Avian influenza H5N1: still a pandemic threat?
Paul F Horwood A BA College of Public Health, Medical and Veterinary Sciences, James Cook University, Townsville, Qld, Australia
B Tel.: +61 7 4781 6106; Email: paul.horwood@jcu.edu.au
Microbiology Australia 42(4) 152-155 https://doi.org/10.1071/MA21044
Submitted: 24 August 2021 Accepted: 11 October 2021 Published: 12 November 2021
Journal Compilation © The Authors 2021 Open Access CC BY-NC-ND, published (by CSIRO Publishing) on behalf of the ASM
Abstract
Highly pathogenic avian influenza H5N1 viruses have become endemic in global poultry populations over the past 25 years and pose an ongoing public health threat. Although the incidence of human cases has declined, viruses from the H5N1 lineage can now be found in poultry throughout Asia, the Middle East and Africa, in addition to causing outbreaks in Europe and the Americas. The recent emergence and spread of reassortant H5Nx viruses, resulting in regional poultry outbreaks, has increased the risk for further evolution of these viruses and possible avian-to-human transmission. Ongoing surveillance and pandemic preparedness for H5N1 and other avian influenza viruses of public health concern are warranted.
Introduction
Influenza A viruses are one of the most important human pathogens. Since the start of the 20th century, these viruses have been responsible for four pandemics in the human population, including the 1918 H1N1 ‘Spanish flu’ pandemic that resulted in the deaths of over 50 million people1. Subsequent influenza pandemics also occurred in 1957 (H2N2, ‘Asian flu’), 1968 (H3N2, ‘Hong Kong flu’) and 2009 (H1N1, ‘Swine flu’)2,3. Following the pandemic period of spread (usually 2–3 years), the virus typically takes on a seasonal pattern of circulation. Currently, the two seasonal influenza viruses circulating in humans emerged during the pandemics in 1968 (H3N2) and 2009 (H1N1).
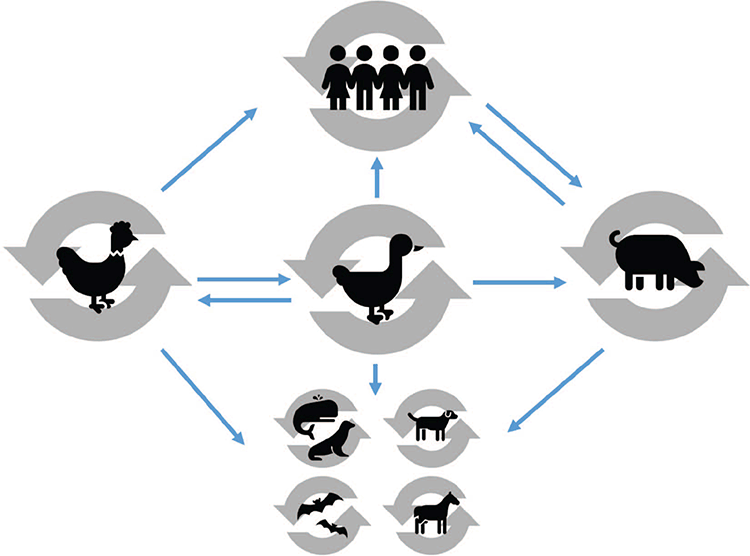
The natural reservoir of influenza A viruses (known as avian influenza viruses, AIVs), are wild waterbirds from the Orders Anseriformes (ducks, geese, swans) and Charadriiformes (waders, gulls, terns)4. There is a large diversity of AIVs amongst their natural avian reservoirs. These viruses replicate in the gastrointestinal tract of their avian hosts which shed large quantities of virus into their aquatic environment. AIV infections in wild waterbirds are generally asymptomatic, or cause mild infections, enabling birds to carry viruses over vast distances during seasonal migrations5. AIVs can jump the species gap from their natural hosts to a wide range of avian and mammalian (e.g. humans, pigs, horses, dogs, cetaceans) hosts (Figure 1). In the context of human exposure risk, the most important of these secondary hosts are domestic poultry and pigs.
Avian influenza ecology
The poultry industry makes important contributions to nutrition, food security and family income in resource-limited settings6. Although some countries have recently made the transition to intensive farming with improved biosecurity practices, in many settings birds are raised in subsistence-level backyard or village farms. In these settings, large flocks of birds are allowed to roam free where there is ample opportunity for them to interact with wild waterbirds or the contaminated aquatic environment, thus introducing AIVs into poultry circulation. Excess birds are commonly transported to central live bird markets (LBMs) where they are sold to consumers and slaughtered by the vendor at the market. LBMs have been associated with avian influenza persistence, distribution and the potential emergence of novel viruses7. The presence of multiple avian species, the lack of sufficient biosecurity measures and the constant introduction of immunologically naïve animals contribute to the threat of the emergence of novel influenza strains with pandemic potential (Figure 2). High levels of avian influenza viruses of public health concern (e.g. H5N1, H7N9, H9N2) have been detected in LBMs throughout Asia, the Middle East and Africa through longitudinal surveillance initiatives8–12. Indeed, many human cases of AIV infection have been associated with contact with poultry at LBMs13,14.

|
Avian influenza evolution
AIVs can be classified into low pathogenic avian influenza viruses (LPAIVs) and highly pathogenic avian influenza avian influenza viruses (HPAIVs), based on their virulence in domestic chickens15. Both LPAIVs and HPAIVs can cross the species gap to infect humans, but adaptation to the new host is required before the virus will be able to spread efficiently amongst this new host. AIVs can evolve this ability through two main mechanisms: (1) due to the segmented RNA genome of AIVs, when two different viruses infect an individual animal at the same time reassortment can occur between the genetic material, potentially resulting in progeny viruses that have the ability to spread in the human population (this process is called antigenic shift); and (2) the high mutation rate of AIVs during replication can lead to the accumulation of mutations that result in an AIV strain that is better adapted to the new host (this process is called antigenic drift)16–18.
A/H5N1 emergence and human cases
Influenza H5N1 emerged as a public health threat in 1997, following the detection of human infections associated with poultry exposure in Hong Kong LBMs. During this outbreak, 18 cases of H5N1 human infection were detected in Hong Kong with a case fatality rate (CFR) of 33%19. The outbreak was contained when Hong Kong authorities closed LBMs and conducted mass culling of poultry19. Unfortunately, the virus subsequently re-emerged in China in 2003, and rapidly spread throughout Southeast Asia during 2004–200520. Poultry outbreaks of influenza A/H5N1 virus have now been reported from many countries in Asia, Europe, the Middle East and Africa21,22. The global spread of H5N1 has been facilitated by the migration of wild waterbirds, the natural host for AIVs. To date, 17 countries have reported human H5N1 infections, with more than 860 confirmed human cases and a CFR of ~53%23. Over the ~25 years since A/H5N1 first emerged, the virus has diversified into multiple HA clades and subclades. Some of these strains have displayed greater fitness for spreading amongst poultry and others have been associated with a higher risk of human infection and disease24,25.
Current influenza A/H5 circulation
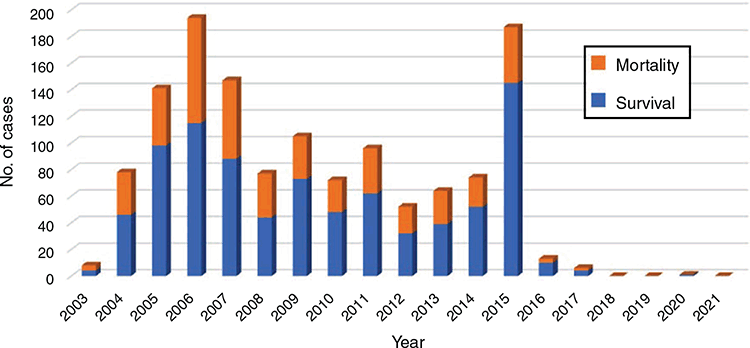
Human H5N1 cases peaked in 2005–2007, followed by a steady decline in cases over the following 15 years, including only one case over the last four years (Figure 3). An obvious break in this trend occurred in 2015 when the emergence of a new strain in Egypt (clade 2.2.1.2), led to an upsurge in human H5N1 cases26. A similar (but not as dramatic) increase in cases occurred in Cambodia in 2013-2014, also linked with the emergence of a new strain (clade 1.1.2 reassortant)27. These cases highlight that although human infections with H5N1 are currently very low this situation can rapidly change following the emergence of novel strains through viral mutations. Indeed, there has been a recent emergence and spread of HPAIV H5 viruses (predominantly clade 2.3.4.4) that have undergone reassortment with various LPAIVs to give rise to a group of viruses known as H5Nx28. The clade 2.3.4.4 H5Nx lineage of viruses first emerged in China around 2008 and have since diversified into numerous novel reassortant viruses, including H5N2, H5N3, H5N5, H5N6 and H5N8. These novel viruses have since spread around the world, causing poultry outbreaks of H5N2 and H5N3 in Taiwan; H5N8 in South Korea, China, Japan and Europe; and H5N2 in USA and Canada29. Of particular concern is the H5N6 virus which has spread through numerous countries in Eurasia, and caused 36 laboratory-confirmed human cases resulting in 21 deaths30. H5N8 viruses have also been associated with seven asymptomatic human infections during a poultry outbreak in Russia30. Reassortment of H5N1 strains with other AIVs of public health concern such as H9N2, have also been reported, prompting concerns for the emergence of other novel lineages31,32.
|
Despite the current low number of human cases, influenza H5N1 circulation in poultry has continued unabated. Viral surveillance in LBMs has established that H5N1 circulation is occurring at levels similar to those detected during the peak period of human infections8–12. The current lack of human H5N1 cases may be linked to the dominant strain currently circulating (clade 2.3.2.1) being more ‘avian-like’ and not as readily able to jump the species gap to infect humans25. However, in some countries, H5N6 reassortant viruses have replaced H5N1 as the dominant virus in circulation, thus highlighting the threat for the emergence of novel AIV viruses33.
Conclusions
For the past ~25 years, AIV H5N1 has been regarded as one of the most pertinent threats for pandemic emergence. Fortunately, spillover of the virus into humans has remained relatively low and the ability to transmit efficiently between humans has not been established. However, the widespread, intense circulation of H5N1 and H5Nx viruses across many regions of the world is an ongoing animal health challenge and a potential threat for public health. The propensity for these viruses to reassort with LPAIVs, particularly in the LBM environment, is a constant source for the introduction of novel viruses into poultry circulation. Surveillance of virus populations in LBMs is an important strategy of monitoring for the emergence of novel strains and may act as an early warning system to prevent major outbreaks of H5N1 (and other H5 viruses) in the human population.
Conflicts of interest
The author declares no conflicts of interest.
Declaration of funding
This research did not receive any specific funding.
References
[1] Morens, D.M. et al. (2018) Influenza cataclysm, 1918. N. Engl. J. Med. 379, 2285–2287.| Influenza cataclysm, 1918.Crossref | GoogleScholarGoogle Scholar | 30575465PubMed |
[2] Webster, R.G. et al. (1992) Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179.
| Evolution and ecology of influenza A viruses.Crossref | GoogleScholarGoogle Scholar | 1579108PubMed |
[3] World Health Organization (WHO) (2009) Outbreak of swine-origin influenza A (H1N1) virus infection-Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58, 467–470.
| 19444150PubMed |
[4] Sonnberg, S. et al. (2013) Natural history of highly pathogenic avian influenza H5N1. Virus Res. 178, 63–77.
| Natural history of highly pathogenic avian influenza H5N1.Crossref | GoogleScholarGoogle Scholar | 23735535PubMed |
[5] Fouchier, R.A. et al. (2009) Epidemiology of low pathogenic avian influenza viruses in wild birds. Rev. Sci. Tech. 28, 49–58.
| Epidemiology of low pathogenic avian influenza viruses in wild birds.Crossref | GoogleScholarGoogle Scholar | 19618618PubMed |
[6] Mottet, A. et al. (2017) Global poultry production: current state and future outlook and challenges. Worlds Poult. Sci. J. 73, 245–256.
| Global poultry production: current state and future outlook and challenges.Crossref | GoogleScholarGoogle Scholar |
[7] Suttie, A. et al. (2019) Avian influenza in the Greater Mekong Subregion. Infect. Genet. Evol. 74, 103920.
| Avian influenza in the Greater Mekong Subregion.Crossref | GoogleScholarGoogle Scholar | 31201870PubMed |
[8] Turner, J.C. et al. (2017) Insight into live bird markets of Bangladesh: an overview of the dynamics of transmission of H5N1 and H9N2 avian influenza viruses. Emerg. Microbes Infect. 6, e12.
| Insight into live bird markets of Bangladesh: an overview of the dynamics of transmission of H5N1 and H9N2 avian influenza viruses.Crossref | GoogleScholarGoogle Scholar | 28270655PubMed |
[9] Horwood, P.F. et al. (2018) Co-circulation of influenza A/H5N1 with H7 and H9 viruses in Cambodian live bird markets with evidence of frequent co-infections in poultry. Emerg. Infect. Dis. 24, 352–355.
| Co-circulation of influenza A/H5N1 with H7 and H9 viruses in Cambodian live bird markets with evidence of frequent co-infections in poultry.Crossref | GoogleScholarGoogle Scholar | 29350140PubMed |
[10] Cheng, K.L. et al. (2020) Avian influenza virus detection rates in poultry and environment at live poultry markets, Guangdong, China. Emerg. Infect. Dis. 26, 591–595.
| Avian influenza virus detection rates in poultry and environment at live poultry markets, Guangdong, China.Crossref | GoogleScholarGoogle Scholar | 31922954PubMed |
[11] Dharmayanti, N.L. et al. (2020) Genetic diversity of the H5N1 viruses in live bird markets, Indonesia. J. Vet. Sci. 21, e56.
| Genetic diversity of the H5N1 viruses in live bird markets, Indonesia.Crossref | GoogleScholarGoogle Scholar | 32735094PubMed |
[12] Sulaiman, L. et al. (2021) Live bird markets in Nigeria: a potential reservoir for H9N2 avian influenza viruses. Viruses 13, 1445.
| Live bird markets in Nigeria: a potential reservoir for H9N2 avian influenza viruses.Crossref | GoogleScholarGoogle Scholar | 34452311PubMed |
[13] Mounts, A.W. et al. (1999) Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997. J. Infect. Dis. 180, 505–508.
| Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997.Crossref | GoogleScholarGoogle Scholar | 10395870PubMed |
[14] Dinh, P.N. et al. (2006) Risk factors for human infection with avian influenza A H5N1, Vietnam, 2004. Emerg. Infect. Dis. 12, 1841–1847.
| Risk factors for human infection with avian influenza A H5N1, Vietnam, 2004.Crossref | GoogleScholarGoogle Scholar | 17326934PubMed |
[15] Deshpande, K.L. et al. (1987) Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc. Natl. Acad. Sci. USA 84, 36–40.
| Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence.Crossref | GoogleScholarGoogle Scholar | 3467357PubMed |
[16] Yoon, S.W. et al. (2014) Evolution and ecology of influenza A viruses. In Influenza Pathogenesis and Control – Volume I. (Compans, R.W. and Oldstone, M.B.A., eds). pp. 359–375. Springer.
[17] Joseph, U. et al. (2017) The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir. Viruses 11, 74–84.
| The ecology and adaptive evolution of influenza A interspecies transmission.Crossref | GoogleScholarGoogle Scholar | 27426214PubMed |
[18] Suttie, A. et al. (2019) Inventory of molecular characteristics for risk assessment of avian influenza viruses. Virus Genes 55, 739–768.
| Inventory of molecular characteristics for risk assessment of avian influenza viruses.Crossref | GoogleScholarGoogle Scholar | 31428925PubMed |
[19] Chan, P.K. (2002) Outbreak of avian influenza A (H5N1) virus infection in Hong Kong in 1997. Clin. Infect. Dis. 34, S58–S64.
| Outbreak of avian influenza A (H5N1) virus infection in Hong Kong in 1997.Crossref | GoogleScholarGoogle Scholar | 11938498PubMed |
[20] Webster, R.G. et al. (2006) H5N1 outbreaks and enzootic influenza. Biodiversity (Nepean) 7, 51–55.
| H5N1 outbreaks and enzootic influenza.Crossref | GoogleScholarGoogle Scholar |
[21] Food and Agriculture Organization of the United Nations (FAO) (2021) Global AIV with Zoonotic Potential: situation update. http://www.fao.org/ag/againfo/programmes/en/empres/Global_AIV_Zoonotic_Update/situation_update.html (accessed August 2021).
[22] World Organisation for Animal Health (OIE) (2021) Highly Pathogenic Avian Influenza (HPAI) Report No. 21: January 15 to February 04, 2021. https://www.oie.int/app/uploads/2021/03/hpai---asof04022021.pdf (accessed August 2021).
[23] World Health Organization (WHO) (2021) Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2021, 15 April 2021. https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who-2003-2021-15-april-2021 (accessed August 2021).
[24] Lai, S. et al. (2016) Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: a systematic review of individual case data. Lancet Infect. Dis. 16, e108–e118.
| Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: a systematic review of individual case data.Crossref | GoogleScholarGoogle Scholar | 27211899PubMed |
[25] Horwood, P.F. et al. (2020) Transmission experiments support clade-level differences in the transmission and pathogenicity of Cambodian influenza A/H5N1 viruses. Emerg. Microbes Infect. 9, 1702–1711.
| Transmission experiments support clade-level differences in the transmission and pathogenicity of Cambodian influenza A/H5N1 viruses.Crossref | GoogleScholarGoogle Scholar | 32666894PubMed |
[26] Refaey, S. et al. (2015) Increased number of human cases of influenza virus A (H5N1) infection, Egypt, 2014–15. Emerg. Infect. Dis. 21, 2171–2173.
| Increased number of human cases of influenza virus A (H5N1) infection, Egypt, 2014–15.Crossref | GoogleScholarGoogle Scholar | 26584397PubMed |
[27] Rith, S. et al. (2014) Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A(H5N1) viruses detected in Cambodia. J. Virol. 88, 13897–13909.
| Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A(H5N1) viruses detected in Cambodia.Crossref | GoogleScholarGoogle Scholar | 25210193PubMed |
[28] Lee, D.H. et al. (2017) Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3. 4.4. J. Vet. Sci. 18, 269–280.
| Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3. 4.4.Crossref | GoogleScholarGoogle Scholar | 28859267PubMed |
[29] Qin, T. et al. (2018) Compatibility between haemagglutinin and neuraminidase drives the recent emergence of novel clade 2.3. 4.4 H5Nx avian influenza viruses in China. Transbound. Emerg. Dis. 65, 1757–1769.
| Compatibility between haemagglutinin and neuraminidase drives the recent emergence of novel clade 2.3. 4.4 H5Nx avian influenza viruses in China.Crossref | GoogleScholarGoogle Scholar | 29999588PubMed |
[30] World Health Organization (WHO) (2021) Avian Influenza Weekly Update Number 803. https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai-20210730.pdf?sfvrsn=223ca73f_136 (accessed August 2021).
[31] Suttie, A. et al. (2018) Influenza A(H5N1) viruses with A(H9N2) single gene (matrix or PB1) reassortment isolated from Cambodian live bird markets. Virology 523, 22–26.
| Influenza A(H5N1) viruses with A(H9N2) single gene (matrix or PB1) reassortment isolated from Cambodian live bird markets.Crossref | GoogleScholarGoogle Scholar | 30075357PubMed |
[32] Gerloff, N.A. et al. (2014) Multiple reassortment events among highly pathogenic avian influenza A (H5N1) viruses detected in Bangladesh. Virology 450–451, 297–307.
| Multiple reassortment events among highly pathogenic avian influenza A (H5N1) viruses detected in Bangladesh.Crossref | GoogleScholarGoogle Scholar | 24503093PubMed |
[33] Nuñez, I.A. et al. (2019) A review of H5Nx avian influenza viruses. Ther. Adv. Vaccines Immunother. 7, 2515135518821625.
| A review of H5Nx avian influenza viruses.Crossref | GoogleScholarGoogle Scholar | 30834359PubMed |
Biography
A/Professor Paul Horwood is an Associate Professor of Virology and Viral Diseases at James Cook University. His research focuses on emerging pathogens and zoonotic diseases, with an emphasis on improving surveillance and laboratory diagnosis in resource-limited settings. He conducts research to better understand zoonotic transmission in high-risk interfaces where humans, domestic animals and wildlife interact.