient medium such as Anacker-Ordals medium supplemented with seawater or artificial sea salts (Fig. 4). Growth does not occur when supplemented with NaCl only, or on blood agar. Growth usually occurs on the commercially available MA 2216 agar (Difco)2.
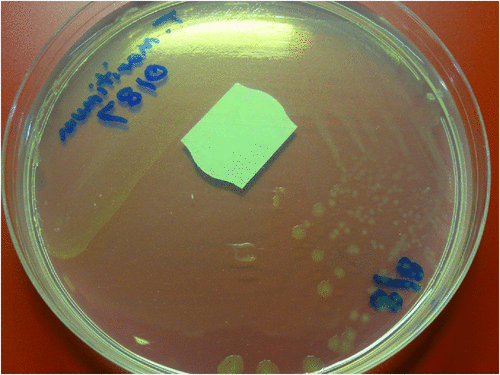
|
Tenacibaculum maritimum does not ferment carbohydrates, is positive for congo red absorption and negative for production of flexirubin pigments as detected by KOH2. Conventional and real time PCRs have been reported for the detection and identification of T. maritimum4,5.
An exotic disease: the not so innocent Nemo
The ornamental or aquarium fish trade in Australia is worth $350 M annually with 15.5 million fish being imported in 2005, and these imported fish are a potential source of exotic pathogens with potential to compromise Australian freshwater and marine aquaculture industries6.
Fish imported into Australia are subject to Australian Quarantine and Inspection Service (AQIS) conditions and on arrival are held in an AQIS-approved quarantine premise for seven days, and if an exotic disease is detected, the consignment is destroyed. In Western Australia (WA), any fish showing signs of disease are sent to the Fish Health Unit, Department of Fisheries WA, and bacteriology is conducted at the Animal Health Laboratories, Department of Agriculture WA.
Edwardsiellaictaluri is the causative agent of enteric septicaemia of catfish (ESC). It is an important infectious disease in the commercial catfish industry in a number of countries including the USA, China, Thailand and Vietnam7,8. E. ictaluri is a potential pathogen of some non-ictalurids, as in experimental challenge of chinook salmon and rainbow trout mortality occurred at high doses9.
E. ictaluri is exotic to Australia and a nationally notifiable disease. Since 1980 the organism has been reported a number of times from imported ornamental fish held in quarantine10, and from quarantine facilities in three states in the past two years. Thus, it is important for Australian laboratories to be vigilant and to have the expertise to detect and identify this organism. A recent detection occurred in imported yoyo loach, so named for the markings on the body (Fig. 5). The remaining fish were destroyed to maintain Australia’s disease-free status for this pathogen.

|
E. ictaluri is an oxidase-negative, Gram-negative rod to coccobacillus. Colonies on blood agar at 48 hours after incubation at 25–30°C are 1–2 mm, round, circular, slightly irregular and pale grey with a butyrous consistency (Fig. 6). It is Strep-like as a greenish-tinge may be seen around the colony and a weak beta-haemolysis underneath the colony. Growth occurs on nutrient agar, MacConkey agar and Salmonella-Shigella agar. Strains are motile at 25°C but non-motile at 37°C. E. ictaluri is not in the API 20E database and results will often give a percentage probability of E. tarda, a common pathogen. API 20E results for E. ictaluri have been reported as 400400057, 410400057 and 410414057 from catfish and rainbow trout isolates1,8. E. ictaluri is differentiated from E. tarda by being negative for indole and H2S, whereas results are positive for E. tarda. As with all exotic pathogens, the identification is confirmed by the Australian Animal Health Laboratories at Geelong, Victoria.
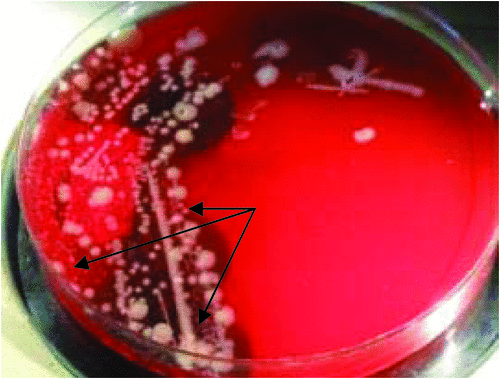
|
A PCR has been reported for the specific detection of E. ictaluri11.
References
[1] Buller, N.B. (2004) Bacteria from Fish and Other Aquatic Animals: a practical identification manual, CABI.[2] Bernardet, J.-F. et al. (1994) Comparative study on Flexibacter maritimus strains isolated from farmed sea bass (Dicentrarchus labrax) in France. Fish Pathol. 29, 105–111.
| Comparative study on Flexibacter maritimus strains isolated from farmed sea bass (Dicentrarchus labrax) in France.Crossref | GoogleScholarGoogle Scholar |
[3] Wakabayashi, H. et al. (1986) Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int. J. Syst. Bacteriol. 36, 396–398.
| Flexibacter maritimus sp. nov., a pathogen of marine fishes.Crossref | GoogleScholarGoogle Scholar |
[4] Bader, J.A. and Shotts, E.B. (1998) Identification of Flavobacterium and Flexibacter species by species-specific polymerase chain reaction primers to the 16S ribosomal RNA gene. J. Aquat. Anim. Health 10, 311–319.
| Identification of Flavobacterium and Flexibacter species by species-specific polymerase chain reaction primers to the 16S ribosomal RNA gene.Crossref | GoogleScholarGoogle Scholar |
[5] Fringuelli, E. et al. (2012) Development of a quantitative real-time PCR for the detection of Tenacibaculum maritimum and its application to field samples. J. Fish Dis. 35, 579–590.
| Development of a quantitative real-time PCR for the detection of Tenacibaculum maritimum and its application to field samples.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFCrsb7L&md5=d171cdacfb4964ec03215a8f8d9e68b9CAS |
[6] Anon. (2006) A strategic approach to the management of ornamental fish in Australia, Natural Resource Management Ministerial Council. Department of Agriculture Fisheries and Forestry.
[7] Hawke, J.P. et al. (1981) Edwardsiella ictaluri sp. nov., the causative agent of enteric septicaemia of catfish. Int. J. Syst. Bacteriol. 31, 396–400.
| Edwardsiella ictaluri sp. nov., the causative agent of enteric septicaemia of catfish.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3MXmtVSns7g%3D&md5=5e35d758544311362be824ecb32c198aCAS |
[8] Crumlish, M. et al. (2002) Identification of Edwardsiella ictaluri from diseased catfish, Pangasius hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam. J. Fish Dis. 25, 733–736.
| Identification of Edwardsiella ictaluri from diseased catfish, Pangasius hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam.Crossref | GoogleScholarGoogle Scholar |
[9] Baxa, D.V. et al. (1990) Susceptibility of nonictalurid fishes to experimental infection with Edwardsiella ictaluri. Dis. Aquat. Organ. 8, 113–117.
| Susceptibility of nonictalurid fishes to experimental infection with Edwardsiella ictaluri.Crossref | GoogleScholarGoogle Scholar |
[10] Humphrey, J.D. et al. (1986) Exotic bacterial pathogens Edwardsiella tarda and Edwardsiella ictaluri from imported ornamental fish Betta splendens and Puntius conchonius, respectively: isolation and quarantine significance. Aust. Vet. J. 63, 369–371.
| Exotic bacterial pathogens Edwardsiella tarda and Edwardsiella ictaluri from imported ornamental fish Betta splendens and Puntius conchonius, respectively: isolation and quarantine significance.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL2s7ls1eqtg%3D%3D&md5=a7fe71f82ab46b917779af395e95c496CAS |
[11] Williams, M.L. and Lawrence, M.L. (2010) Verification of an Edwardsiella ictaluri-specific diagnostic PCR. Lett. Appl. Microbiol. 50, 153–157.
| Verification of an Edwardsiella ictaluri-specific diagnostic PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitVSktbY%3D&md5=9230ce03e624c998382fe5f0ff7de473CAS |
Biography
Dr Nicky Buller is a Senior Microbiologist in the Department of Agriculture and Food Western Australia, and Group Leader of the Bacteriology and Mycobacteriology Sections of the Animal Health Laboratories. She has spent most of her career at AHL in the diagnostic bacteriology laboratory and completed a PhD in molecular epidemiology of Dichelobacternodosus, the causative organism of ovine footrot. She has a particular interest in the bacteriology of fish and aquatic animals and is the author of Bacteria From Fish and Other Aquatic Animals: A Practical Identification Manual, published in 2004 by CABI, UK.


