Listeria monocytogenes: illuminating adaptation with proteomics
John P Bowman , Rolf E Nilsson , Chawalit Kocharunchitt and Tom RossFood Safety Centre
Tasmanian Institute of Agriculture
University of Tasmania, College Road
Sandy Bay, Tas 7005, Australia
Tel: +61 3 6226 6380
Fax: +61 3 6226 2642
Email: john.bowman@utas.edu.au
Microbiology Australia 34(2) 75-77 https://doi.org/10.1071/MA13026
Published: 13 May 2013
With increased consumption of minimally processed ready-to-eat foods the potential for exposure to Listeria monocytogenes has increased. Thus, there is a need to maintain a balance between food convenience and safety. L. monocytogenes is not a homogenous species; certain strains are more resilient to stressful conditions while others are potentially more virulent. To understand the basis of these differences we are applying proteomics to determine the molecular mechanism of adaptations of L. monocytogenes in food-relevant environments. The goal is to define how this species grows, behaves and survives thus allowing us to fine tune food safety risk management, especially when developing new minimal food processes or considering introduction of unpasteurised food such as raw milk cheeses.
Listeria monocytogenes causes listeriosis, a serious disease primarily acquired by food consumption, and that mainly impacts immunocompromised people, the elderly, and neonates. Listeriosis also occurs in livestock and was originally discovered in animals. Though the prevalence of listeriosis in Australia is relatively low it remains a major concern for the food industry. A case in point is the January 2013 brie and camembert cheese-associated outbreak that occurred in south-east Australia, associated with 3 deaths and one miscarriage. L. monocytogenes mainly occurs as an environmental contaminant and can enter food anywhere along the industrial food supply chain including within domestic settings. Despite being relatively nutritionally fastidious this tendency to be a frequent contaminant is due to the species inherent hardiness since it is low water activity and acid tolerant and able to grow at refrigeration temperatures. Fortunately it is readily eliminated by standard pasteurisation or by cooking so that the threat is limited to certain ready-to-eat foods, typically those with long refrigerated shelf-lives.
L. monocytogenes mainly causes disease by invading gastrointestinal epithelial cells by encouraging endocytosis1. Once inside host cells other proteins aid intracellular survival, mobility, and cell-to-cell spread. L. monocytogenes is adept at evading and surviving within cellular and humoral immune systems. A fascinating aspect of L. monocytogenes is its ability to shift from an environmental saprophytic state where virulence genes are turned off to a parasitic state within animal or human hosts2. At 37°C, if carbohydrate levels are low, many virulence genes are activated. This switch is mainly controlled by temperature sensitive small RNAs3. The ability to respond to stress is also intertwined in this transitive process. The many sequenced genomes of L. monocytogenes are rich in transcriptional regulators controlled in overarching regulons by various “master” regulators, which functionally overlap in a complex network. This network allows L. monocytogenes to rapidly respond to changing environments, including switching on and off stress defence and virulence genes4.
Using comprehensive proteomics we are attempting to understand more holistically the mechanistic basis of L. monocytogenes’ adaptation to different situations. State-of the art proteomics is now a very powerful tool and is becoming more cost-effective. Employing gel-free and label-free liquid chromatography (operated in either one or two dimensional modes) and sensitive, high resolution ion trap mass spectrometry, it is possible to take complex protein mixtures digested by a peptidase such as trypsin and identify and quantify peptides en-masse after bioinformatic comparison to proteome databases. This is possible due to better separation of individual peptides and highly accurate mass estimations to error levels of less than 1 part per 2 million. By counting each individual peptide spectrum (a spectral count) one can estimate the protein abundance of most proteins detectable within the proteome of a bacteria. This has major cost and labour advantages over gel-based proteomics. For L. monocytogenes which encodes some 2900–3000 proteins such an approach is very efficient with moderate depth peptide surveys able to detect >40% of its proteome. Though some limitations occur with this approach, such as accurately determining the abundance of inefficiently extracted proteins (e.g. proteins with several transmembrane helical domains) it is still readily possible to generate a large amount of data that can be used comparatively to “dissect” specific genomic functions and phenotypes. Details on the typical LC/MS methodology used has been reviewed5. A number of software- and statistical approaches for assessing protein abundance via spectral counting have been devised that have improved validation of sample comparisons6,7 and also have improved absolute protein quantitation in highly complex samples8.
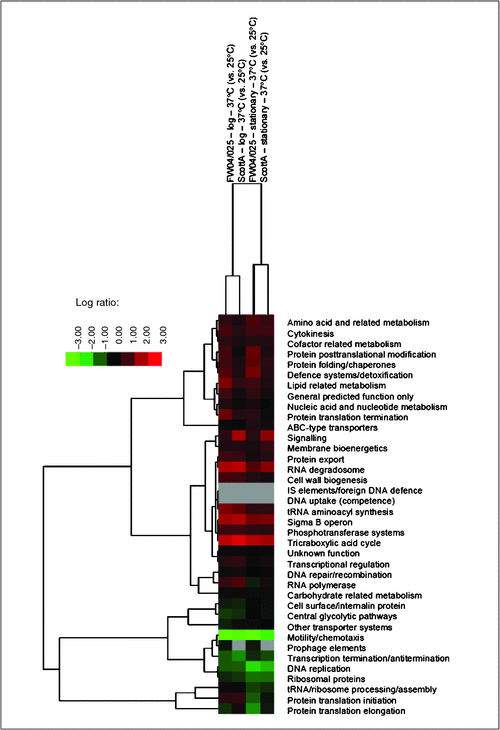
We have used gel-free proteomics to investigate a variety of stresses and phenotypes of L. monocytogenes and E. coli O157:H7 strains, including water activity, cold, acid and alkaline stresses all relevant to either food or the food processing environment9–11. Essentially any prokaryote or eukaryote for which a proteome is available could be studied in a similar fashion across a myriad of scenarios as long as the proteins can be obtained in sufficient quantities. As mentioned above L. monocytogenes pathobiology is strongly controlled by temperature. As an example, we compared the proteomes of genetically different strains at two different temperature, 25°C and 37°C. Cells at these temperatures behave quite differently. At 25°C cells are actively swimming via peritrichous flagella while at 37°C cells lack flagella, become hydrophobic and better attach to surfaces. As can be seen in the accompanying heat map (Figure 1) when the proteins (covering 40-42% of the strain proteomes) are organised on the basis of their essential cellular function large differences between the temperatures occur, above and beyond growth phase- and strain-dependent changes. One of the obvious hallmarks of the temperature effect at 37°C is the suppression of motility and chemotaxis proteins with the most suppressed protein at 37°C being flagellin (reduced >300-fold). The most induced protein (40-60 fold) in both strains at 37°C is an OsmC family protein similar to the organic hydroperoxide resistance protein OhrA of Bacillus subtilis. Organic peroxides are toxic metabolites that accumulate during metabolism12 thus it makes sense that at a more rapid rate of metabolism enhancement of peroxide detoxification is needed. Several other stress response proteins are also enhanced at 37°C including glutamate decarboxylases required for survival against acid shocks, such as gastric passage, as well as superoxide dismutase, which protects against reactive oxygen radicals. Such protein changes likely also contribute to gastrointestinal, intracellular and external environmental survival. Hundreds of other protein changes also occur, including that of many involving uncharacterised proteins. Individual protein abundances within a given proteome differ by 4-5 orders of magnitude and, thus, defining and especially interpreting proteomic-level responses consistent at the species level as well as between strains represent substantial challenges to overcome.
|
In summary, proteomics is rapidly emerging as an accessible approach that can capture large amounts of functionally relevant proteomic, and by inference genomic information. The range of applications within microbiology itself is enormous in terms of understanding bacterial behaviour, physiology and pathogenesis.
Acknowledgements
Strain FW04/0025 shown in Figure 1 was obtained from the Medical Diagnostic Unit Public Health Laboratory University of Melbourne with special thanks to Ms Agnes Tan and Dr Geoff Hogg.
References
[1] Pentecost, M. et al. (2010) Listeria monocytogenes Internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 6, e1000900.| Listeria monocytogenes Internalin B activates junctional endocytosis to accelerate intestinal invasion.Crossref | GoogleScholarGoogle Scholar | 20485518PubMed |
[2] Toledo-Arana, A. et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956.
| The Listeria transcriptional landscape from saprophytism to virulence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtVOrsbg%3D&md5=86c4fd5aceb7afa05699ea025c589d87CAS | 19448609PubMed |
[3] Loh, E. et al. (2009) A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779.
| A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFylsbbK&md5=045fb764cf1765db5a6b77bf6894ae9dCAS | 19914169PubMed |
[4] Chaturongakul, S. et al. (2011) Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200.
| Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXisVOqur4%3D&md5=0bdf30852de7169c6e97a4814d2916daCAS | 21037293PubMed |
[5] Yates, J.R. et al. (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11, 49–79.
| Proteomics by mass spectrometry: approaches, advances, and applications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVKmt7fP&md5=52b2696cb23d7b4e8699a6e1b45e72fdCAS | 19400705PubMed |
[6] Pham, T.V. et al. (2010) On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26, 363–369.
| On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhs1Onsr4%3D&md5=5a8858c6d577211f8f8cbd5c8836cef0CAS | 20007255PubMed |
[7] Li, M. et al. (2010) Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J. Proteome Res. 9, 4295–4305.
| Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXovFems7s%3D&md5=b80209ba5f85b1587ad3641c1cca471fCAS | 20586475PubMed |
[8] Griffin, N.M. et al. (2010) Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89.
| Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFGjsLvL&md5=3a055d2b82c3ecb7289fff517a896eafCAS | 20010810PubMed |
[9] Kocharunchitt, C. et al. (2012) Integrated transcriptomic and proteomic analysis of the physiological response of Escherichia coli O157:H7 Sakai to steady-state conditions of cold and water activity stress. Mol. Cell. Proteomics 11, M111.0091019.
[10] Bowman, J.P. et al. (2012) Investigation of the Listeria monocytogenes Scott A acid tolerance response and associated physiological and phenotypic features via whole proteome analysis. J. Proteome Res. 11, 2409–2426.
| Investigation of the Listeria monocytogenes Scott A acid tolerance response and associated physiological and phenotypic features via whole proteome analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtVWhsLs%3D&md5=d94ec310d6330b3765eef36a4d439926CAS | 22372944PubMed |
[11] Nilsson, R.E. et al. (2013) MudPIT profiling reveals a link between anaerobic metabolism and the alkaline adaptive response of Listeria monocytogenes EGD-e. PLoS ONE 8, e54157.
| MudPIT profiling reveals a link between anaerobic metabolism and the alkaline adaptive response of Listeria monocytogenes EGD-e.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVGks7Y%3D&md5=2675b29a98f5b4a46d919b3c020f49b0CAS | 23342094PubMed |
[12] Cussiol, J.R. et al. (2010) Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 285, 21943–21950.
| Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXos1Wnsr0%3D&md5=6fe4f654b28eca522432433f5780d432CAS | 20463026PubMed |
Biographies
Tom Ross is the Associate Professor in Food Microbiology and Graduate Research Coordinator at the Food Safety Centre, Tasmanian Institute of Agriculture at the University of Tasmania.
John Bowman is the Associate Professor in Microbiology at the Food Safety Centre, Tasmanian Institute of Agriculture at the University of Tasmania.
Chawalit Kocharunchitt is a postdoctoral research associate at the Food Safety Centre, Tasmanian Institute of Agriculture at the University of Tasmania.
Rolf Nilsson is a postdoctoral research associate at the Food Safety Centre, Tasmanian Institute of Agriculture at the University of Tasmania.


