The iGEM competition: research-led teaching in microbiology
Nicholas V ColemanSchool of Life and Environmental Sciences
Building G08, Maze Crescent
University of Sydney
Darlington, NSW 2006, Australia
Tel: +61 2 9351 6047
Fax: +61 2 9351 5858
Email: nicholas.coleman@sydney.edu.au
Microbiology Australia 37(2) 81-83 https://doi.org/10.1071/MA16028
Published: 18 April 2016
The International Genetically Engineered Machine Competition (iGEM) is a global science fair in synthetic biology (SynBio). The relatively new discipline of SynBio is distinguished from ‘genetic engineering’ in its more systematic approach, and its focus on understanding life via creation, rather than dissection1. Microbiology is central to SynBio, which usually relies on Escherichia coli or yeast as model systems.
The iGEM competition is structured around teams of undergraduates, who compete to make the most novel and/or useful genetically modified organisms (GMOs). The teams work for 6 months in the lab, and present their work at the iGEM Jamboree (Boston, USA) in October. The competition now includes approximately 250 teams from around the world, including five from Australia (Macquarie University, University of Sydney, University of New South Wales, University of Melbourne, Australian National University). iGEM projects are diverse, and reflect the astounding possibilities of SynBio, such as biocomputers, bioplastics, biosensors, and much more. Perhaps the most famous example is ‘bacterial photography’, in which an E. coli strain was created that deposited a black pigment in response to light2.
iGEM offers a complete and authentic scientific experience to undergraduates. iGEM promotes a culture of open source science and transparency of research. All teams publish their research in an online wiki that is open to the public, and all new genetic constructs generated are freely shared via the Parts Registry3. The skills needed for a good team include microbiology, molecular biology, biochemistry, mathematical modelling, web design and educational outreach, and thus teams typically consist of students from several different degree programs, who must learn to work together.
I have been the supervisor of iGEM teams from The University of Sydney for the past three years. I believe this competition offers a unique research-led teaching opportunity, which develops graduate attributes which are often neglected in our core teaching program, as summarised in the following sections.
Inquiry and problem solving
Most undergraduate practical classes in microbiology involve experiments where the outcome is known prior to starting work. While such practical classes are useful for teaching important techniques, they are not reflective of a real scientific investigation. In the iGEM competition, each team is aiming to construct new GMOs with novel properties, and although much effort goes into design and modelling, ultimately the outcomes are not predictable, necessitating troubleshooting, problem-solving, improvisation, and critical reading of the literature to achieve the desired goals.
As an example, our 2013 team attempted to reconstruct a pathway for biodegradation of 1,2-dichloroethane in E.coli. Only four genes are required, and on paper, it seemed simple to move these genes into a plasmid in E.coli. However, about half-way through our project, the team realised that one enzyme required an unusual cofactor not found in E. coli, necessitating a rapid re-think and redesign of our plasmid construct. We learned an important lesson that year, i.e. read the literature thoroughly before getting into the lab!
Communication
iGEM teams need to communicate their results in several different formats, some of which are ‘non-standard’ in microbiology education. The most important document that the teams generate is a web page (wiki). This webpage is open to the public, and is maintained in perpetuity by the iGEM organisation; as a result, the students feel that they are part of the global scientific community, and that their work ‘matters’ – this sense of belonging and significance is central to a career as a professional scientist, but is not commonly experienced by undergraduates. Please visit our USyd iGEM wiki pages to see some examples4–6.
Teams must also be able to effectively communicate their work to the competition judges and to other teams as talks and posters. The intensely competitive environment of the jamboree leads to outstanding performances in the talks, with their engaging style, professional use of modern technology, and creative thinking, including the use of theatre, costumes, comedy, time-lapse animations, etc.
iGEM teams must engage with the wider community, and express complex concepts in simple language; such skills are not usually taught in undergraduate science degrees. Our iGEM students have helped to run hands-on microbiology and molecular biology workshops for school students, and they have participated in Science Week activities at the Australian Museum. We have collaborated in the setup of a community-based lab7, which aims to enable public access to SynBio technologies. An analysis of our 2015 outreach activities is shown in Figure 1; note the use of diverse media (newspaper, radio, social media, in-person activities) that enabled us to reach out and engage with diverse age-groups.
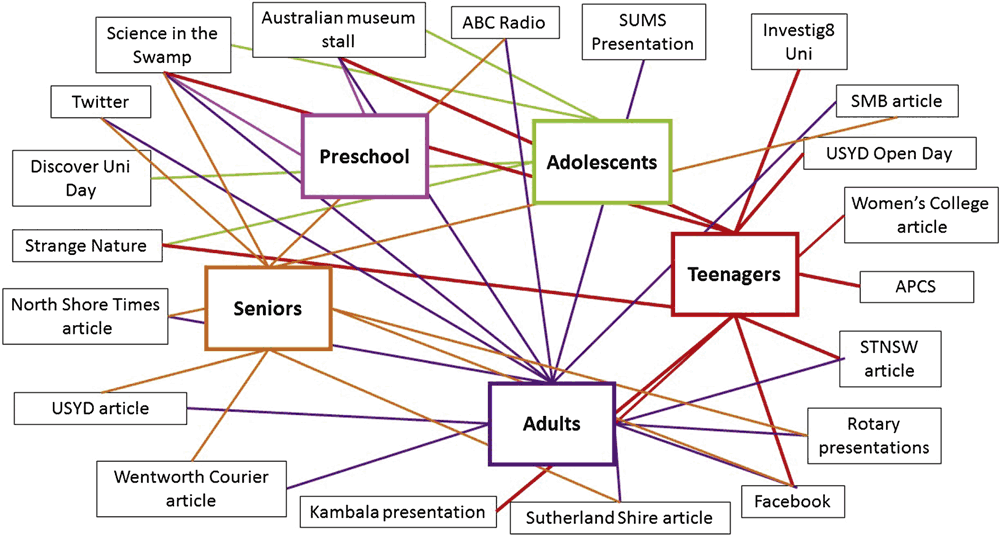
|
Personal and professional responsibility
Graduate attributes relating to ethical conduct are critical for employers, but are only very rarely addressed in undergraduate curricula. iGEM is unique in its focus on safety and ethics, which can be quite confronting for students who have not had to think about these issues before. All teams in the competition have to document in considerable detail the biosafety issues relating to their project, and how any risks will be managed; this is especially important since almost all iGEM projects revolve around the creation of new GMOs.
Our USyd iGEM teams undergo extensive safety training before they are allowed into the lab, and they assist in preparation of documents for our Institutional Biosafety Committee. Our teams have embraced the bioethics issues, merging this with their outreach activities by running a writing competition aimed at high school students, which encourages them to think critically about the impact of genetic technologies on society8.
Conclusions
The iGEM competition develops valuable graduate attributes and skills which are often neglected in science undergraduate degrees, such as inquiry-based problem solving, communication, professional responsibility, fundraising, outreach, and media engagement. I would also add ‘resilience’ to this list, since these students learn to grapple with real-world problems, which require repeated efforts and many failures before eventual success (if any). I believe these attributes and skills greatly enhance the employability of graduates from this program. Informal feedback from USyd students indicates that all believed iGEM was a valuable part of their education, and half indicated that the iGEM experience was the most valuable part of their university education to date. My lab will once again host an iGEM team in 2016; I encourage you to get involved, and join our growing local synthetic biology community.
Acknowledgements
Thanks to all the members of USyd iGEM teams from 2013–2015 for their enthusiasm and effort: Robbie Oppenheimer, Andrew Tuckwell, Desmond Li, Vivian Li, Shuravi Paul, Cyril Tang, Hugh Ford, James Bergfield, Rokiah Alford, Abigail Sison, Jeanne Zhang, Andy Bachler, Callum Grey, Tom Geddes, Lizzie Richardson, Harrison Steel, Mark Somerville, Gaia Hermann, Sandi Bo, Mahiar Mahjoub.
References
[1] Levskaya, A. et al. (2005) Synthetic biology: engineering Escherichia coli to see light. Nature 438, 441–442.| Synthetic biology: engineering Escherichia coli to see light.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1Gis7nF&md5=ec87c48f69093a3a9a3f1f38dc8fd157CAS | 16306980PubMed |
[2] Shetty, R. et al. (2011) Assembly of biobrick standard biological parts using three antibiotic assembly. In: Synthetic Biology, Pt B: Computer Aided Design and DNA Assembly. (Voigt, C., ed), pp. 311–326, Elsevier Press.
[3] http://parts.igem.org
[4] http://2013.igem.org/Team:SydneyUni_Australia
[5] http://2014.igem.org/Team:USyd-Australia
[6] http://2015.igem.org/Team:Sydney_Australia
[7] http://www.foundry.bio
[8] http://www.strangenature.org
Biography
Dr Nick Coleman is a Senior Lecturer in Microbiology at the University of Sydney. Nick teaches 2nd and 3rd year microbiology to students in science, medical science, and agriculture degree programs. His research interests include bioremediation, biocatalysis, bacterial evolution, and synthetic biology. He is a member of the ASM NSW/ACT Branch Committee.


