A sea of microbes: the diversity and activity of marine microorganisms
Justin R SeymourPlant Functional Biology & Climate Change Cluster
University of Technology Sydney
GPO Box 123, Broadway
Sydney, NSW 2007, Australia
Tel: +61 2 9514 1776
Email: Justin.Seymour@uts.edu.au
Microbiology Australia 35(4) 183-187 https://doi.org/10.1071/MA14060
Published: 30 October 2014
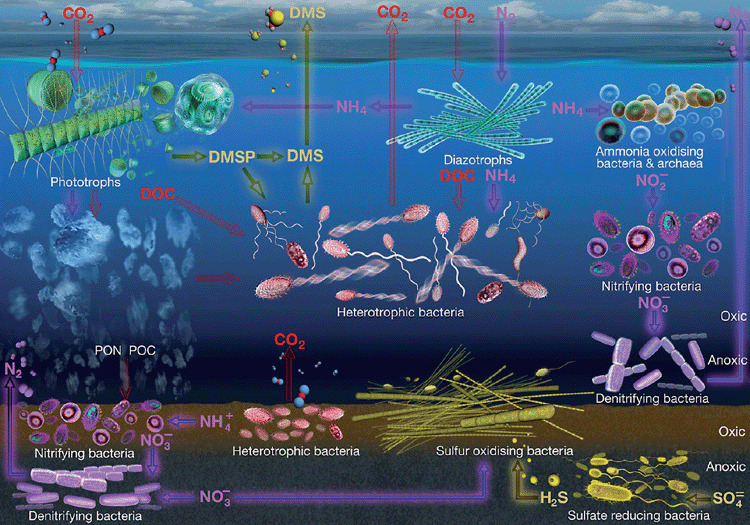
Covering 70% of the earth’s surface, with an average depth of 3.6 km, the ocean’s total volume of 1.3 billion cubic kilometres represents perhaps the largest inhabitable space in the biosphere. Within this vast ecosystem, 90% of all living biomass is microbial. Indeed, seawater from all marine environments, ranging from the warm and sunlit upper ocean to the cold, dark and anoxic deep sea floor, and from the tropics to the arctic, is teeming with microbial life. A single teaspoon of seawater typically contains over 50 million viruses, 5 million Bacteria, 100,000 Archaea and 50,000 eukaryotic microbes. The numerical importance of these microbes is matched only by their ecological and biogeochemical significance. By performing the bulk of oceanic primary production and mediating key chemical transformation processes, planktonic microbes form the base of the marine food-web and are the engines that drive the ocean’s major biogeochemical cycles (Figure 1). While marine microbes are the dominant biological feature throughout the entire water column and within ocean sediments, as well as being important symbionts and pathogens of marine animals and plants, this review will focus on the activity and diversity of microbes inhabiting seawater in the upper sun-lit depths of the global ocean.
‘Keystone microbes’ in a sea of diversity
The ecological and biogeochemical importance of marine microbes is underpinned by the staggering diversity of microbial communities within the ocean. This has only become apparent relatively recently, as the advent of molecular microbiological and genomic approaches has revolutionised the field of marine microbiology. Deep 16S rRNA amplicon sequencing approaches have led to estimates that a single litre of seawater can contain tens of thousands of microbial ‘species’1. More recent approaches using single cell genomics have revealed that even a single marine bacterial species is comprised of hundreds of discrete co-existing sub-populations, which differ in flexible gene content and exhibit different ecological characteristics and environmental tolerances and preferences2. These and other observations of the adaptive divergence of distinct ecotypes among marine bacterial species3 indicate that there is likely to be a microbe to exploit every niche in the ocean. However, embedded within this striking biodiversity are some star players that can be considered ‘keystone organisms’ among the ocean’s microbiota.
Between 25–50% of all prokaryotic cells in the ocean belong to a single clade of Alphaproteobacteria, known as SAR114. Originally named after clone 11 from a clone library derived from seawater collected in the Sargasso Sea5, the SAR11 clade is now recognised as perhaps the most abundant group of closely related organisms on earth. While SAR11 dominates microbial assemblages in ocean surface water across the globe4, bacteria belonging to this clade are classic oligotrophs and are most successful in the open ocean, which from a microbe’s perspective is akin to a desert, where organic resources are significantly below the levels required by most other heterotrophic bacteria. SAR11 can thrive under these conditions with the aid of genome streamlining, whereby their extremely small genomes lack many of the cellular functions thought to be required by free-living bacteria6. As a consequence, SAR11 bacteria utilise atypical nutrient acquisition strategies that appear to minimise the cells’ requirements for organic substrates and allow replication in the most nutrient deplete conditions6.
Not all important marine bacteria are oligotrophs like SAR11. Several copiotrophic species, which prefer the nutrient rich waters found near to the coast or in association with phytoplankton blooms, also often represent a significant fraction of marine bacterial assemblages. Genera including the Roseobacters and Flavobacteria are ecologically important groups that commonly occur in high numbers in seawater7–9. Unlike the highly streamlined genome of the specialist oligotroph SAR11, the genomes of these organisms are often large, providing the cells with significant metabolic flexibility underpinned by a diverse repertoire of energy and carbon acquisition strategies, which allows them to exploit a wide variety of marine niches and thrive under a range of environmental conditions9.
While spatial partitioning of the key oligotrophic and copiotrophic bacteria occurs across geographic boundaries10, within a given location ocean microbial communities also undergo constant substrate controlled successional shifts. During periods of low seawater nutrient concentrations and reduced primary production, oligotrophs such as SAR11 dominate the microbial community. However, following nutrient input events, like those that occur during and immediately after the spring phytoplankton bloom, bacterial communities rapidly shift to become dominated by copiototrophic organisms, including the Roseobacters and Flavobacteria, which can efficiently degrade phytoplankton derived carbohydrates11. These successional processes are often repeatable and predictable, with highly synchronous seasonal shifts in the composition of microbial communities observed over multiple years12, indicating the presence of well-defined microbial niches and unique functional properties of the organisms that inhabit them13.
Other important marine bacteria include members of the cyanobacteria. Two particularly numerically and ecologically important genera are Prochlorococcous and Synechococcus, which often comprise a significant fraction of the total bacterial community in the sunlit ocean surface waters14. Commonly occurring in concentrations of 105 cells/mL, Prochlorococcus is the most abundant phototrophic organism on earth and is responsible for a significant fraction of global photosynthesis15. While occurring in latitudes as high as 40°, Prochlorococcus dominates in tropical oligotrophic regions14. On the other hand, the closely related genus Synechococcus is more cosmopolitan, occurring in high abundances in temperate coastal waters as well as in the open ocean14. In some coastal environments Synechococcus can represent the most abundant bacterial genus16.
Once thought to be primarily extremophiles inhabiting only harsh environments like the extremely hot and anoxic waters surrounding deep sea hydrothermal vents, the Archaea are now recognised as another ubiquitous and important group of marine microbes17. The two main groups of marine Archaea, the Marine Group I Thaumarchaeota and Marine Group II Euryarchaeota, are in fact estimated to account for more than 20% of all prokaryotic cells in the global ocean18. Indeed, below depths of 100 m, the Group I Thaumarchaeota are often the dominant group of microbes in seawater. A significant representative from this group, Nitrosopumilus maritimus is globally abundant and due to its unique mechanisms for nitrification and autotrophy is believed to play an important role in marine carbon and nitrogen biogeochemical cycling19.
The most abundant and diverse of all marine microbes are viruses. There are 1031 viruses in the global ocean and it has been estimated that every second 1023 viral infections occur in the sea20. Marine viruses are known to infect most marine organisms from seaweeds to whales, but as a simple consequence of available host density, most viruses in seawater are bacteriophages20. By manipulating community composition through the selective killing of dominant organisms and facilitating the exchange of genetic material via horizontal gene transfer, marine viruses are a fundamental structuring agent within marine microbial foodwebs21. Moreover, by lysing microbial cells, viruses can strongly alter marine chemical cycling pathways, which has a substantial impact on ocean biogeochemistry22.
Marine microbial activities and their influence on ocean biogeochemistry
Marine biogeochemical cycling processes, driven by the activities of microbes, control the rates and directions of ocean-atmosphere gas exchange, which strongly influences global climate23. However, marine microbial activities and growth rates are highly variable in space and time and across species. On average, marine bacterial growth rates are relatively low, with cells in the open ocean dividing only 0.2 times per day24. However, under optimum conditions, some marine bacteria have the capacity for staggering metabolic rates. For instance, Pseudomonas natrigens is capable of dividing once every 10 minutes24, a likely adaptation for the opportunistic exploitation of intermittent and ephemeral substrate pulses in a dynamic ocean environment. The sum of this heterogeneous pool of marine microbial activities controls the turnover of labile organic substrates and inorganic nutrients in seawater, which ultimately regulates the ocean’s major biogeochemical cycles23 (Figure 1).
Within the ocean’s carbon cycle, phototrophic microbes including cyanobacteria like Prochlorocococus and Synechococcus and a diverse assemblage of eukaryotic phytoplankton use sunlight to fix CO2 into living biomass. Approximately 60 billion tonnes of carbon are fixed each year by these phototrophic microbes25. For perspective, this equates to 40% of total global carbon fixation, yet the biomass of marine microbial phototrophs is equivalent to only about 1% of terrestrial plant biomass25. The carbon fixed by marine photosynthetic microbes has several fates, which are largely determined by the activities of other microbes within seawater, and the balance of these processes profoundly influences global carbon budgets.
Some of the photosynthetically fixed carbon is transferred from phototrophic microbial biomass directly into the marine foodweb via zooplankton grazing. In addition, up to 50% of fixed carbon is exuded by phytoplankton back into the surrounding seawater in the form of dissolved organic carbon (DOC)26. This carbon is ultimately transferred to the higher food-web via a trophic pathway called the ‘Microbial Loop’, whereby heterotrophic bacteria rapidly assimilate DOC from the water column and are subsequently consumed by bacterivorous protists, which are then eaten by larger zooplankton27.
In addition to the fixed carbon that is channeled into the foodweb either by zooplankton grazing or the microbial loop, another significant fraction of C sinks out of the photic zone as dead or senescent phytoplankton biomass, in the form of Particulate Organic Carbon (POC). This POC is transported to the deep ocean sediments and effectively removed from the carbon cycle for millennia, via a process known as the ‘Biological Carbon Pump’. This downward flux of POC leads to the sequestration of up to 300 million tonnes of C to the deep sea each year. However, before all sinking POC is sequestered to the deep sea floor, a significant amount is metabolised by heterotrophic bacteria in the water column, leading to the return of the sinking C to the food web and its subsequent conversion to CO2 through respiration28. Importantly though, recent evidence indicates that not all of the sinking POC that is metabolised by bacteria is respired back as CO2. A potentially large proportion is instead converted into refractory (non-bioavailable) DOC, or RDOC, which remains in the water column. This process, known as the Microbial Carbon Pump, leads to the sequestration of biologically unavailable RDOC in the water column, where it may be stored for thousands of years29.
The balance of these microbiologically mediated carbon cycling processes ultimately determines whether regions of the ocean act as sources or sinks of CO230. Indeed, given that marine carbon cycling processes are so tightly coupled to microbiological activity within seawater, and the oceanic pools and fluxes of carbon are among the largest on earth, even seemingly subtle changes in the composition and activity of microbial assemblages have the potential to profoundly influence the global carbon cycle.
The role of planktonic microbes in the marine sulphur cycle is largely tied to the production and transformation of an organic sulphur compound called dimethylsulphoniopropionate, or DMSP, which is produced by several species of phytoplankton. DMSP is believed to act as an antioxidant, cryoprotectant or osmolyte for phytoplankton31, but also represents an important microbial growth resource, contributing to up to 10% of the carbon demand and over 40% of the sulphur requirements for heterotrophic bacteria in ocean environments32. However, not all marine bacteria use, and cycle, DMSP in the same way, and this has important implications for marine sulphur cycling.
Many marine bacteria demethylate DMSP and subsequently assimilate the sulphur into proteins33. However, others cleave DMSP in a manner that liberates the volatile compound dimethyl sulphide, or DMS34. This is significant because DMS is the major vehicle for ocean to atmosphere sulphur efflux and once in the atmosphere, DMS is rapidly oxidised into aerosol sulphates, which act as cloud condensation nuclei (CCN)35. The balance between the competing demethylation and cleavage pathways of DMSP degradation, which is determined by the composition and activities of marine bacterial populations, influences the amount of DMS that is released from the ocean into the atmosphere and subsequently influences regional climate.
Nitrogen is a key limiting nutrient in many parts of the ocean, so its input and cycling by microbial activity fundamentally shapes the fertility and biology of the global ocean. Diverse populations of bacteria, archaea and eukaryotic phytoplankton are responsible for performing and mediating the key nitrogen cycling steps of fixation, assimilation, nitrification and denitrification36. Analogous to soil environments, where discrete nitrogen cycling microbes and transformation processes are vertically partitioned across oxic niches, different depths of the ocean water column play host to specific microbiological modules of the marine nitrogen cycle37. Additionally, there is strong geographic partitioning of some key nitrogen cycling microbes and processes across the global ocean, with, for instance, much of cyanobacterial nitrogen fixation concentrated within the warm, well-lit oligotrophic regions of the ocean38. Among all of the ocean’s biogeochemical cycles, our understanding of the nitrogen cycle has perhaps been most radically re-shaped with the advent of molecular microbiological approaches36. The resultant discoveries of new groups of nitrogen cycling microbes, including unicellular nitrogen fixing cyanobacteria and large populations of ammonia oxidising archaea, have fundamentally shifted our understanding of marine nitrogen cycling and the microbiological processes that regulate global nitrogen budgets.
New directions: dynamic microbial networks control ocean function
Our view of the identities and roles of microbes in the ocean continues to rapidly expand. Microbial oceanography is currently transitioning from a fruitful era of discovery where molecular tools allowed us to uncover the diversity of marine microbial communities and their functions, to an exciting new phase where this information is feeding sophisticated new ecological questions and concepts. For instance, recent evidence suggests that rather than consisting of a soup of loosely associated populations, the microbial communities in seawater are comprised of highly interconnected ecological units that function in tight synchrony12,13,39,40. It is becoming apparent that networks of key groups of marine microbes consistently co-occur non-randomly, indicating the existence of community assembly rules among the microbial plankton41, while the occurrence of multi-species timing of gene regulation in seawater40,42 likely permits coordination of complex metabolic processes. With the aid of a suite of new automated sampling technologies42 and rapidly advancing ecogenomic approaches2, exploration of these new ecological concepts promises to deliver unprecedented insights into the lives of the ocean’s smallest, but arguably most important inhabitants.
References
[1] Sogin, M.L. et al. (2006) Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl. Acad. Sci. USA 103, 12 115–12 120.| Microbial diversity in the deep sea and the underexplored ‘rare biosphere’.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xotlyisb4%3D&md5=e48217c292428ed02d8fda935a6f8e71CAS |
[2] Kashtan, N. et al. (2014) Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344, 416–420.
| Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXmsFWmtLo%3D&md5=75998280e1824d97b27f95e93f81469fCAS | 24763590PubMed |
[3] Brown, M.V. et al. (2005) Coupling 16S-ITS clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 7, 1466–1479.
| Coupling 16S-ITS clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVGnurvP&md5=c8016da1b2f03640399179fc32f6eb33CAS | 16104869PubMed |
[4] Morris, R.M. et al. (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810.
| SAR11 clade dominates ocean surface bacterioplankton communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XpsFygt7Y%3D&md5=07fbc4e7cce1367578d801cc545f8885CAS | 12490947PubMed |
[5] Giovannoni, S.J. et al. (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63.
| Genetic diversity in Sargasso Sea bacterioplankton.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXktFymu7s%3D&md5=2e7cf1b9d2cc7c23645b03efd472cabcCAS | 2330053PubMed |
[6] Giovannoni, S.J. et al. (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245.
| Genome streamlining in a cosmopolitan oceanic bacterium.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXnvFyls7k%3D&md5=ad1bceaa927ea40806238a3feeefe642CAS | 16109880PubMed |
[7] Moran, M.A. et al. (2007) Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73, 4559–4569.
| Ecological genomics of marine Roseobacters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXot1egs7o%3D&md5=01ee7b760843bdfef68c90c2094bd2e0CAS | 17526795PubMed |
[8] Kirchman, D.L. (2002) The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100.
| 1:CAS:528:DC%2BD38XitFaqt7k%3D&md5=52ed8e9ad908e46520d8b31b99b7341cCAS | 19709188PubMed |
[9] Newton, R.J. et al. (2010) Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798.
| Genome characteristics of a generalist marine bacterial lineage.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmtFCns78%3D&md5=092ebc2216f3d9f6dc2113bdbf670da2CAS | 20072162PubMed |
[10] Lauro, F.M. et al. (2009) The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. USA 106, 15527–15533.
| The genomic basis of trophic strategy in marine bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtFyjsLrM&md5=2ba31127ccb449d857ffa3ab37e671b3CAS | 19805210PubMed |
[11] Teeling, H. et al. (2012) Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611.
| Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xmt1Gntrw%3D&md5=44f843f38a0782acc32f133234848549CAS | 22556258PubMed |
[12] Fuhrman, J.A. et al. (2006) Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. USA 103, 13104–13109.
| Annually reoccurring bacterial communities are predictable from ocean conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xptlemu7k%3D&md5=3140d3d36fd2163b64e1df90050d35faCAS | 16938845PubMed |
[13] Fuhrman, J.A. (2009) Microbial community structure and its functional implications. Nature 459, 193–199.
| Microbial community structure and its functional implications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXlvFShu70%3D&md5=381461ce7a56e0f400af462341db06c2CAS | 19444205PubMed |
[14] Partensky, F. et al. (1999a) Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters. In: Marine Cyanobacteria, no. spécial 19 (Charpy, L. and Larkum, A.W.D., eds), Bulletin de l’Institut oceanographique, pp. 457–475.
[15] Partensky, F. et al. (1999)b Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127.
| 1:CAS:528:DyaK1MXitVert78%3D&md5=5efdb84b30efab320a832a510993145cCAS | 10066832PubMed |
[16] Messer, L.F. et al. (2014) Prokaryotic and diazotrophic population dynamics within a large oligotrophic inverse estuary. Aquat. Microb. Ecol. , .
[17] DeLong, E.F. (2003) Oceans of Archaea. Microbe Magazine 69, 503–511.
[18] Karner, M. et al. (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510.
| Archaeal dominance in the mesopelagic zone of the Pacific Ocean.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M7hvFOqsw%3D%3D&md5=1823cf59e20345a786b139cd9e59796fCAS | 11206545PubMed |
[19] Walker, C.B.A. et al. (2010) Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 107, 8818–8823.
| Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmsVOrt7Y%3D&md5=5b1c490c8c803e84b864aefaf530d220CAS |
[20] Suttle, C. (2007) Marine viruses – major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812.
| Marine viruses – major players in the global ecosystem.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVeis7nK&md5=223d5fd74df2ff2d7a01ebedcccbf96fCAS | 17853907PubMed |
[21] Breitbart, M. (2012) Marine viruses: truth or dare. Ann. Rev. Mar. Sci. 4, 425–448.
| Marine viruses: truth or dare.Crossref | GoogleScholarGoogle Scholar | 22457982PubMed |
[22] Fuhrman, J.A. (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548.
| Marine viruses and their biogeochemical and ecological effects.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXjvFeiurk%3D&md5=fa93b89510baa1896ca3a5f71d0c6d0aCAS | 10376593PubMed |
[23] Falkowski, P.G. et al. (2008) The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039.
| The microbial engines that drive Earth’s biogeochemical cycles.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXmtVKnsbo%3D&md5=3b510518275205ce7807c177aa2ee960CAS | 18497287PubMed |
[24] Pomeroy, L.R. et al. (2007) The microbial loop. Oceanography (Wash. D.C.) 20, 28–33.
| The microbial loop.Crossref | GoogleScholarGoogle Scholar |
[25] Falkowski, P.G. (1994) The role of phytoplankton photosynthesis in global biogeochemical cycles. Photosynth. Res. 39, 235–258.
| The role of phytoplankton photosynthesis in global biogeochemical cycles.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXksFertb8%3D&md5=d78b50ae9f60ca36eadde77c6f916a72CAS | 24311124PubMed |
[26] Aaronson, S. (1978) Excretion of organic matter by phytoplankton in vitro. Limnol. Oceanogr. 23, 838.
| Excretion of organic matter by phytoplankton in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE1cXlsVaqtb0%3D&md5=60c5fe35215fb899eaa49c423eb37adbCAS |
[27] Azam, F. et al. (1983) The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263.
| The ecological role of water-column microbes in the sea.Crossref | GoogleScholarGoogle Scholar |
[28] Azam, F. and Long, R.A. (2001) Sea snow microcosms. Nature 414, 495–498.
| Sea snow microcosms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXptFeksLk%3D&md5=9f9f6730d761108d37152d0371d124f8CAS | 11734832PubMed |
[29] Jiao, N. and Azam, F. (2011) Microbial carbon pump and its significance for carbon sequestration in the ocean. In: Microbial Carbon Pump in the Ocean (Jiao N., et al., eds.), Science/AAAS, Washington, DC, pp. 43–45.
[30] Ducklow, H.W. and Doney, S.C. (2013) What is the metabolic state of the oligotrophic ocean? Ann. Rev. Mar. Sci. 5, 525–533.
| What is the metabolic state of the oligotrophic ocean?Crossref | GoogleScholarGoogle Scholar | 22809191PubMed |
[31] Kiene, R.P. et al., eds (1996) Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, Plenum Press.
[32] Kiene, R.P. et al. (2000) New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209.
| New and important roles for DMSP in marine microbial communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXms1Wrtbw%3D&md5=5acdd8140b795d3465f4ebf3e94c3d79CAS |
[33] Howard, E.C. et al. (2006) Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652.
| Bacterial taxa that limit sulfur flux from the ocean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFeitr7M&md5=93a767ee0f19bd102fecb8ac7bcc506fCAS | 17068264PubMed |
[34] Curson, A.R.J. et al. (2011) Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859.
| Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht12iurbE&md5=36a8e01b664edf186953ec66c2719178CAS |
[35] Simó, R. (2001) Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287.
| Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links.Crossref | GoogleScholarGoogle Scholar | 11369106PubMed |
[36] Zehr, J.P. and Kudela, R.M. (2011) Nitrogen cycle of the open ocean: from genes to ecosystems. Ann. Rev. Mar. Sci. 3, 197–225.
| Nitrogen cycle of the open ocean: from genes to ecosystems.Crossref | GoogleScholarGoogle Scholar | 21329204PubMed |
[37] Gruber, N. (2008) The marine nitrogen cycle: overview and challenges. In: Nitrogen in the Marine Environment (Capone, D.G., et al., eds), Elsevier.
[38] Ward, B.A. et al. (2013) Iron, phosphorus and nitrogen supply ratios define the biogeography of nitrogen fixation. Limnol. Oceanogr. 58, 2059–2075.
| Iron, phosphorus and nitrogen supply ratios define the biogeography of nitrogen fixation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvVyit7%2FP&md5=7cf0dc42cd090178d1f60d29f7c9742eCAS |
[39] Steele, J.A. et al. (2011) Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425.
| Marine bacterial, archaeal and protistan association networks reveal ecological linkages.Crossref | GoogleScholarGoogle Scholar | 21430787PubMed |
[40] Ottesen, E.A. et al. (2013) Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc. Natl. Acad. Sci. USA 110, E488–E497.
| Pattern and synchrony of gene expression among sympatric marine microbial populations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjtVertLs%3D&md5=17e572d0098a460cef0990ec69ab8e93CAS | 23345438PubMed |
[41] Beman, J.M. et al. (2011) Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. ISME J. 5, 1077–1085.
| Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnvVaqsbo%3D&md5=ec5e6b21a8169e762d3862f0978901f6CAS | 21228895PubMed |
[42] Ottesen, E.A. et al. (2014) Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 345, 207.
| Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtFWjtLjO&md5=c4a5a79e3b545ffbf401f94d476a6747CAS | 25013074PubMed |
Biography
Justin Seymour is an ARC Future Fellow, based at the University of Technology, Sydney and his research interests incorporate aquatic microbial ecology and biological oceanography. His research involves examinations of the ecology of microbes across a range of marine environments, spanning tropical coral reefs to Antarctica, and a continuum of spatiotemporal scales. At the ocean-basin scale he is interested in how large-scale oceanographic processes influence microbial community dynamics and functionality, while at the scale of individual drops of seawater he studies the foraging behaviours of individual marine microbes living within a patchy chemical seascape.