Evaluating exotic predator control programs using non-invasive genetic tagging
Maxine P. Piggott A E , Rebecca Wilson B , Sam C. Banks C , Clive A. Marks D , Frank Gigliotti B and Andrea C. Taylor AA Australian Centre for Biodiversity, School of Biological Sciences, Monash University, Clayton,Victoria 3800, Australia.
B Vertebrate Pest Research Unit, Department of Primary Industries (Victoria), PO Box 49, Frankston, Victoria 3199, Australia.
C The Fenner School of Environment and Society, The Australian National University, Canberra, ACT 0200, Australia.
D Nocturnal Wildlife Research Pty Ltd, PO Box 2126, Wattletree Road Post Office, East Malvern, Victoria 3145, Australia.
E Corresponding author. Email: mpiggott@bio.mq.edu.au
Wildlife Research 35(7) 617-624 https://doi.org/10.1071/WR08040
Submitted: 13 March 2008 Accepted: 17 June 2008 Published: 17 November 2008
Abstract
Carnivorous predators are difficult to detect using conventional survey methods, especially at low levels of abundance. The introduced red fox (Vulpes vulpes) in Australia is monitored to determine the effectiveness of control programs, but assessing population parameters such as abundance and recruitment is difficult. We carried out a feasibility study to determine the effectiveness of using faecal DNA analysis methods to identify individual foxes and to assess abundance before and after lethal control. Fox faeces were collected in two sampling periods over four separate transects, and genotyped at five microsatellite loci. Two transects were subject to lethal control between collection periods. DNA was extracted from 170 fox faeces and, in total, 54 unique genotypes were identified. Fifteen biopsy genotypes from 30 foxes killed during lethal control were detected among the faecal genotypes. Overall, a similar number of genotypes were detected in both sampling periods. The number of individuals sampled in both periods was low (n = 6) and new individuals (n = 24) were detected in the second collection period. We were also able to detect animals that avoided lethal control, and movement of individuals between transects. The ability to identify individual foxes using these DNA techniques highlighted the shortcomings of the sample design, in particular the spatial scale and distances between transects. This study shows that non-invasive DNA sampling can provide valuable insight into pre and post fox abundance in relation to lethal control, individual behaviour and movement, as well as sample design. The information gained from this study will contribute to the design of future studies and, ultimately, control strategies.
Acknowledgements
This trial was funded by the Catchment and Water Division of the Department of Sustainability and Environment (Victoria). All research procedures were carried out in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (1997) and were approved by the Victorian Institute of Animal Science Animal Experimentation Committee (AEC) as protocol 2399. We thank Rebecca Norris, Brad Scott, Frank Busana, Steve McPhee, Michael Johnston and Michael Lindeman for assisting in the collection of faecal samples. The Department of Justice (Victoria) granted an exemption from the Control of Weapons Act 1990 to allow the importation, possession and use of M-44 ejectors and baits in accordance with the Australian Pesticides and Veterinary Medicines Authority Permit No. TPM0015A.
Adams, J. R. , and Waits, L. P. (2007). An efficient method for screening faecal DNA genotypes and detecting new individuals and hybrids in the red wolf (Canis rufus) experimental population area. Conservation Genetics 8, 123–131.
| Crossref | GoogleScholarGoogle Scholar | CAS |
Busana, F. , Gigliotti, F. , and Marks, C. A. (1998). Modified M-44 ejector for the baiting of red foxes. Wildlife Research 25, 209–215.
| Crossref | GoogleScholarGoogle Scholar |
Marks, C. A. , Gigliotti, F. , and Busana, F. (2003). Field performance of the M-44 ejector for red fox (Vulpes vulpes) control. Wildlife Research 30, 601–609.
| Crossref | GoogleScholarGoogle Scholar |
Sharp, A. , Norton, M. , Marks, A. , and Holmes, K. (2001). An evaluation of two indices of red fox (Vulpes vulpes) abundance in an arid environment. Wildlife Research 28, 419–424.
| Crossref | GoogleScholarGoogle Scholar |

Smith, D. A. , Ralls, K. , Hurt, A. , Adams, B. , Parker, M. , and Maldonado, J. E. (2006). Assessing reliability of microsatellite genotypes from kit fox faecal samples using genetic and GIS analyses. Molecular Ecology 15, 387–406.
| Crossref | GoogleScholarGoogle Scholar | CAS | PubMed |

Sunnucks, P. , and Hales, D. F. (1996). Numerous transposed sequences of mitochondrial cytochrome oxidase I–II in aphids of the genus Sitobion (Hemiptera: Aphididae). Molecular Biology and Evolution 13, 510–524.
| CAS | PubMed |

Thompson, J. A. , and Fleming, P. J. S. (1994). Evaluation of the efficacy of 1080 poisoning of red foxes using visitations to non-toxic baits as an index of fox abundance. Wildlife Research 21, 27–39.
| Crossref | GoogleScholarGoogle Scholar |

Valière, N. , Berthier, P. , Mouchiroud, D. , and Pontier, D. (2002). GEMINI: software for testing the effects of genotyping errors and multitubes approach for individual identification. Molecular Ecology Notes 2, 83–86.

Waits, L. P. , Luikart, G. , and Taberlet, P. (2001). Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology 10, 249–256.
| Crossref | GoogleScholarGoogle Scholar | CAS | PubMed |

Wandeler, P. , and Funk, S. M. (2006). Short microsatellite DNA markers for the red fox (Vulpes vulpes). Molecular Ecology Notes 6, 98–100.
| Crossref | GoogleScholarGoogle Scholar | CAS |

Wandeler, P. , Funk, S. M. , Largiadér, C. R. , Gloor, S. , and Breitenmoser, U. (2003). The city-fox phenomenon: genetic consequences of a recent colonization of urban habitat. Molecular Ecology 12, 647–656.
| Crossref | GoogleScholarGoogle Scholar | CAS | PubMed |

White, G. C. , and Burnham, K. P. (1999). Program MARK: Survival estimation from populations of marked animals. Bird Study 46(Suppl.), 120–138.

White, J. G. , Gubiani, R. , Smallman, N. , Snell, K. , and Morton, A. (2006). Home range, habitat selection and diet of foxes (Vulpes vulpes) in a semi-urban riparian environment. Wildlife Research 33, 175–180.
| Crossref | GoogleScholarGoogle Scholar |

Witmer, G. W. (2005). Wildlife population monitoring: some practical considerations. Wildlife Research 32, 259–263.
| Crossref | GoogleScholarGoogle Scholar |

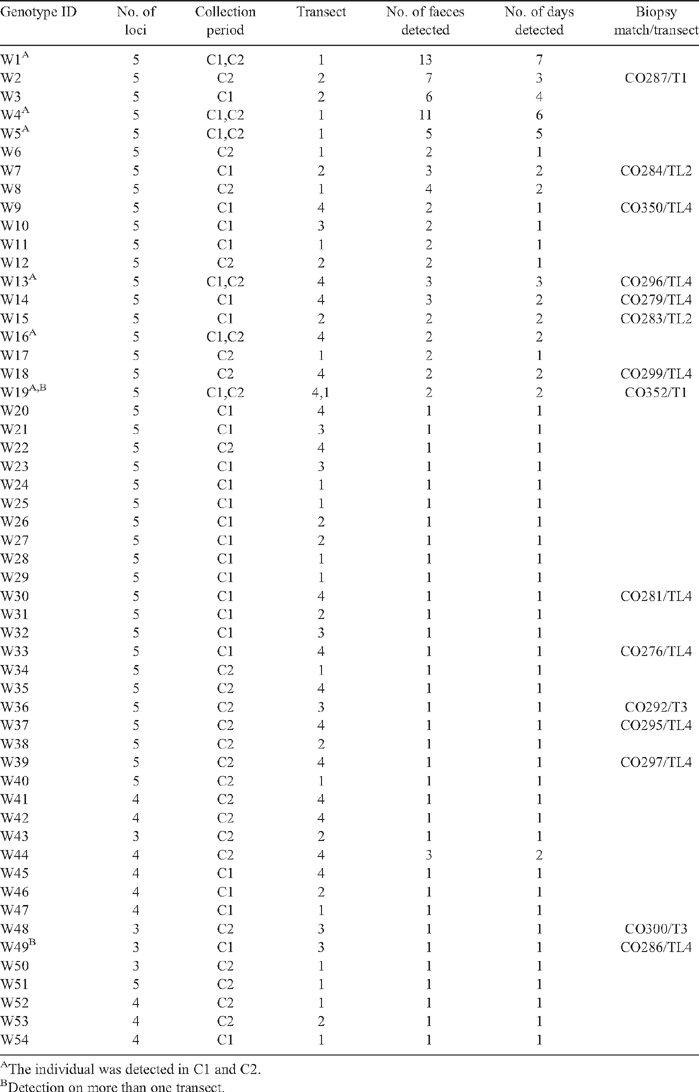
Appendix 1. Faecal Vulpes vulpes genotypes detected during the study and faecal genotype matches with biopsy genotypes from dead animals
|


