Biogeography of vine thickets and open woodland in subtropical eastern Australia: a case study of three camaenid land snail genera
Lorelle Stanisic A B * , Carmel McDougall B C and Paul Oliver
A B * , Carmel McDougall B C and Paul Oliver  A D
A D
A
B
C
D
Handling Editor: Steven Cooper
Abstract
Dry rainforests and open woodlands occur across much of eastern Australia. However, the biogeographic history of these habitats remains poorly known, especially when compared to nearby moist rainforest areas. Land snails are commonly used as model organisms to understand patterns of origins of regional endemism due to their low vagility. Here we present an analysis of patterns of mitochondrial genetic diversity in three camaenid snail lineages with distributions centred on vine-thicket and open woodland habitats of eastern Queensland, specifically Euryladra from open woodlands, Brigaladra from inland semievergreen vine thickets, and Figuladra from coastal vine thickets. Lineages from habitats west of the Great Dividing Range show relatively low genetic divergence between localities, with particularly low structuring in the open woodland taxon Euryladra. Figuladra from vine-thicket habitats closer to the coast shows relatively deeper genetic divergence, with marked divergences between some upland and lowland areas in south-east Queensland, and across the St Lawrence Gap. This structuring suggests that taxa associated with vine thicket habitats have had a more discernible history of isolation than open woodlands. This said, genetic divergence across many vine thickets patches in lowland coastal regions is also shallow, suggesting many apparently disjunct vine thicket habitats and their associated species also have a recent history of connectivity.
Keywords: biogeography, camaenids, dry rainforest, genetic diversity, fire, land snails, open woodland, vine thicket.
Introduction
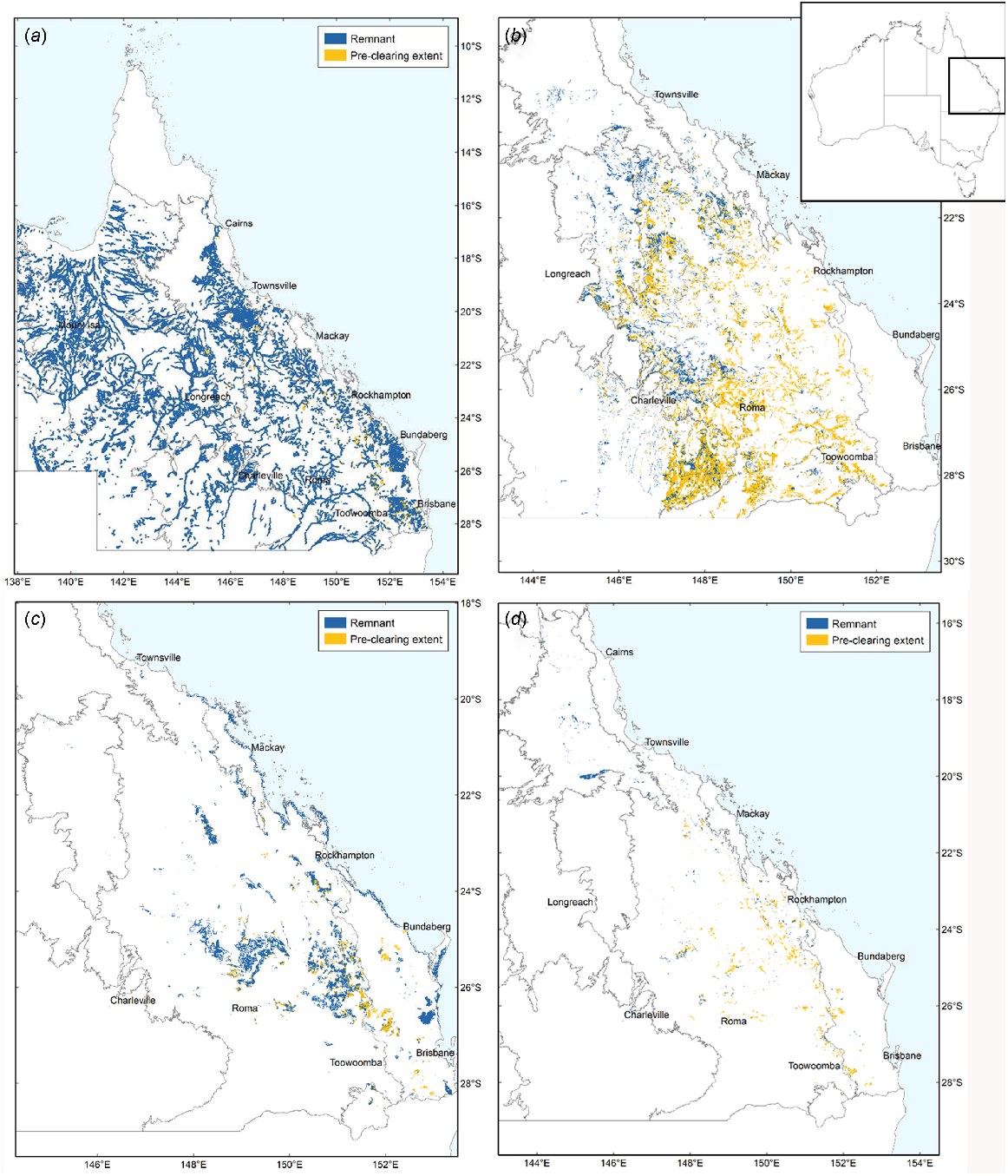
The tropical rainforest biome spans both moist, humid and evergreen forests, and more seasonally dry forests and with typically lower overall rainfall and deciduous vegetation (Pennington et al. 2018). Whereas moist rainforests are relatively well researched, the conservation status and biogeographic history of seasonally dry forests are more poorly known (Webb and Tracey 1994; Dexter et al. 2018; Pennington et al. 2018). Australia is a diverse landmass that features areas of moist, humid rainforest and savannah, but is dominated by arid and semiarid habitats (Fig. 1) (Murphy and Lugo 1986; Byrne et al. 2011; Catullo and Scott Keogh 2014; Monroe 2015) (Table 1). However, along the eastern coast of Australia there are many localised areas of seasonally dry and fire-sensitive rainforest habitats, typically interspersed with areas of more fire-tolerant open woodland and savannah (Figs 1 and 2; Neldner et al. 2019a). The term ‘vine thicket’ is a term widely used to describe these partly deciduous dry forests in Australia (McDonald 2006; Grimshaw 2017). The deciduous nature of the vegetation, a strategy to survive long periods of drought, is one factor separating vine thickets from moist, humid rainforests (Murphy and Lugo 1986; Dexter et al. 2018). Variants of vine thickets include microphyll vine forest (MVF) and coastal microphyll vine thicket (Fig. 2e, g). At the driest limit, dry rainforest communities sometimes grade into other habitats such as semievergreen vine thicket (SEVT) and brigalow scrub (Fig. 2c) (McDonald 1996).
Key vegetation groups in this study in Queensland, eastern Australia. (a) Open forests and woodland. (b) Poplar box and silver-leafed ironbark woodland. (c) Vine thicket (notophyll vine thicket, microphyll vine forest and coastal vine thicket). (d) Semievergreen vine thicket (SEVT). Inset: Map of Australia. Legend indicates the current remnant forests in blue and the cleared forests in yellow. Source Neldner et al. (2019a). Creative Commons Attribution 3.0 Australia (CC BY) licence.
Biome | Vegetation group | Terminology | Description | Study area | |
|---|---|---|---|---|---|
Subtropical forest: a closed-canopy rainforest and dry rainforest. | Dry rainforests and vine thickets | Complex microphyll vine forest (CMVF) Semideciduous microphyll vine forest (MVF) | Complex to simple, semideciduous microphyll sometimes with Araucaria cunninghamii (hoop-pine). | SEQ, MEQ | |
Microphyll vine forest/thicket on coastal dunes and sand masses. | SEQ, MEQ | ||||
Microphyll vine forest with feather or fan palms on alluvia, along streamlines and in swamps on ranges or within coastal sand masses. | SEQ, MEQ | ||||
Notophyll vine forest (NVF) | Notophyll vine forests, frequently with Araucaria spp. or Agathis spp. (kauri pines) | SEQ (high summits only) | |||
Notophyll vine forest and microphyll fern forest to thicket on high peaks and plateaus | SEQ, MEQ | ||||
Semievergreen vine thicket (SEVT) | Semievergreen to deciduous microphyll vine thicket. | SEQ, MEQ SCQ | |||
Woodland and savanna | Eucalypt Open Forests | Wet eucalypt tall open forest | Wet eucalypt tall open forest on uplands and alluvia. | SEQ, MEQ SCQ | |
Eastern eucalypt woodlands | Moist to dry eucalypt open forests to woodlands usually on coastal lowlands and ranges. | SEQ, MEQ | |||
Corymbia citriodora (spotted gum) dominated open forests to woodlands on undulating to hilly terrain. | SCQ | ||||
Moist to dry eucalypt open forests to woodlands mainly on basalt area | SEQ SCQ | ||||
Dry eucalypt woodlands to open woodlands, mostly on shallow soils in hilly terrain | SEQ SCQ | ||||
Dry to moist eucalypt woodlands and open forests, mainly on undulating to hilly terrain | SEQ, MEQ SCQ | ||||
Temperate eucalypt woodlands. | SCQ | ||||
Eucalypt open forests to woodlands on floodplain | Eucalyptus spp. dominated open forest and woodlands drainage lines and alluvial plains | SCQ, MEQ | |||
Melaleuca Forests and Woodlands | Melaleuca open woodlands | Melaleuca spp. dry woodlands to open woodlands on sandplains or depositional plains. | SEQ, MEQ SCQ | ||
Melaleuca spp. on seasonally inundated open forests and woodlands of lowland coastal swamps and fringing drainage lines. | SEQ, MEQ SCQ | ||||
Callitris Forests and Woodlands | Callitris woodland–open forests | Callitris glaucophylla (white cypress pine) or C. intratropica (northern cypress pine) woodlands to open forests | SCQ | ||
Acacia Forests and Woodlands | Mulga | Acacia aneura (mulga) dominated associations on red earth plains, sandplains or residuals | SCQ | ||
Brigalow | Acacia harpophylla (brigalow) sometimes with Casuarina cristata (belah) open forests to woodlands on heavy clay soils. | SCQ | |||
Shrubland In Australia, precipitation is widely considered | Complex of open shrubland to closed shrubland, grassland, low woodland and open forest, on strand and foredunes. Includes pure stands of Casuarina equisetifolia (coastal she oak). | MEQ, SEQ | |||
Acacia open forest | Open forest to woodland dominated by Acacia | MEQ, SCQ |
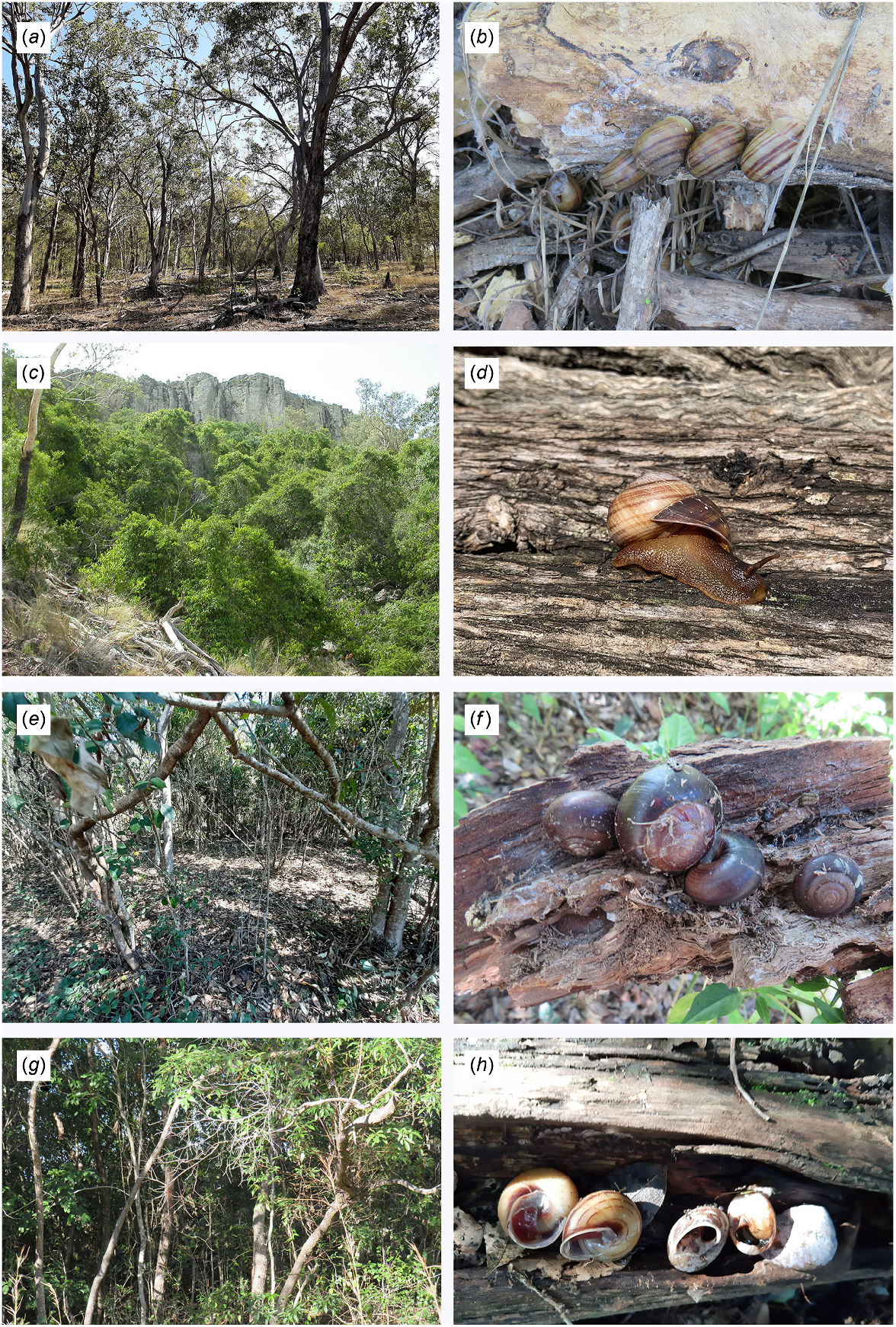
Exemplars of habitat types and related land snail species. (a) Open woodland, Rodds Peninsula, SEQ. (b) Euryladra mattea on roots of open Eucalyptus woodland, Horrigan Rd, Raglan, SEQ. (c) SEVT (dry vine thicket) on rocky scree, Lord’s Table, Peak Range, SCQ. (d) Brigaladra volgiola on log in semievergreen vine thicket, Foleyvale, Duaringa, SCQ. (e) Riparian vine thicket, McClure Rd, Mt Coulston, SEQ. (f) Figuladra lessoni, under log in coastal vine thicket, Boyne I., SEQ. (g) Upland microphyll vine forest, Connors Hump, MEQ. (h) Figuladra bayensis in aestivation inside log, microphyll vine forest, at Mt Mudlo, SEQ. Image d: Craig Eddie.
Mesic ecosystems are central to understanding the evolution of Australia’s terrestrial biota (Byrne et al. 2011). The distribution of genetic diversity and biogeographic history of Australia’s vine thickets remains very poorly understood, especially when compared against the biota of Australia’s wet rainforests, which have been extensively studied for many decades (for example: Webb and Tracey 1981; Hugall et al. 2003; Byrne et al. 2011; Weber et al. 2014; Bryant and Krosch 2016). This knowledge gap is particularly concerning because vine thickets are more localised and have been more extensively cleared than moist, humid rainforests (Metcalfe and Bui 2016; Accad et al. 2017).
Land snails are key environmental indicators and biodiversity predictors that enable monitoring of the health of terrestrial ecosystems and identification of possible pressures or threats (Stanisic 1998, 1999; Stanisic and Ponder 2004). Camaenid land snails are a family of forest and woodland snails that are often closely associated with vine thicket habitats in Australia (Stanisic 1996; Stanisic and Ponder 2004; Stanisic et al. 2022), suggesting they are an excellent model group for the biogeographic study of this vegetation assemblage. Figuladra Köhler & Bouchet 2020 is a genus of camaenid occurring predominately in vine thickets of Queensland in eastern Australia, ranging from south-eastern Queensland (SEQ) to mid-eastern Queensland (MEQ) (Stanisic et al. 2010) (Figs 2 and 3). The distribution of Figuladra also spans the St Lawrence Gap, a stretch of low-lying open woodland and savanna between St Lawrence and Rockhampton in mid-eastern Queensland, which has been identified as an important barrier for many mesic taxa (Bryant and Krosch 2016; Simpson et al. 2018). The habitat in this area is dominated by dry, open sclerophyll woodland and savanna, with no significant stands of closed forest (Bryant and Krosch 2016), so it would be predicted that dry rainforest associated Figuladra might also show turnover at this zone.
Map of study area showing localities for three land snail genera included in this study (Brigaladra, Euryladra and Figuladra) based on distributions estimated using specimen records in the Queensland Museum.
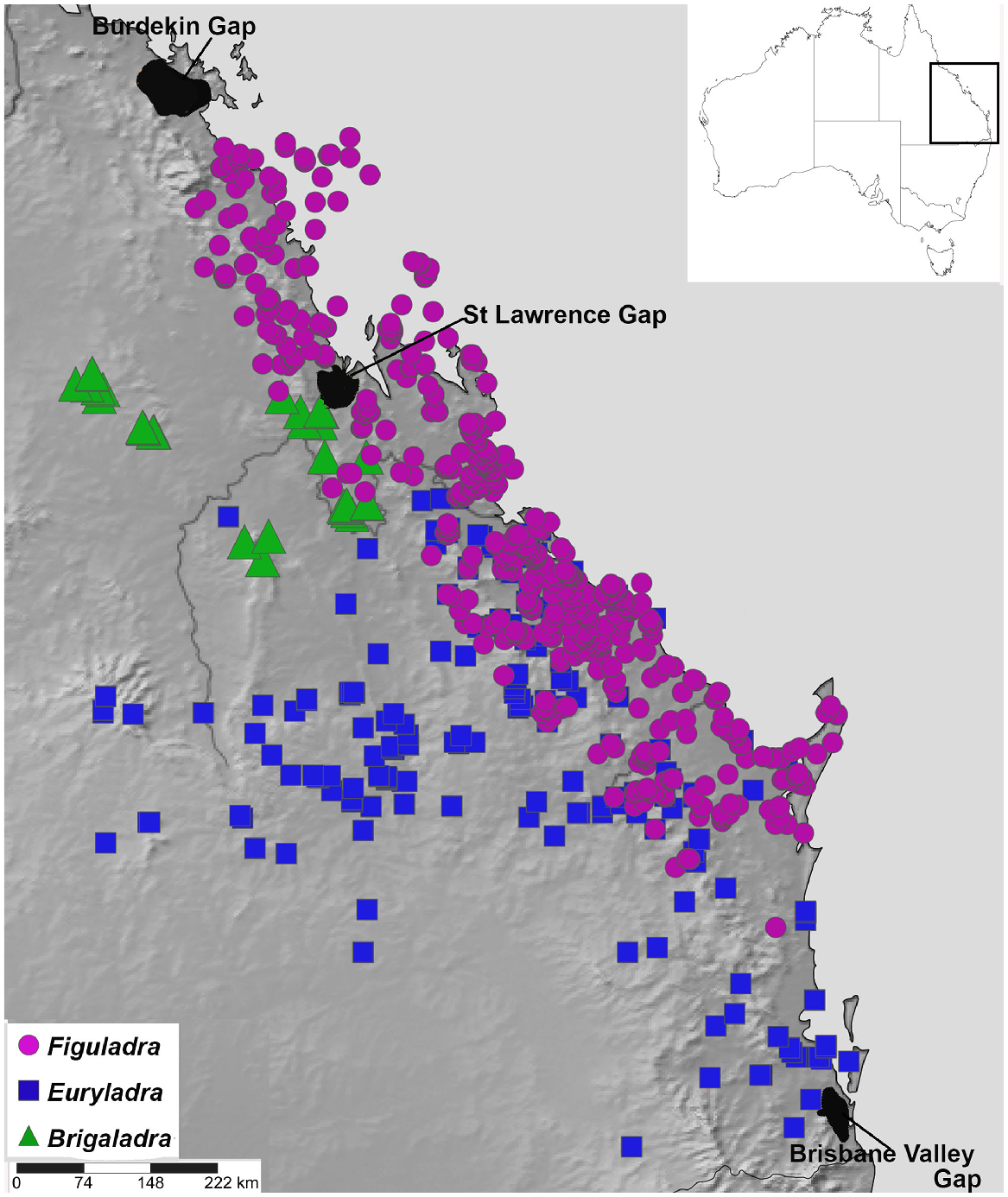
Although Figuladra species are closely associated with vine thickets, two other closely related and recently described monotypic genera that occur in the same general region range more widely into open woodland habitats where fire is more frequent. Euryladra mattea can be found in the open woodlands and edges of vine thickets in SEQ and west of the Great Dividing Range as far as Injune, south central Queensland (SCQ). Brigaladra volgiola is found in upland vine thickets in MEQ (Boomer Range, Broadsound Range, Peak Range and Expedition Range) and riparian lines in the adjacent woodland. Based on the much wider and more continuous nature of open woodland habitats (Fig. 1a, b), one prediction would be that E. mattea may show less of a signature of isolation than species associated with more restricted vine-thicket habitats.
Comparison of patterns of genetic diversity and structuring can be particularly useful for understanding if ecologically varying species show evidence of similar or different responses to environmental variation. With respect to vine thickets and nearby woodlands, genetic analyses are few. Lizards from the Brigalow Belt have been found to show moderate diversity in a species associated with upland areas, but very shallow diversity in a more widespread lowland species associated with brigalow forests (Wilmer et al. 2020). Genetic analyses of specialised fossorial skinks in vine thickets has revealed higher levels of localised diversity and endemism (Amey et al. 2018). Biogeographical studies of the terrestrial invertebrate taxa in the dry rainforests of Queensland are even more sparse. Stanisic (1999) represents one of the few studies of invertebrate taxa utilising land snails of the Brigalow Lands. Land snails are often endemic to very localised vine thicket patches (Solem 1979; Stanisic 1990, 1994, 1998). However, this work has not so far incorporated genetic data and we have no understanding of the timing of divergence between these isolated snail populations.
Here, we use these three camaenid snail lineages (Figuladra, Euryladra, and Brigaladra) to address the following questions. First, do lineages associated with seemingly isolated contemporary vine thicket habitats show deep genetic structuring and diversity, indicative of histories of isolation and diversification within this habitat, or, conversely, has there been extensive recent connectivity? We pay particular attention to patterns of genetic divergence at potential barriers or turnover points in central Queensland that have been identified for other taxa, expressly the St Lawrence Gap. Second, we consider whether species associated with vine thickets show higher levels of genetic diversity and structuring when compared with species in nearby, and seemingly more continuous, open woodland habitats.
Materials and methods
Sampling
We obtained genetic data from 93 specimens of land snails in the genera Brigaladra, Euryladra and Figuladra, largely collected by the first author and all now registered into the wet collection of the Queensland Museum (Supplementary Table S1). Eight additional sequences from previous phylogenetic analyses of camaenids (Clade 1, Hugall and Stanisic 2011: fig. 7; Stanisic et al. 2022) were downloaded from Genbank (Table 2). We sampled across the full geographic range of all recognised and candidate taxa. The final alignment included one outgroup sample from araucarian dry rainforest in north-eastern Queensland, Monteithosites heliostracum Stanisic, 1996. Codes used in haplotype networks and phylogenetic analyses represent nominal or putative species (identified in Stanisic et al. 2022, or proposed based on morphology and collection locality) and are as follows: AP, Figuladra appendiculata; AU, F. aureedensis; B, F. barneyae; BR, camaenid SQ16; C, F. challisi; F, camaenid BL47; FB, F. bayensis; FN, F. narelleae; IN, F. incei incei; L, F. incei curtisiana (lessoni); M, F. muirorum; MAT, Euryladra mattea; P, F. pallida and camaenid BL45; RD, F. reducta; RI, camaenid SQ15; V, Brigaladra volgiola; VL, camaenid SQ17.
| Species | GenBank Id | Locality | |
|---|---|---|---|
| Billordia nicoletteae | GQ851018.1 | Sarina MEQ | |
| Camaenid BL47 | G851068.1 | Seaforth, MEQ | |
| Camaenid SQ17 | G851073.1 | Bulburin State Forest, SEQ | |
| Figuladra barneyae | G851100.1 | Connors Hump, MEQ | |
| Figuladra incei incei | G851011.1 | Cullen I, MEQ | |
| Euryladra mattea | G851106.1 | Taroom, SCQ | |
| Brigaladra volgiola | GQ851328 | Broadsound Range, SEQ | |
| Monteithosites heliostracum | GQ851007.1 | Bakers Blue Mt, NEQ |
Vegetation classifications
To understand how genetic diversity in Figuladra snails was distributed across habitats, we used a system of representative broad vegetation groups (BVG) that has been established for the Australian state of Queensland. Neldner and colleagues (2019b) categorise Queensland’s broad vegetation groups to reflect the vegetation structure along a mesic gradient from wet closed forests (rainforests) of the coast and north-east, to the arid spinifex hummock grasslands of the south-west. In eastern Queensland south of the Wet Tropics, the BVGs with their biogeographic and landscape attributes comprise subtropical rainforest, dry rainforest, open woodland and savanna encompassing three regions, MEQ, SEQ and SCQ (Table 1). The vast majority of snails included in the study were collected from dry rainforests (vine thickets) or open woodlands.
DNA extraction, amplification and alignment
DNA was extracted from the foot muscle using the QIAGEN DNeasy tissue kit with the Spin Column protocol. Fragments of one partial mitochondrial gene (16S) were amplified using 16SCS1 (Chiba 1999) and 16SBD1 (Sutcharit et al. 2007) primers. Each 25 μL reaction was run using 12.5 μL Phusion High-Fidelity DNA Polymerase with 0.625 μL of each primer and 0.3 μL of DNA. Reactions were performed under standard thermoprofile conditions (ThermoFisher Scientific) viz 16S mtDNA: 98°C for 2 min, followed by 35 cycles of 98°C for 30 s, 48°C for 30 s, and 72°C for 30 s and a final elongation step at 72°C for 5 min. The size of the PCR products was checked under UV transillumination after gel electrophoresis in a 1.5% agarose gel and TAE buffer solution. The PCR products were sent for purification and commercial sequencing at Macrogen Inc (Geumchum = Gu, Korea). Chromatographs were checked and aligned using the MAFFT v7.308 (Katoh and Standley 2013) plugin in Geneious 9.1.8 (Kearse et al. 2012) with default settings. We used Guidance2 (Sela et al. 2015) to identify and remove unreliably aligned regions in the 16S alignment by employing default settings.
Phylogenetic analysis
Phylogenetic analyses were performed using Maximum Likelihood (ML) and Bayesian Inference (BI). A Maximum Likelihood phylogeny was reconstructed using IQ-Tree 1.6 (Nguyen et al. 2015, univie.ac.at) under the best-fit model using ModelFinder in the software. Nodal support was inferred by performing 1000 ultrafast bootstrap replicates. Bayesian posterior probabilities were estimated by running a 1 000 000 generation Metropolis-coupled Markov Chain Monte Carlo (four chains, one heated, sampling rate 1000 generations) with a burn-in of 100 000 as implemented by MrBayes (Ronquist et al. 2012) plug-in in Geneious 9.1.8.
Population genetic summaries and demographic analysis
For each major lineage, the number of mutations, distinct haplotypes, haplotypic diversity and nucleotide diversity was determined using DnaSP v6.12.03 (Rozas et al. 2017; www.ub.edu). The number of variable and parsimony-informative sites in the alignment and pairwise sequence divergences (uncorrected-p distance) within and between target species groups were calculated using MEGA6 v10.2.6 (Tamura et al. 2013; https://www.megasoftware.net). TCS haplotype networks (Clements and Northrop 2002) for lineages were inferred using the 16S data with the program PopART (Leigh and Bryant 2015; http://popart.otago.ac.nz).
We tested for varying signatures of range expansion in each of the major lineages using two neutrality tests: Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) performed in DnaSP v6 using the 16S alignment data. In the absence of selection, these indices will reflect the demographic history of the populations: significant negative values are interpreted as evidence of demographic expansion and significant positive values suggest population structure. Estimated values are compared against a null distribution to test whether they are significantly different from populations where genetic drift and mutation are at equilibrium (Fig. 3).
Results
Sequence alignment and summary details
The final alignment comprised 101 individuals, including the single outgroup sample from north-eastern Queensland (NEQ), Monteithosites heliostracum Stanisic, 1996. Modeltest, as implemented in IQ-Tree, identified TN + F + I + G4 as the best-fit model of molecular evolution. The ML alignment of all 101 sequences had 771 columns of which 267 were parsimony-informative, 52 were singleton sites and 452 were invariant sites.
Major lineages
The ML and BI analyses produced largely identical topologies (Fig. 4). In all analyses, five major strongly supported and largely geographically disjunct lineages were identified. Three of these lineages are in the genus Figuladra. The first major lineage was found in the MEQ region and is thus called the MEQ lineage. It comprises three recognised species and one putative species in mid-eastern Queensland, ranging from St Helen’s Beach in the north to Stanage Bay in the south, and adjacent islands off the coast. Species in this group occur in seasonally dry tropical forest habitats at typically low elevation, ranging from microphyll vine forests to dry araucarian rainforests. The second lineage was found in SEQ and is named as such. This lineage contains five recognised species and four putative taxa from riparian and coastal vine thickets between the Boomer Range and Inskip Point south of Fraser Island. A final lineage within Figuladra (SEQ Uplands) occurs in the foothills and low ranges of the eastern escarpments of the Great Dividing Range (GDR) and contains two recognised species occupying microphyll vine forest habitat. The range of the two species in SEQ uplands was disjunct, and seemingly interdigitated by the distribution of the SEQ clade. The MEQ and SEQ clades occur within 100 km of each other in the region corresponding to the St Lawrence Gap in mid-eastern Queensland. The other two major lineages correspond to Brigaladra from SCQ and Euryladra from SEQ and SCQ.
Phylogram of Figuladra, Euryladra and Brigaladra from eastern Queensland based on ML and BI phylogenetic analyses of 16S sequences. Numbers on top of branches represent bootstrap values, below branches posterior probabilities. Each clade is related to its associated habitat and vegetation type. Numbering and colour correspond to haplotype networks. Tip labels represent individual specimens and codes relate to species groups; these are also used in Figs 5 and 6. Scale bar indicates 3% of modelled sequence divergence.
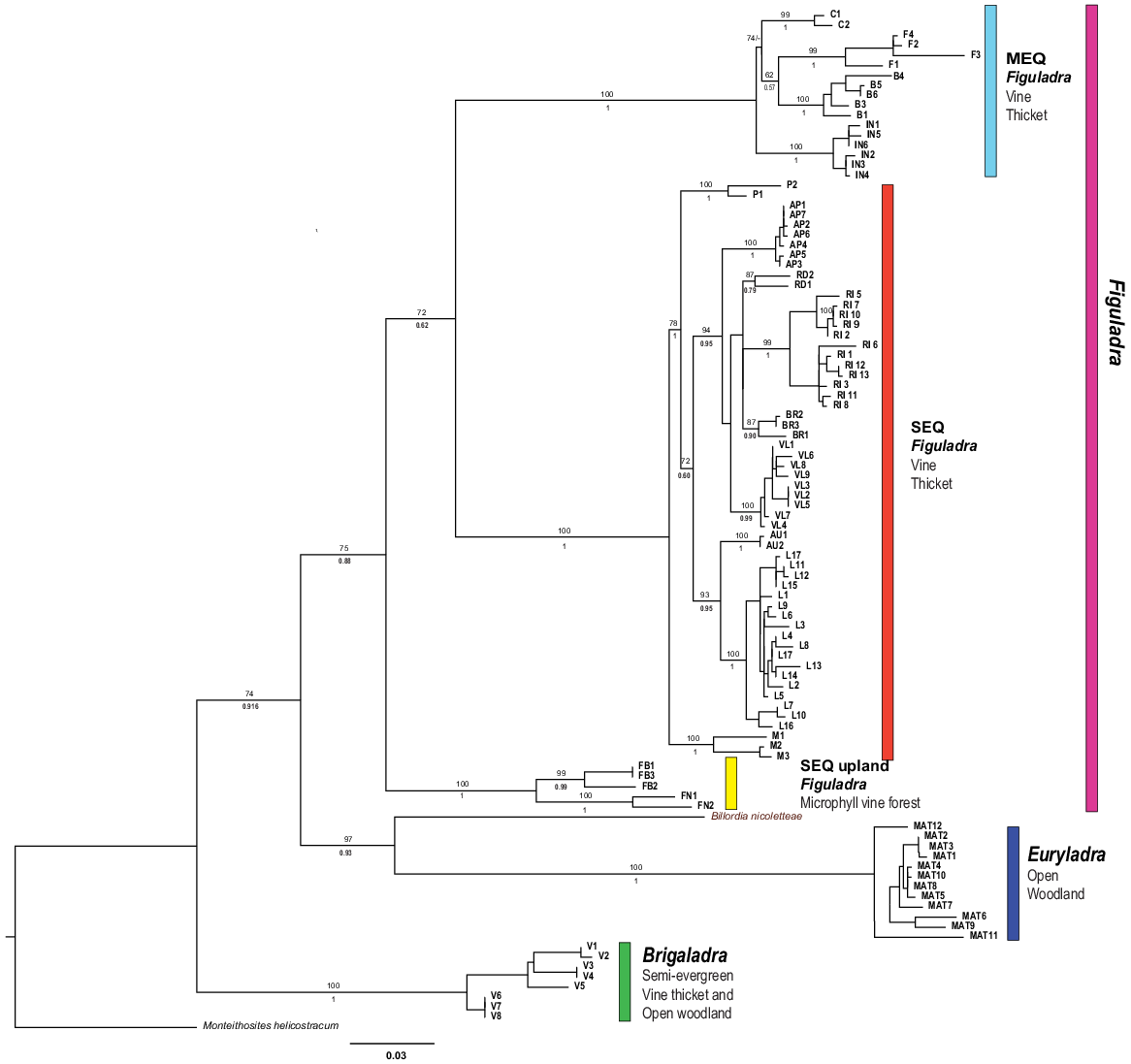
Pairwise sequence divergences (p-distances) between the five main lineages ranged from 9.7 to 17% (Tables 3 and S2). The lowest divergences were found between the three lineages of Figuladra (9.7–13.3%); the two monotypic genera were more divergent (15.4–17%). The St Lawrence Gap marked a geographic point that separates the SEQ and MEQ lineages, and substantial genetic divergence was seen between species living on either side (11.4–13.3%) of the gap. The SEQ and SEQ upland clades are similarly divergent (9.7–13.3%), and as noted above appear to show some elevational segregation where they come into close contact, with SEQ upland being restricted to higher elevations, and SEQ more widespread in lowlands.
| Clades | SEQ | MEQ | SEQ upland | Euryladra | |
|---|---|---|---|---|---|
| SEQ | |||||
| MEQ | 12.0 (11.4–13.3) | ||||
| SEQ upland | 10.0 (9.7–13.3) | 12.5 (11.4–13.6) | |||
| Euryladra | 16.2 (15.1–16.5) | 15.9 (15.4–17) | 15.4 (15–15.6) | ||
| Brigaladra | 14.9 (14–15.1) | 13.9 (13.3–14.6) | 14.0 (13.8–14) | 16.5 (14–16.7) |
Genetic diversity within lineages
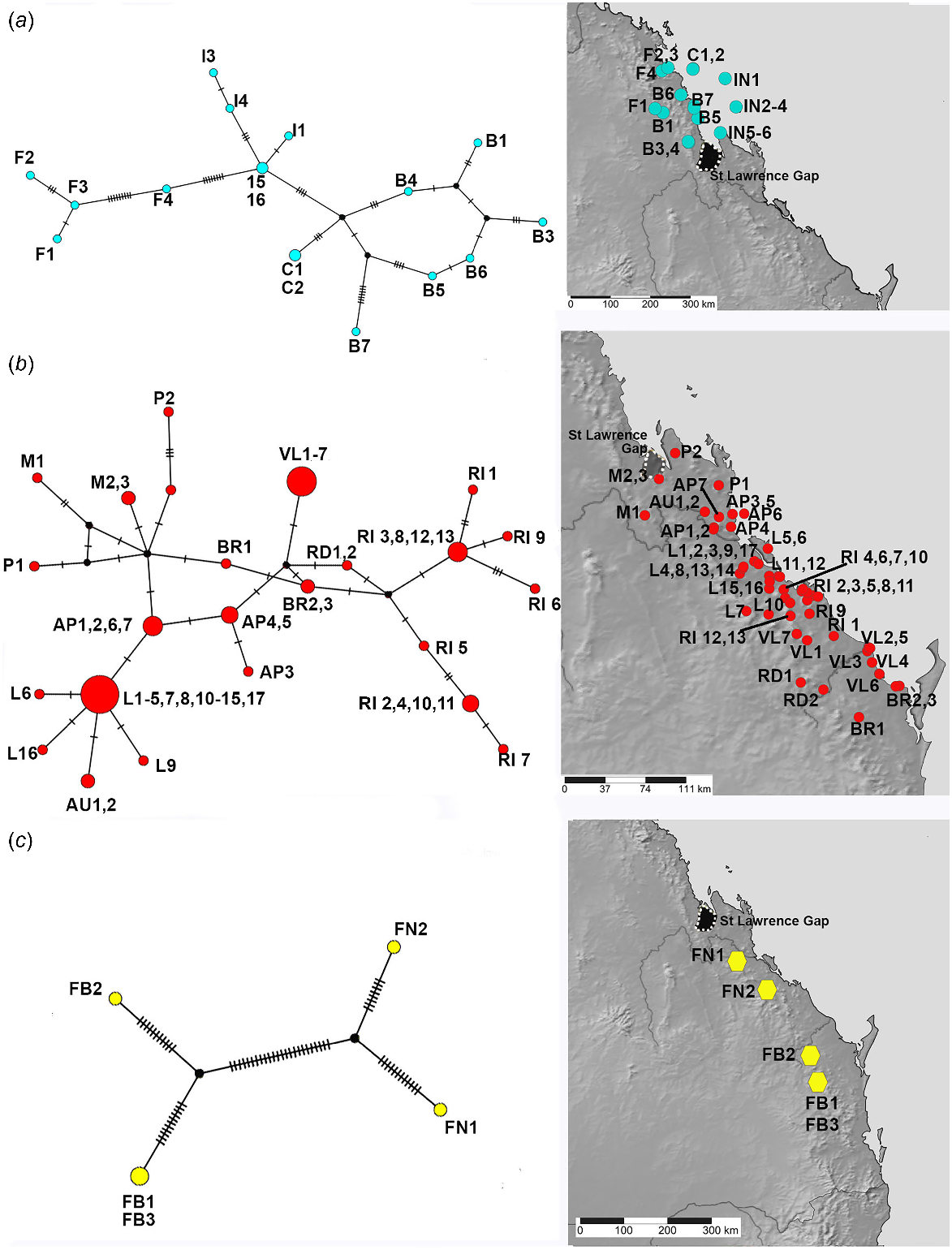
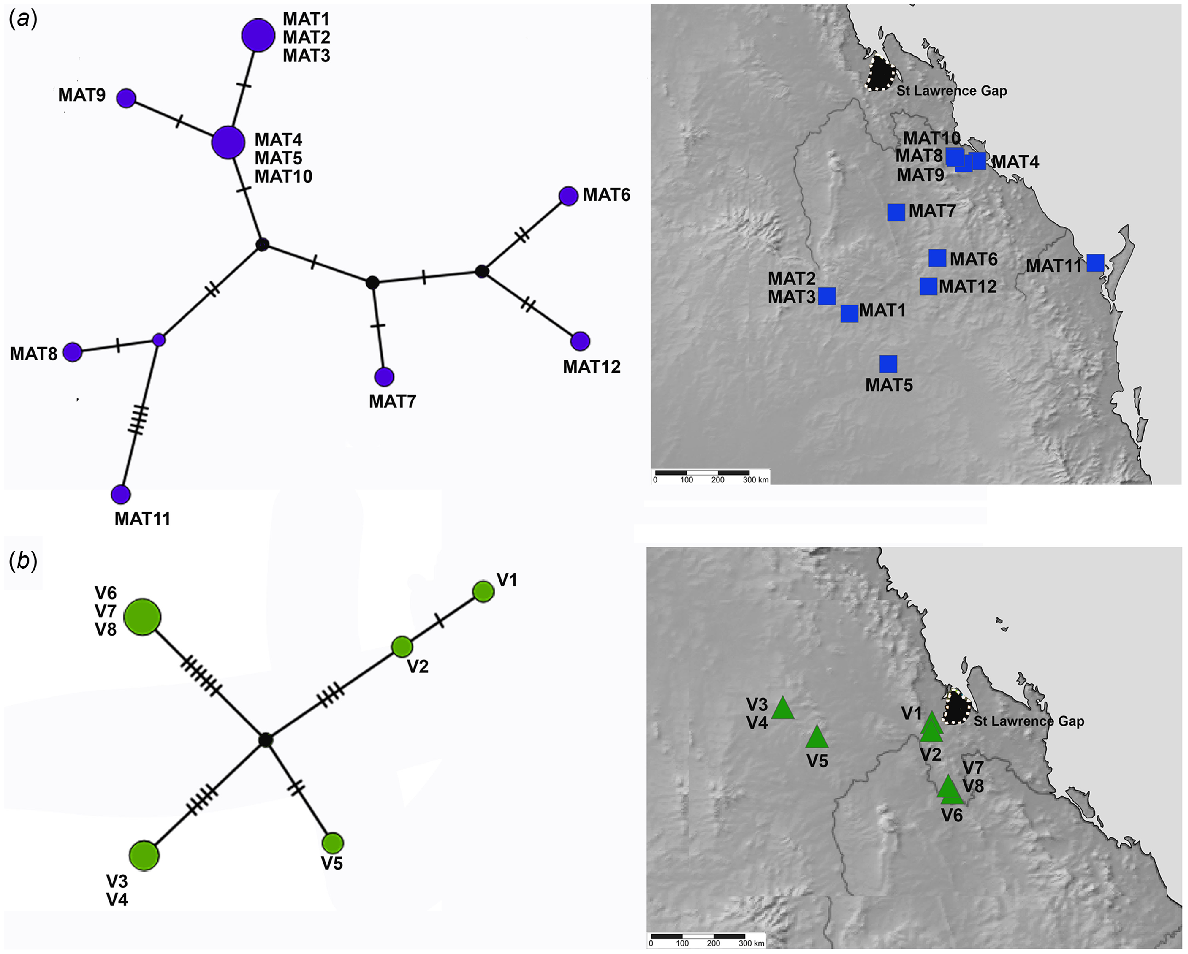
The better sampled and more widespread coastal and lowlands vine thicket associated SEQ and MEQ lineages of Figuladra show the highest number of distinct haplotypes, but lower values for the overall number of mutations, average pairwise differences and nucleotide diversity (Table 4, Fig. 5a, b). The MEQ lineages also show the highest values for haplotypic diversity (Hd). In contrast, seemingly disjunct populations of the SEQ upland clade from isolated patches of microphyll vine forest were sparsely sampled (n = 5) and showed the lowest number of distinct haplotypes (n = 4), but showed the highest pairwise differences (k), highest nucleotide diversity (π) and the greatest number of mutations (M) (Table 4, Fig. 5c). The widespread open woodland lineage Euryladra shows a relatively low number of distinct haplotypes, mutations and haplotypic diversity, and the lowest observed average pairwise differences and nucleotide diversity (Table 4, Fig. 6a). Finally, when compared against other lineages sampled, apparently disjunct populations of the more sparsely sampled inland vine thicket lineage Brigaladra show low numbers of distinct haplotypes and mutations, and moderate nucleotide diversity and average pairwise differences (Table 4). This lineage also shows the lowest observed values for haplotypic diversity (Table 4, Fig. 6b).
| Clades | n | M | h | Hd | π | k | Tajima’s D (p) | Fu’s F test | Fu’s FS (p) | |
|---|---|---|---|---|---|---|---|---|---|---|
| MEQ vine thicket species | 18 | 42 | 15 | 0.9800 | 0.03148 | 11.33 | −0.29384 | −3.340 | −0.32090 | |
| SEQ vine thicket species | 60 | 29 | 24 | 0.9034 | 0.02048 | 3.850 | −1.23155 | −10.927 | −2.90365 | |
| SEQ upland vine forest species | 5 | 66 | 4 | 0.9000 | 0.04746 | 32.70 | 0.24380 | 3.643 | 0.36827 | |
| Euryladra SEQ/SCQ open woodland species | 12 | 17 | 8 | 0.9091 | 0.01102 | 4.045 | −1.22092 | −1.645 | −1.37907 | |
| Brigaladra SCQ SEVT and open woodland species | 8 | 18 | 5 | 0.8571 | 0.02061 | 8.143 | 0.89521 | 1.890 | 1.23763 | |
| All groups | 102 | 0.09336 | 0.63257 |
n, number of samples; M, number of mutations; h, number of unique haplotypes; Hd, haplotypic diversity; π, nucleotide diversity; k, average number of pairwise differences (for details see Table S3).
TCS haplotype network and associated maps inferred from mitochondrial 16S sequences of species associated with vine thickets. (a) MEQ lineage. (b) SEQ lineage. (c) SEQ upland with Figuladra narelleae (FN) and F. bayensis (FB). Sizes of the circles are proportional to the number of individuals with that haplotype. The number of mutational steps along the branches is indicated.
TCS haplotype network and associated maps inferred from mitochondrial 16S sequences of species associated with more open and inland woodlands. (a) Euryladra mattea (MAT). (b) Brigaladra volgiola (V) (both open woodland and semievergreen vine thicket). Sizes of the circles are proportional to the number of individuals with that haplotype. The number of mutational steps along the branches is indicated.
A negative value of Tajima’s D was inferred for the MEQ and SEQ coastal lineages as well as the wide-ranging Euryladra, suggesting recent expansion. Conversely, the two upland groups, SEQ upland and Brigaladra, show a positive Tajima’s D value, suggesting balancing selection and no recent genetic sweeps. However, for most clades these values are not statistically significant, with only the SEQ lineage having a significant negative value of the Fu’s F test (indicative of recent population expansion). Several of these lineages are considered to be complexes of several species, violating one assumption of the range expansion test (single interbreeding population) and accordingly some of these results may have to be treated with caution.
Patterns within key vegetation communities
Figuladra species from vine thicket environments on the east coast of Queensland show moderate genetic diversity with an average genetic diversity between samples of 4.6% (Table S3). Particularly high diversity with an average p-distance of 6.4%, is evident in the SEQ upland clade which mostly inhabits microphyll vine forest. The Euryladra clade from open woodlands shows lower diversities (1.9%). The upland Brigaladra, which inhabits open woodland and vine thicket on rocky scree in SCQ, shows average genetic diversity intermediate between the other two lineages (2.8%).
Discussion
Here we present a broadscale analysis of comparative mitochondrial diversity patterns in a suite of related snail lineages associated with vine thicket and open woodland habitats in southern to central Queensland. We hypothesised that taxa associated with continuous, drier and more widespread open woodland habitats would show lower genetic diversity than taxa associated with comparatively mesic and seemingly disjunct patches of vine thicket. Our data broadly support this hypothesis and reveal particularly deep divergences at the St Lawrence Gap and in one clade associated with upland areas. However, even within vine thickets, some apparently disjunct populations in upland areas west of the GDR, and also within the SEQ and MEQ ranges, showed relatively shallow divergences.
Upland refugia
Altitudinal endemism and biotic refugia in eastern Australia have highlighted key areas of persistence in wetter rainforests in higher montane elevations such as Eungella (Ashton et al. 2020), the Wet Tropics (Yeates and Monteith 2008) and Border Ranges (Ashton et al. 2016). Here we found that the SEQ upland lineage showed very high nucleotide diversity and p-distances, despite only five samples being available. This genetic structuring and apparently disjunct distribution suggest this lineage has a history of localised persistence in relatively upland habitats, but in an area of eastern Australia that has not, as far we are aware, previously been highlighted as a refugium. The ranges inhabited by the two species in the SEQ upland clade (100–750 m above sea level) are relatively low and characterised by araucarian microphyll vine forest, as opposed to coastal vine thicket which is often associated with slightly drier conditions (Fig. 7). Our results, and similar discoveries of potentially localised and undescribed upland endemic mygalomorph spiders (Rix et al. 2020), highlight the probability that even relatively low ranges in south-eastern Queensland may have enabled the persistence of restricted-range taxa. Other taxa characterised by localised endemism such as mygalomorph spiders remain poorly sampled in this area, emphasising a need for further surveys.
Detailed map of the distribution of the SEQ upland clade of Figuladra distribution showing specimen records for Figuladra narelleae and F. bayensis embedded within the distribution of the SEQ clade species, F. reducta.
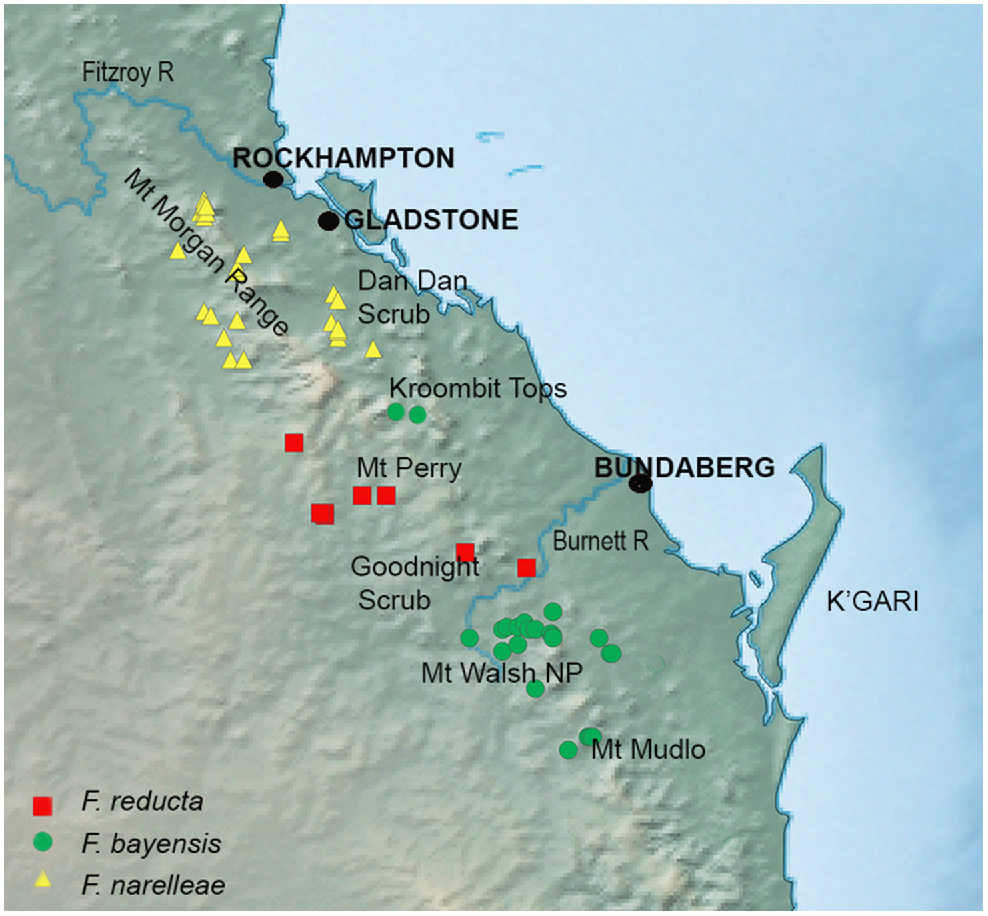
The ranges of the two recognised species in the SEQ upland clade (F. bayensis and F. narelleae) are intersected by outwardly similar upland areas (Mt Perry and the Goodnight Scrub) that are home to Figuladra reducta, which is embedded within the more widespread SEQ clade rather than the SEQ upland lineage. There are no localities where the species within the SEQ uplands and SEQ clades occur together, with the closest region being Mt Woowoonga National Park (microphyll vine forest, F. bayensis, SEQ upland clade) and the Goodnight Scrub (dry vine thicket, F. reducta, SEQ clade) west of Gin Gin, SEQ. These two upland areas are separated by the Burnett River, a major river system in SEQ and differing vine thicket habitats. Although the exact historical processes that have shaped the distribution of SEQ upland lineages are unknown, the existence of a wide gap suggests former connectivity and then extinction in the intervening areas – perhaps driven either by climatic change, or potentially even interactions with the more widespread SEQ clade. There are no known areas of sympatry between Figuladra species, providing some support for the idea that competition or displacement may well play a role in maintaining the boundaries of populations in the SEQ and SEQ upland lineages. More geographically comprehensive and larger sample sizes coupled with genomic analyses could provide a means to further understand the embedded and contrasting distributions of the SEQ upland and SEQ lineages, with the prediction being that the former will show evidence of range contraction and bottlenecking, and the latter will show evidence of range expansion.
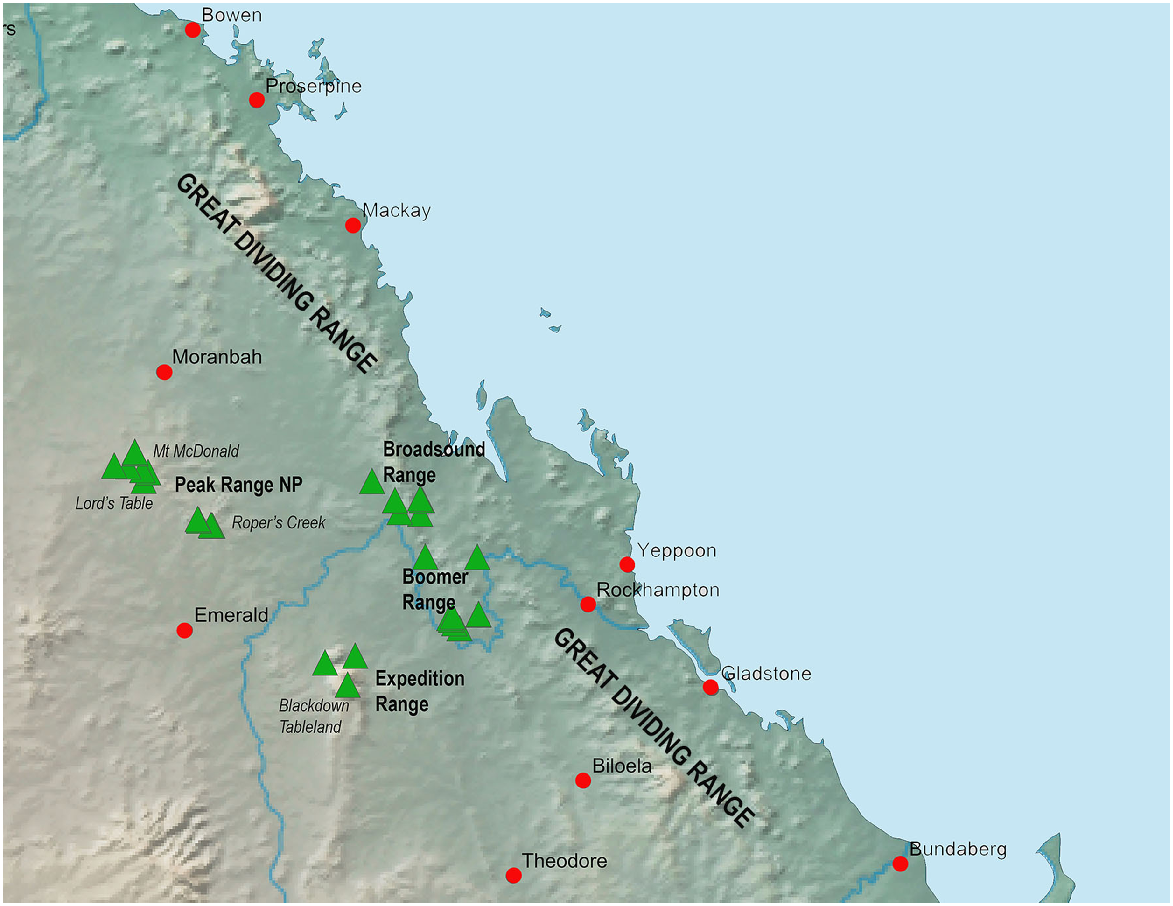
The Brigaladra lineage is also associated with seemingly isolated low ranges with extensive rocky habitat west of the GDR, typically, but not always, in areas of drier semievergreen vine thicket (SEVT) on scree slopes. This lineage occurs across the Broadsound, Boomer, Expedition and Peak Ranges (Fig. 8), most of which are separated by very dry lowlands dominated by open woodland in which this taxon has not been observed. Populations sampled from across the range of this taxon show genetic divergence much lower than the SEQ upland clade (average intraspecific p-distance 2.8%), and also lower than the lower elevation woodland and vine thicket clades of Figuladra. This suggests relatively recent connectivity between these inland upland areas. Profound environmental change through major glacial cycling may have influenced the distribution of the Brigaladra, and the current distribution may be relictual. This lineage also shows an ability to use open woodland environments on the boundaries of vine thickets, suggesting ecological plasticity, which may enable dispersal across apparent lowland barriers.
Genetic patterns across the lowlands
The St Lawrence Gap is an area of relatively low and dry open woodland that is an important biogeographic barrier for many taxa, especially those associated with relatively wet habitats (Burke et al. 2013; Bryant and Krosch 2016). However, there is striking variation in both temporal and distributional divergence patterns across this region. Some wet forest taxa show very deep structuring indicative of millions of years of isolation (Hugall et al. 2003; Bryant and Krosch 2016), whereas others, especially those associated with slightly drier habitats, show shallower divergence, indicative of effectively no, or at least more recent, isolation (Bryant and Krosch 2016; Hyman and Köhler 2019). Here, we found evidence that this region is also an important turnover point for major lineages of Figuladra (MEQ and SEQ, this study) that respectively occur widely in lowland vine thickets to the north and south. Figuladra is the second lineage of large camaenid to show divergence across this gap, the other being Sphaerospira (Hugall et al. 2003). In Figuladra, this turnover occurs on both the Torilla Peninsula (Byfield NP and Stanage Bay) and adjacent areas of the mainland to the west. This suggests that this barrier of dry and seemingly unsuitable habitat was also continuous during glacial lowstands.
In contrast to the relatively deep genetic diversity and structuring in the SEQ upland clade and the St Lawrence Gap, within the two lowland clades of SEQ and MEQ we found typically low nucleotide distances between sampled populations. The haplotype network for the SEQ clade in particular tends to show only a few base pair differences between sampled haplotypes. This is also the clade that shows the strongest signature of range expansion, suggesting that through recent glacial cycles, and despite their apparently disjunct contemporary distribution, there has been quite significant gene flow between many areas of vine thicket and only recent isolation. These observations indicate that many patches of lowland vine thicket habitat have a relatively dynamic recent evolutionary history of connectivity between patches. Comparative phylogeography based on genomic datasets (i.e. SNPs) combined with demographic modelling could be undertaken to test this hypothesis further. These observations also suggest that the contemporary taxonomy of these groups, which recognise multiple species, requires further work to identify characters to support the isolation of these shallowly divergent and parapatric species.
Patterns of phylogeographic structuring in the extensive and seemingly more continuous areas of open Eucalyptus dominated open woodland in southern and central Queensland are also poorly known. The Euryladra clade is closely associated with this habitat and shows lower nucleotide diversity and average pairwise divergence than any of the other four lineages analysed, despite its wider range. One haplotype (Fig. 6a: MAT4, MAT 5, MAT10) occurs across the extreme limits of its distribution – from Targinnie near Gladstone, SEQ, and Dulacca east of Roma, SCQ, a distance of approximately 480 km. Low genetic diversity in this open woodland group indicates that there has been more extensive recent geneflow, suggesting contemporary connectivity of this habitat type (or at least at European settlement) mirrors the past distribution, with no evidence of major historical barriers.
It is widely hypothesised that fire has played an important role in shaping the distribution of open woodland and vine thicket vegetation, with the latter considered to be negatively affected by fire (Bond and Midgley 2012). Low genetic divergences between many snail populations in disjunct vine thickets suggests that the intensification of fire may have isolated many of these relatively recently in their evolutionary history. Fire also continues to pose a major contemporary threat to many vine thicket associated land snails (Stanisic and Ponder 2004), both directly and indirectly through the destruction of microhabitat. There is an urgent need to both understand biodiversity patterns in dry rainforests, and to better understand the evolutionary and ecological impacts of fire.
Conclusions
We found that ecologically differing snail species from subtropical areas of eastern Australia show contrasting patterns of mitochondrial genetic diversity. Snail lineages associated with insular, fragmented and relatively mesic habitat vine thickets show more pronounced genetic structure than related taxa inhabiting contiguous and relatively widespread open woodland habitats. Our study also highlights upland vine thickets inhabited by the SEQ upland lineage of Figuladra as a potential ecosystem of high evolutionary diversity and endemism that should be prioritised for conservation and further survey, and the St Lawrence Gap as a potentially important barrier to vine thicket taxa. Finally, our results suggest lowland vine thicket and open woodland associated species in eastern Australia may have dynamic histories of recent expansion and isolation and highlight the need for further studies to better understand potential past and future impacts of fire in these systems.
Data availability
All data generated or analysed during this study are included in this article and its Supplementary Information. Sequence information generated in this study are available at GenBank, accession numbers OQ676310 – OQ676369.
Declaration of funding
This research was funded by HDR candidate support funding from Griffith University.
Permits
In order to allay any conservation concerns, the majority of specimens were collected in areas outside National Parks and State Forests. Where specimens were taken from within these gazetted jurisdictions they were carried out under Scientific Purposes permit number WlTK16810116.
Acknowledgements
We acknowledge the Queensland Museum and Griffith University for allowing us to access facilities and collections for this study. We sincerely thank John Stanisic from the Queensland Museum for his help in fieldwork and for his input to the understanding of the biogeography of Queensland land snails. A special thanks to Sandy Pollock from the Queensland Herbarium for his discussion on vegetation and sources for the maps in Fig. 1. We also extend our sincere appreciation to Craig Eddie for his assistance with the collection of some specimens from SCQ for this project in times when museum fieldwork was unable to be conducted due to COVID and Frank Köhler from the Australian Museum for his help with the molecular analysis. We thank the reviewers for their constructive comments.
References
Ashton LA, Odell EH, Burwell CJ, Maunsell SC, Nakamura A, McDonald WJF, Kitching RL (2016) Altitudinal patterns of moth diversity in tropical and subtropical Australian rainforests. Austral Ecology 41, 197-208.
| Crossref | Google Scholar |
Ashton LA, Leach EC, Odell EH, McDonald WJ, Arvidsson D, Kitching RL (2020) The Eungella biodiversity study: filling the knowledge gap. Proceedings of the Royal Society of Queensland 125, 11-21.
| Google Scholar |
Bond WJ, Midgley JJ (2012) Fire and the angiosperm revolutions. International Journal of Plant Sciences 173, 569-583.
| Crossref | Google Scholar |
Bryant LM, Krosch MN (2016) Lines in the land: a review of evidence for eastern Australia’s major biogeographical barriers to closed forest taxa. Biological Journal of the Linnean Society 119, 238-264.
| Crossref | Google Scholar |
Burke JM, Ladiges PY, Batty EL, Adams PB, Bayly MJ (2013) Divergent lineages in two species of Dendrobium orchids (D. speciosum and D. tetragonum) correspond to major geographical breaks in eastern Australia. Journal of Biogeography 40, 2071-2081.
| Crossref | Google Scholar |
Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D, Aplin K, Cantrill DJ, Cook LG, Crisp MD, Keogh JS, Melville J, Moritz C, Porch N, Sniderman JMK, Sunnucks P, Weston PH (2011) Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38, 1635-1656.
| Crossref | Google Scholar |
Catullo RA, Scott Keogh J (2014) Aridification drove repeated episodes of diversification between Australian biomes: evidence from a multi-locus phylogeny of Australian toadlets (Uperoleia: Myobatrachidae). Molecular Phylogenetics and Evolution 79, 106-117.
| Crossref | Google Scholar | PubMed |
Chiba S (1999) Accelerated evolution of land snails Mandarina in the oceanic Bonin Islands: evidence from mitochondrial DNA sequences. Evolution 53, 460-471.
| Crossref | Google Scholar | PubMed |
Dexter KG, Pennington RT, Oliveira-Filho AT, Bueno ML, Silva de Miranda PL, Neves DM (2018) Inserting tropical dry forests into the discussion on biome transitions in the tropics. Frontiers in Ecology and Evolution 6, 104.
| Crossref | Google Scholar |
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915-925.
| Crossref | Google Scholar | PubMed |
Grimshaw P (2017) Dry rainforests of SEQ. Available at https://www.lfwseq.org.au/dry-rainforests-seq/ [Accessed 8 June 2021]
Hugall A, Stanisic J (2011) Beyond the prolegomenon: a molecular phylogeny of the Australian camaenid land snail radiation. Zoological Journal of the Linnean Society 161, 531-572.
| Crossref | Google Scholar |
Hugall A, Stanisic J, Moritz C (2003) Phylogeography of terrestrial gastropods: the case of the Sphaerospira lineage and history of Queensland rainforests. In ‘Molecular systematics and phylogeography of Mollusks’. (Eds C Lydeard, R David) pp. 270–301. (Smithsonian Institution Press: Washington, DC)
Hyman IT, Köhler F (2019) Phylogeny and systematic revision of the helicarionid semislugs of eastern Queensland (Stylommatophora, Helicarionidae). Contributions to Zoology 88, 351-451.
| Crossref | Google Scholar |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649.
| Crossref | Google Scholar | PubMed |
Leigh JW, Bryant D (2015) Popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6, 1110-1116.
| Crossref | Google Scholar |
McDonald WJF (2006) Vine thicket communities of the Brigalow Belt Bioregion of Queensland. Australasian Plant Conservation: Journal of the Australian Network for Plant Conservation 14, 11-12.
| Crossref | Google Scholar |
Metcalfe D, Bui E (2016) Land: Regional and landscape-scale pressures: Land clearing. Australia state of the environment 2016. Available at https://soe.environment.gov.au/theme/land/topic/2016/regional-and-landscape-scale-pressures-land-clearing.
Monroe MH (2015) Australia: the land where time began. A biography of the Australian continent. Available at https://austhrutime.com/woodlands_australia.htm [Accessed 8 June 2021]
Murphy PG, Lugo AE (1986) Ecology of tropical dry forest. Annual Review of Ecology and Systematics 17, 67-88.
| Crossref | Google Scholar |
Neldner VJ, Niehus RE, Wilson BA, McDonald WJF, Ford AJ, Accad A (2019b) The vegetation of Queensland. Descriptions of Broad Vegetation Groups V 4. Available at https://www.des.qld.gov.au/__data/assets/pdf_file/0029/81929/descriptions-of-broad-vegetation-groups.pdf [Accessed 28 April 2022]
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268-274.
| Crossref | Google Scholar | PubMed |
Pennington RT, Lehmann CER, Rowland LM (2018) Tropical savannas and dry forests. Current Biology 28, R541-R545.
| Crossref | Google Scholar | PubMed |
Rix MG, Wilson JD, Harvey MS (2020) First phylogenetic assessment and taxonomic synopsis of the open-holed trapdoor spider genus Namea (Mygalomorphae: Anamidae): a highly diverse mygalomorph lineage from Australia’s tropical eastern rainforests. Invertebrate Systematics 34, 679-726.
| Crossref | Google Scholar |
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539-542.
| Crossref | Google Scholar | PubMed |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34, 3299-3302.
| Crossref | Google Scholar | PubMed |
Sela I, Ashkenazy H, Katoh K, Pupko T (2015) GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43, W7-W14.
| Crossref | Google Scholar | PubMed |
Simpson L, Clements MA, Crayn DM, Nargar K (2018) Evolution in Australia’s mesic biome under past and future climates: insights from a phylogenetic study of the Australian Rock Orchids (Dendrobium speciosum complex, Orchidaceae). Molecular Phylogenetics and Evolution 118, 32-46.
| Crossref | Google Scholar | PubMed |
Stanisic J (1990) Systematics and biogeography of eastern Australian Charopidae (Mollusca, Pulmonata) from subtropical rainforests. Memoirs of the Queensland Museum 30, 1-241.
| Google Scholar |
Stanisic J (1994) The distribution and patterns of species diversity of land snails in eastern Australia. Memoirs of the Queensland Museum 36, 207-214.
| Google Scholar |
Stanisic J (1996) The large camaenids of Mideastern Queensland. Australasian Shell News 92, 1-2.
| Google Scholar |
Stanisic L, Köhler F, McDougall C (2022) Two new genera of land snail from dry subtropical forests of eastern Australia: Brigaladra gen. nov. and Euryladra gen. nov. (Eupulmonata: Camaenidae). Invertebrate Systematics 36, 506-532.
| Crossref | Google Scholar |
Sutcharit C, Asami T, Panha S (2007) Evolution of whole-body enantiomorphy in the tree snail genus Amphidromus. Journal of Evolutionary Biology 20, 661-672.
| Crossref | Google Scholar | PubMed |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585-595.
| Crossref | Google Scholar | PubMed |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725-2729.
| Crossref | Google Scholar | PubMed |
Weber LC, Vanderwal J, Schmidt S, McDonald WJF, Shoo LP (2014) Patterns of rain forest plant endemism in subtropical Australia relate to stable mesic refugia and species dispersal limitations. Journal of Biogeography 41, 222-238.
| Crossref | Google Scholar |
Wilmer JW, Amey AP, McDougall C, Venz M, Peck S, Oliver PM (2020) Comparative mitochondrial phylogeography of two legless lizards (Pygopodidae) from Queensland’s fragmented woodlands. Israel Journal of Ecology and Evolution 66, 142-150.
| Crossref | Google Scholar |