The use of mycobacterial interspersed repetitive unit typing and whole genome sequencing to inform tuberculosis prevention and control activities
Gwendolyn L. Gilbert A B C and Vitali Sintchenko A BA Centre for Infectious Diseases and Microbiology, Westmead Hospital
B Sydney Institute for Emerging Infectious Diseases and Biosecurity, The University of Sydney
C Corresponding author. Email: lyn.gilbert@sydney.edu.au
NSW Public Health Bulletin 24(1) 10-14 https://doi.org/10.1071/NB12106
Published: 15 July 2013
Abstract
Molecular strain typing of Mycobacterium tuberculosis has been possible for only about 20 years; it has significantly improved our understanding of the evolution and epidemiology of Mycobacterium tuberculosis and tuberculosis disease. Mycobacterial interspersed repetitive unit typing, based on 24 variable number tandem repeat unit loci, is highly discriminatory, relatively easy to perform and interpret and is currently the most widely used molecular typing system for tuberculosis surveillance. Nevertheless, clusters identified by mycobacterial interspersed repetitive unit typing sometimes cannot be confirmed or adequately defined by contact tracing and additional methods are needed. Recently, whole genome sequencing has been used to identify single nucleotide polymorphisms and other mutations, between genotypically indistinguishable isolates from the same cluster, to more accurately trace transmission pathways. Rapidly increasing speed and quality and reduced costs will soon make large scale whole genome sequencing feasible, combined with the use of sophisticated bioinformatics tools, for epidemiological surveillance of tuberculosis.
Unlike many other bacterial pathogens, different strains of Mycobacterium tuberculosis complex cannot be easily distinguished by the use of phenotypic methods. It was not until the 1990s, when M. tuberculosis complex molecular typing methods were developed, that it became possible to accurately identify tuberculosis (TB) transmission and study the origins and global distribution of M. tuberculosis complex genotypes. M. tuberculosis complex is a clonal species, in which horizontal transmission (recombination) of genetic material is rare. Genetic variation results from: (a) point mutations or single nucleotide polymorphisms (SNPs); (b) small DNA insertions or deletions, often mediated by mobile genetic elements, such as insertion sequences (IS); (c) larger DNA deletions, resulting in regions of difference (RDs) between strains, major enough to determine subspecies within the M. tuberculosis complex; and (d) variation in numbers of repeat sequences at various genomic loci. All of these are potential genotyping targets.
The appropriate choice of targets depends on the purpose of genotyping. For example, monitoring TB transmission requires targets that change rapidly enough to distinguish epidemiologically unrelated strains (such as IS or repeat sequences), but may not provide accurate information about lineage, whereas studies of long-term epidemiological trends and lineages require more stable targets reflecting rare genetic events (such as SNPs or RDs) that can trace the origins and global spread of different strains.
The rationale for molecular typing depends on the setting. In low-incidence settings it is often used, routinely, to identify case clusters (and, potentially, their sources) that are not apparent by conventional contact tracing. Routine typing can also identify laboratory contamination and false-positive culture results and, so, prevent unnecessary treatment. In high-prevalence settings, molecular typing is more likely to be performed, selectively, to investigate the evolution and transmission of M. tuberculosis to inform and improve TB control programs.
Strain typing methods
One of the first genotyping methods described was IS6110 restriction fragment length polymorphism (RFLP) typing which, until recently, was the most discriminatory method available.1 IS6110 is found only in the M. tuberculosis complex, usually in multiple copies; the number of copies and the sizes of restriction fragments that contain them distinguish different strains, except those with few, or sometimes no, IS6110 copies. The method involves cutting DNA with a restriction enzyme, identifying fragments containing IS6110 with a labelled molecular probe and separating them on a gel to produce a strain-specific banding pattern. Its disadvantages are that it requires prolonged incubation of isolates to produce a large enough quantity of DNA and the banding patterns can be difficult to interpret and reproduce. However, it has been used successfully, for many years, to provide insights into M. tuberculosis transmission, distinguish reactivation from reinfection, and identify laboratory cross-contamination.2
Spoligotyping detects the presence or absence of 43 non-repetitive spacer oligonucleotides, which are variably interspersed between short chromosomal repeats at the direct repeat locus of the M. tuberculosis genome.3,4 The direct repeat locus is amplified by polymerase chain reaction (PCR) and the product is hybridized to a series of probes on a strip to identify the presence of spacer sequences. It requires relatively little DNA and can be used on early cultures or clinical specimens.3 The results are objective and reproducible but it is less discriminatory than IS6110-RFLP and so less suitable for transmission studies. However, it differentiates M. tuberculosis strains into broad families that generally correspond with geographic areas5 and, sometimes, with characteristics such as virulence, transmissibility and resistance.6
Recently, mycobacterial interspersed repetitive unit (MIRU) typing has largely overtaken both IS6110 and spoligotyping, although they are sometimes used together. MIRUs are tandem repeat sequences, dispersed among intergenic regions of the M. tuberculosis genome, which vary between M. tuberculosis strains according to the number of repeats at each locus. They are one of a group of variable number tandem repeat (VNTR) sequences, which are the basis of multilocus VNTR analysis (MLVA), a common bacterial genotyping method.
M. tuberculosis MIRU-VNTR typing, originally based on 12 loci,7 is rapid, objective and reproducible. It was rapidly adopted as the basis for large-scale, high-throughput M. tuberculosis genotyping8–10 but, even in combination with spoligotyping, was less discriminatory than IS6110 fingerprinting, especially for isolates of the widespread Beijing spoligotypes.11 Because of the potential for false clusters, identified by MIRU-12 and spoligotyping, IS6110-RFLP typing of clustered isolates was often needed to more accurately define outbreaks,12 especially if there were no obvious epidemiological links between cases.
Subsequently, a more discriminatory set of 24-MIRU-VNTR loci, including the original 12, was identified. MIRU-24 typing of more than 800 geographically diverse isolates, representing all major lineages, identified 40% more genotypes and demonstrated a fourfold lower rate of clustering than MIRU-12 typing.13 Even a smaller subset of 15 loci, with the highest evolutionary rates, was as discriminatory as IS6110-RFLP typing. Thus, MIRU-15 typing was proposed as a new standard for routine use and MIRU-24 as a high-resolution alternative for phylogenetic studies.11,13 Others have confirmed the higher discriminatory power of MIRU-24, compared with MIRU-12.14
MIRU-24 typing
MIRU-24 (or -15) is now the new ‘gold standard’ for routine M. tuberculosis typing. To facilitate global epidemiological studies, a web-based database (http://www.miru-vntrplus.org/) has been established, based on a collection of 186 reference strains, representing the primary M. tuberculosis complex lineages.15,16 The database includes geographic origin, drug susceptibility profile and comprehensive genetic lineage information, including the MIRU-24, SNP and RD profiles, spoligotype and IS6110-RFLP fingerprint of each strain. MIRU-24 analysis reliably predicted the correct lineage of greater than 99% of nearly 700 external strains.15,16 The database allows users to compare strains with the reference set, search for similar strains and map geographic information. An expanding genotype nomenclature (MLVA MtbC15-9 type) has been implemented to facilitate comparison between laboratories.16
MIRU typing can be performed either by separate PCRs for each locus and measurement of amplicon sizes by gel electrophoresis or by several multiplex PCRs, using fluorescently labeled primers, and determination of amplicon sizes by capillary electrophoresis in a DNA analyser. The number of tandem repeats (of known length) at each locus is calculated from the amplicon size and results are expressed as a numerical code corresponding to the numbers of repeats at each locus, which is the strain ‘fingerprint’.
In a recent international proficiency study17 of MIRU-24 typing, a panel of 30 DNA extracts, from 20 well-characterised M. tuberculosis strains, was tested in 37 laboratories. There was wide variation in reproducibility of results between laboratories (average 60%); it was better (88%) when commercial kits were used than when in-house-adapted methods using capillary (70%) or gel (50%) electrophoresis were used. However, many in-house-adapted methods achieved greater than 80% reproducibility, suggesting that, with care, they can be used reliably.17 The study demonstrated potential pitfalls of comparing genotypes between laboratories and the importance of quality assurance programs.
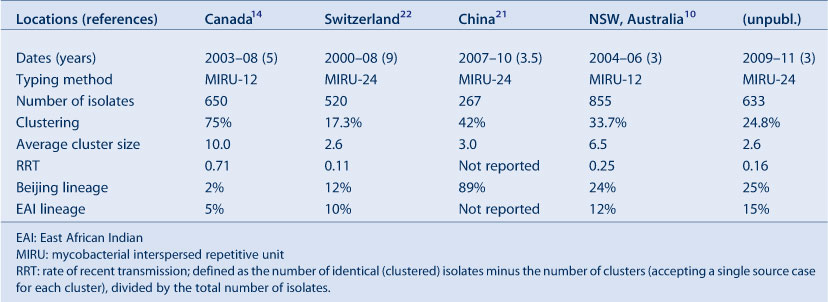
The New South Wales (NSW) Mycobacterium Reference Laboratory has used IS6110-RFLP and spoligotyping since 2004, MIRU-12 since 2005 and MIRU-24 since 2010, to confirm suspected transmission between epidemiologically-related TB cases,18 investigate outbreaks,12 differentiate TB reactivation from reinfection,19 identify otherwise unsuspected case clusters or laboratory contamination and estimate rates of recent transmission.10 The most common spoligotypes in NSW belong to the Beijing family (more than one-quarter of all isolates examined), which was first described in China and neighbouring countries in 1995, and has since been reported in many parts of the world, especially Asia and the former Soviet states. We have tracked an expanding cluster of cases, due to a strain with a unique MIRU-24 profile, among Aboriginal communities in the NSW North Coast region, which has involved at least 30 cases since 2000 (unpublished data; see also Devlin and Passmore20). Table 1 compares our MIRU-typing results with those of several studies showing rates of recent transmission (RRT) using this method.14,21,22 It is worth noting that a low RRT reflects the degree of success of the local TB control program.
|
Despite the improved discriminatory power of MIRU-24 typing, apparent clusters cannot always be confirmed, even by the most detailed contact tracing, suggesting some false clustering, especially in settings with high rates of transmission of closely related M. tuberculosis strains. In China, a high proportion of cases are caused by the so-called ‘modern’ Beijing lineage. Isolates belonging to this lineage have only one or two IS6110 copies and, generally, similar MIRU-24 patterns. False clustering of MIRU patterns occurs due to homoplasy or convergent evolution (independent mutations resulting in the same genotypes among isolates with different ancestry). This can mean, for example, that reinfection with a similar strain could be misinterpreted as reactivation of infection due to treatment failure. In future, problems like this could be resolved by whole genome sequencing, or inclusion of additional targets in typing methods. For example, SNPs have been identified, which can reliably differentiate these otherwise homogeneous ‘modern’ Beijing strains, with the same or very similar, MIRU-24 types, into several lineages. In a recent study, the combination of MIRU-15 typing plus PCR amplification and sequence typing of three hypervariable regions and eight SNPs overcame false clustering; it was therefore proposed for second-line typing of clustered isolates in settings of high prevalence of Beijing strains.21
Whole genome sequencing
The ultimate strain typing system is whole genome sequencing which, until recently, seemed fanciful because of the cost, time taken and requirement for sophisticated bioinformatics tools for sequence analysis. However, recent advances in sequencing technology have dramatically reduced the cost and turnaround time and improved the quality of sequence data and several recent investigations have demonstrated its potential.
Whole genome sequencing was used in the Netherlands to investigate a well-characterised M. tuberculosis transmission chain (confirmed by genotyping and contact tracing) involving five patients aged over 12 years, for which no source had been identified.23 Four SNPs, a tandem-repeat polymorphism and an IS6110 transposition were identified between the first and last isolates of this cluster. The remaining isolates were screened for these six polymorphisms and five were found in isolates from one patient, an alcoholic who had been non-compliant with treatment. It was hypothesised that molecular evolution of M. tuberculosis in vivo may be driven by environmental factors such as intermittent antibiotic use leading to successive bursts of multiplication, or by genomic stress due to alcohol abuse. Identifying factors that stimulate mutational bursts could have major implications for TB management.23 In a Canadian study,24 whole genome sequencing of 32 isolates from a large 3-year community outbreak and four unrelated historical isolates, all with identical MIRU-24 and IS6110-RFLP types, was combined with social network analysis based on patient interviews, to determine the origin and transmission dynamics of the outbreak. Analysis of more than 200 SNPs revealed two genetically distinct lineages, both represented among historical isolates, indicating that they had separated before the outbreak started. Additional, targeted social network and epidemiological analyses identified three co-primary case-patients who had been symptomatic for prolonged periods before diagnosis. The outbreak coincided with an increase in crack cocaine use in the community.24 Whole genome sequencing implicated socio-environmental factors, not identified by conventional genotyping and contact tracing, in triggering simultaneous expansion of two pre-existing M. tuberculosis lineages that were subsequently sustained within a high-risk social network.
The exponential fall in cost and increased speed and quality of whole genome sequencing will continue.25 Only the availability of appropriate tools to analyse and interpret the results will limit its use. It has the potential to rapidly track M. tuberculosis transmission and microevolution and identify virulent and antibiotic resistant strains. When combined with appropriate clinical data this will improve our understanding of disease risks and how to prevent it; allow more timely and individually targeted therapy; and identify new vaccine, antibiotic and diagnostic targets.
Conclusion
In the relatively short period since molecular strain typing of M. tuberculosis has been available and widely used, it has considerably increased our understanding of the evolution and modes of transmission of this ubiquitous and important pathogen. It has demonstrated that transmission can occur as a result of apparently casual, transient contact by leading to more detailed investigation of possible common sources of exposure of apparently unrelated individuals who share the same infecting strain. It has allowed comparison of rates of clustering between different population groups and communities, which reflects and can lead to improvement in the efficacy of TB control programs. Finally, it has provided new insights into the evolution of M. tuberculosis over long periods of time and widely distant geographic regions and also during the course of a single case cluster. As methods have improved from the highly discriminatory but slow and technically demanding IS6110 RFLP to the simpler, but equally discriminatory MIRU-24, genotyping has become more accessible, which means that our understanding of M. tuberculosis evolution and epidemiology has correspondingly increased. Whole genome sequencing promises to provide further insights and even more discriminatory and informative strain typing.
References
[1] van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993; 31 406–9.| 1:STN:280:DyaK3s7mtlaruw%3D%3D&md5=33ae001261a72346b2990f42aed3e699CAS | 8381814PubMed |
[2] Houben RMGJ, Glynn JR. A systematic review and meta-analysis of molecular epidemiological studies of tuberculosis: development of a new tool to aid interpretation. Trop Med Int Health 2009; 14 892–909.
| A systematic review and meta-analysis of molecular epidemiological studies of tuberculosis: development of a new tool to aid interpretation.Crossref | GoogleScholarGoogle Scholar |
[3] Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 1997; 35 907–14.
| 1:CAS:528:DyaK2sXitlartL8%3D&md5=ec9159d5e3ed9af0811990cab8a93c4cCAS | 9157152PubMed |
[4] Soini H, Pan X, Teeter L, Musser JM, Graviss EA. Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J Clin Microbiol 2001; 39 217–21.
| Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXotlCktA%3D%3D&md5=16892a0cac75c0566b99bc9c31a71880CAS | 11136774PubMed |
[5] Filliol I, Driscoll JR, van Soolingen D, Kreiswirth BN, Kremer K, Valetudie G, et al. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J Clin Microbiol 2003; 41 1963–70.
| Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study.Crossref | GoogleScholarGoogle Scholar | 12734235PubMed |
[6] Buu TN, Huyen MN, Lan NTN, Quy HT, Hen NV, Zignol M, et al. The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int J Tuberc Lung Dis 2009; 13 900–6.
| 1:STN:280:DC%2BD1MvksFSmsA%3D%3D&md5=756c19a522551709e75c71180bbf20a2CAS | 19555542PubMed |
[7] Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol 2000; 36 762–71.
| Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXjs1Gmtbk%3D&md5=80b72a0dd3ae9d61da2b5e8865b155c2CAS | 10844663PubMed |
[8] Cowan LS, Diem L, Monson T, Wand P, Temporado D, Oemig TV, et al. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol 2005; 43 688–95.
| Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXit1aqs78%3D&md5=dc43b261ad8088dbafd7ee40bc2b6af6CAS | 15695665PubMed |
[9] van Deutekom H, Supply P, de Haas PEW, Willery E, Hoijng SP, Locht C, et al. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J Clin Microbiol 2005; 43 4473–9.
| Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVOrtLvM&md5=e7aff9e55901a6ae8d6a8a3eac2cdb6dCAS | 16145094PubMed |
[10] Gallego B, Sintchenko V, Jelfs P, Coiera E, Gilbert GL. Three-year longitudinal study of genotypes of Mycobacterium tuberculosis in a low prevalence population. Pathology 2010; 42 267–72.
| Three-year longitudinal study of genotypes of Mycobacterium tuberculosis in a low prevalence population.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXktVSgtLs%3D&md5=af382540d2be1647ad2521d0e829630dCAS | 20350221PubMed |
[11] Allix-Béguec C, Fauville-Dufaux M, Supply P. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2008; 46 1398–406.
| Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis.Crossref | GoogleScholarGoogle Scholar | 18234864PubMed |
[12] Merritt TD, Sintchenko V, Jelfs P, Worthing M, Robinson B, Durrheim DN, et al. An outbreak of pulmonary tuberculosis in young Australians. Med J Aust 2007; 186 240–2.
| 17391086PubMed |
[13] Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006; 44 4498–510.
| Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmslKjsQ%3D%3D&md5=b34947693766b94f9b9b98be3ab0b905CAS | 17005759PubMed |
[14] Christianson S, Wolfe J, Orr P, Karlowsky J, Levett PN, Horsman GB, et al. Evaluation of 24 locus MIRU-VNTR genotyping of Mycobacterium tuberculosis isolates in Canada. Tuberculosis (Edinb) 2010; 90 31–8.
| Evaluation of 24 locus MIRU-VNTR genotyping of Mycobacterium tuberculosis isolates in Canada.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmt1KgsLs%3D&md5=c5b55eb75694b9d997a73b7b354d2609CAS | 20056488PubMed |
[15] Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 2008; 46 2692–9.
| Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates.Crossref | GoogleScholarGoogle Scholar | 18550737PubMed |
[16] Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res 2010; 38 W326–31.
| MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXotVSru7c%3D&md5=a030a0e3e9f74f8f9c97d011cbd259f4CAS | 20457747PubMed |
[17] de Beer JL, Kremer K, Kodmon C, Supply P, van Soolingen D. Global Network for the Molecular Surveillance of Tuberculosis 2009. First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains. J Clin Microbiol 2012; 50 662–9.
| Global Network for the Molecular Surveillance of Tuberculosis 2009. First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains.Crossref | GoogleScholarGoogle Scholar | 22170917PubMed |
[18] Jelfs P, Sintchenko V, Gilbert GL. Genotyping of Mycobacterium tuberculosis in New South Wales: results from 18 months of a statewide trial. N S W Public Health Bull 2006; 17 81–5.
| Genotyping of Mycobacterium tuberculosis in New South Wales: results from 18 months of a statewide trial.Crossref | GoogleScholarGoogle Scholar | 17036086PubMed |
[19] Dobler CC, Crawford ABH, Jelfs PJ, Gilbert GL, Marks GB. Recurrence of tuberculosis in a low-incidence setting. Eur Respir J 2009; 33 160–7.
| Recurrence of tuberculosis in a low-incidence setting.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1M%2Fks1Sktw%3D%3D&md5=8d8c5867963f0b8afe0274bc672bb448CAS | 18829676PubMed |
[20] Devlin S, Passmore E. Ongoing transmission of tuberculosis in Aboriginal communities in NSW. N S W Public Health Bull 2013; 24 38–42.
[21] Luo T, Yang C, Gagneux S, Gicquel B, Mei J, Gao Q. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. J Clin Microbiol 2012; 50 633–9.
| Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xls1eqsbc%3D&md5=56ad842ba4e96e6e17f8ed78ef41498cCAS | 22205801PubMed |
[22] Fenner L, Gagneux S, Helbling P, Battegay M, Rieder HL, Pfyffer GE, et al. Mycobacterium tuberculosis transmission in a country with low tuberculosis incidence: role of immigration and HIV infection. J Clin Microbiol 2012; 50 388–95.
| Mycobacterium tuberculosis transmission in a country with low tuberculosis incidence: role of immigration and HIV infection.Crossref | GoogleScholarGoogle Scholar | 22116153PubMed |
[23] Schürch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, Siezen RJ, et al. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect Genet Evol 2010; 10 108–14.
| The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale.Crossref | GoogleScholarGoogle Scholar | 19835997PubMed |
[24] Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 2011; 364 730–9.
| Whole-genome sequencing and social-network analysis of a tuberculosis outbreak.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXis1ymsb0%3D&md5=99afe3a799c92692740b825382df1139CAS | 21345102PubMed |
[25] National Human Genome Research Institute, National Institutes of Health. DNA sequencing costs. Available at: www.genome.gov/sequencingcosts (Cited 7 August 2012).

