Resolving distribution and population fragmentation in two leaf-tailed gecko species of north-east Australia: key steps in the conservation of microendemic species
Lorenzo V. Bertola A , Megan Higgie A and Conrad J. Hoskin
A , Megan Higgie A and Conrad J. Hoskin  A B
A B
A College of Science and Engineering, James Cook University, Townsville, Qld 4811, Australia.
B Corresponding author. Email: conrad.hoskin@jcu.edu.au
Australian Journal of Zoology 66(2) 152-166 https://doi.org/10.1071/ZO18036
Submitted: 21 May 2018 Accepted: 4 October 2018 Published: 2 November 2018
Abstract
North Queensland harbours many microendemic species. These species are of conservation concern due to their small and fragmented populations, coupled with threats such as fire and climate change. We aimed to resolve the distribution and population genetic structure in two localised Phyllurus leaf-tailed geckos: P. gulbaru and P. amnicola. We conducted field surveys to better resolve distributions, used Species Distribution Models (SDMs) to assess the potential distribution, and then used the SDMs to target further surveys. We also sequenced all populations for a mitochondrial gene to assess population genetic structure. Our surveys found additional small, isolated populations of both species, including significant range extensions. SDMs revealed the climatic and non-climatic variables that best predict the distribution of these species. Targeted surveys based on the SDMs found P. gulbaru at an additional two sites but failed to find either species at other sites, suggesting that we have broadly resolved their distributions. Genetic analysis revealed population genetic structuring in both species, including deeply divergent mitochondrial lineages. Current and potential threats are overlain on these results to determine conservation listings and identify management actions. More broadly, this study highlights how targeted surveys, SDMs, and genetic data can rapidly increase our knowledge of microendemic species, and direct management.
Introduction
Some vertebrate species have remarkably small natural distributions. These species can be particularly interesting biogeographically, and are often of conservation significance due to small and fragmented populations. Microendemic lizards and frogs characterise vertebrate diversity in the rainforests of Queensland, in north-eastern Australia. Tropical and subtropical rainforests have a broken distribution along the coastal mountains of Queensland and microendemic lizards and frogs occur in these isolates (Moritz et al. 2005; Rosauer et al. 2017). As a broad-scale example, 13 leaf-tailed geckos species (Orraya, Saltuarius, Phyllurus) occur allopatrically across these rainforest areas (Hoskin and Couper 2013). As a finer-scale example, 16 species of microhylid frogs (Cophixalus, Austrochaperina) occur across the mountains of the largest of these areas, the Wet Tropics (Hoskin 2004). Even small isolates harbour considerable endemic diversity. For example, seven highly distinct rainforest-associated reptiles and frogs are restricted to the Melville Range, isolated from relatives in rainforest patches to the north and south for millions of years (Hoskin 2014; Hoskin and Couper 2014). Many microendemics are restricted to a single mountain or range, while others occur as distinct subpopulations separated by areas of unsuitable habitat (e.g. Borsboom et al. 2010; Couper and Hoskin 2013). The biogeographic insights and conservation challenges for these vertebrates are similar to those for ‘short-range endemic’ invertebrates (Harvey et al. 2011).
Species may be highly localised and fragmented naturally due to long-term processes, or may have declined to small distributions in recent times due to human-associated impacts, or a combination of both. The small distributions of the Queensland rainforest lizards and frogs can generally be considered to be natural – the result of long-term contraction and fragmentation of rainforests over millions of years to mesic refugia in the east-coast mountains (Moritz et al. 2005). These refugia have not been static in recent time, expanding and contracting through the Pleistocene glacial cycles (Hugall et al. 2002). To persist in an area, low-vagility rainforest-associated species had to survive through the globally dry glacial periods, when Australian east-coast rainforests had their most restricted known distribution (Hugall et al. 2002; Moritz et al. 2005; Bell et al. 2010; Rosauer et al. 2017). In many areas from where rainforest largely disappeared, rainforest-associated reptile and frog lineages persisted in deeply piled rocky areas that acted as mesic refugia (‘lithorefugia’: Couper and Hoskin 2008). In coastal Queensland these lithorefugia range from large boulder-fields to small areas of piled boulders on slopes or in individual gullies (Couper and Hoskin 2008). Therefore, climate, topography and geology have all played key roles in persistence of rainforest lineages. Competition and other species interactions also play a role in shaping distributions, but resolving these interactions is complex (Cunningham et al. 2016).
Although these species appear to have persisted in small areas for millennia (Moritz et al. 2005; Hoskin et al. 2011; Rosauer et al. 2017), they are still of conservation concern. Taxa with small distributions are among the most vulnerable to extinction (MacArthur and Wilson 1967; Terborgh and Winter 1980; Gilpin and Soule 1986; Harvey et al. 2011). Stochastic events (e.g. fire or drought) can affect the entire species, whereas for widespread species these events may affect some populations only. For this reason, distribution area is one of the key factors in threatened species listing, even without any evidence of population decline (e.g. Vulnerable D2: IUCN 2012). Small distributions also offer little buffering from human-induced change in the environment, such as clearing, invasive species, novel disease and climate change. Even seemingly localised threats such as illegal collecting or hunting, or changes in fire regime, may impact the entire population of a highly localised species. Therefore, threatened species categories are based largely around the combination of distributional area/population size and observed or projected decline due to an identified threat (e.g. IUCN 2012).
Many of the highly localised Queensland rainforest lizards and frogs are listed as threatened species under federal (Environment Protection and Biodiversity Conservation Act, EPBC Act, 1999) and state (Nature Conservation Act, NCA, 1992) legislation, based on small distributional area in combination with observed or projected threats such as disease, climate change, and changes in fire regime (Curtis 2012). Illegal collecting is an additional, currently unquantified, threat to many of these species due to their rarity and spectacular appearance (e.g. leaf-tailed geckos) (Auliya et al. 2016). While many species are listed, many others have not been assessed or are considered Data Deficient due to a lack of information. Data Deficient species, and indeed most of the listed species, have received very little targeted survey effort and are easily overlooked in general fauna surveys due to cryptic appearance and habits, and specific habitat requirements. Regardless of whether species are listed or not, management is generally limited by a lack of detailed information on fine-scale distribution, population genetic structuring and connectivity, and identification and quantification of threats.
Management of Queensland’s microendemic reptiles and frogs requires the following broad steps: (1) resolve the distribution in detail, (2) identify any distinct lineages reflecting historical isolation and/or local adaptation (Crandall et al. 2000), (3) determine current isolation and connectivity between populations, (4) estimate population sizes, (5) resolve threats, and (6) propose and test management actions. These steps have been addressed for some high-profile species in Queensland, particularly birds and mammals (e.g. the northern bettong of the Wet Tropics: Laurance 1997; Pope 2000; Vernes and Pope 2006; Bateman et al. 2012) but rarely for highly localised reptile and frog species. An exception is the Critically Endangered skink Nangura spinosa, which has received considerable attention in regard to distribution, phylogeographic structure, abundance and threats (e.g. Borsboom et al. 2010).
Here we attempt to resolve the fine-scale distribution and population genetic structure of two highly localised leaf-tailed geckos in north-east Queensland. Leaf-tailed geckos, the carphodactyline clade comprising the genera Phyllurus, Saltuarius and Orraya, exemplify the pattern of microendemism in moist refugia along the east coast of Australia (Couper et al. 2000; Moritz et al. 2005; Hoskin and Couper 2013). This is particularly the case for the genus Phyllurus, which comprises nine species, eight of which are highly localised and distributed contiguously in rainforest isolates along the coastal ranges of Queensland (Couper and Hoskin 2013; Wilson and Swan 2013). Most Phyllurus occur in mid-east Queensland but two species occur to the north, on the northern side of a long-term biogeographic barrier, the Burdekin Gap (Hoskin et al. 2003). These two species, P. amnicola and P. gulbaru, occur in the Townsville region, at the far southern end of the Australian Wet Tropics (Fig. 1). The Wet Tropics is a fairly continuous area of tropical rainforest along the mountains and associated coastal lowlands between Townsville, Cairns and Cooktown. At its southern extent, in the Townsville region, rainforest is very patchy and confined to uplands, gullies and rocky slopes, in a matrix of open, fire-prone eucalyptus woodland with a grassy understorey.
Phyllurus gulbaru was discovered in Patterson’s Gorge in the southern Paluma Range in 2001 and was described in 2003 (Hoskin et al. 2003). Surveys around the time of discovery found the species at two sites within the gorge, but not in adjacent areas, and the species was assumed to be restricted to the rocky habitats of Patterson’s Gorge (Hoskin et al. 2003). P. amnicola was discovered on Mt Elliot, a southern upland outlier to the Wet Tropics region, in 1998 and was described in 2000 (Couper et al. 2000). Surveys at the time of discovery, and in the few years following, found the species in two areas on Mt Elliot and the species was thus assumed to be restricted to rocky habitats on Mt Elliot (Couper et al. 2000; Hoskin et al. 2003). Both species were found to be restricted to rocky habitats associated with rainforest, particularly rainforest gullies and boulder slopes, with the geckos emerging from rock crevices to hunt for invertebrates on rock surfaces at night (Couper et al. 2000; Hoskin et al. 2003). Since the early 2000s there have been no surveys targeting these species, with the only additional records coming from the known sites and a serendipitous recent discovery of a population of P. gulbaru in Hervey Range, 3.5 km south of the closest population in Patterson’s Gorge (Murray and de Jong 2017).
Phyllurus gulbaru is listed as Critically Endangered at the international (IUCN) and Australian (EPBC) levels, and listed as Endangered by the Queensland Government (NCA). These listings reflect the small and fragmented known range (two disjunct areas of habitat in Patterson’s Gorge) and potential threats, including fire, illegal collecting, climate change, and nearby urbanisation that may introduce threats such as invasive species (Hoskin et al. 2003; Barnett et al. 2017). Fire has been particularly highlighted as a threat (Hoskin et al. 2003; Curtis 2012), as the frequency, intensity and timing of fires have changed in the last century due to burning for cattle grazing, fire management through high-frequency late-dry-season burning, and due to the introduction of highly flammable grasses. P. amnicola is not listed at the international, federal or state levels because rocky rainforest habitat (and hence, it is assumed, the gecko) is widespread on Mt Elliot, and the entire known distribution falls within a protected area. The species is nonetheless also highly localised and potentially impacted by the same threats associated with small population size: illegal collecting, altered fire regimes, invasive species, and climate change.
In this study, we aimed to resolve the geographic distribution of these species and test for genetic structuring among populations. We conducted targeted surveys for these species in the southern Wet Tropics/Townsville region based on expert knowledge. We then used the distribution data to resolve the key environmental attributes determining the distribution of P. gulbaru and P. amnicola, and modelled the potential distributions of both species. In doing this, we adopt the hierarchical stepwise Species Distribution Model (SDM) approach of van Gils et al. (2014) to model highly localised species that have small numbers of unique occurrence points. We then conducted further targeted surveys in key areas of the potential distributions predicted by the SDMs. Additionally, we sequenced a mitochondrial gene for individuals from all known populations of both species to assess historical isolation and broad-scale genetic structuring within each species. From these results we derive management recommendations for P. gulbaru and P. amnicola, and comment more broadly on the knowledge requirements for management of Queensland’s many microendemic reptile and frog species.
Materials and methods
Field surveys – Round 1
Surveys were conducted between September 2015 and September 2016. Survey sites were selected on Google Earth by Conrad Hoskin, an expert on leaf-tailed geckos (e.g. Couper et al. 2000; Hoskin et al. 2003; Couper and Hoskin 2013; Hoskin and Couper 2013). Surveys targeted areas of layered rock associated with rainforest in the broad Townsville region. Surveys were conducted on foot at night using head torches. GPS coordinates were recorded for each observed individual.
Species distribution modelling
We adopted a hierarchical, stepwise species distribution modelling approach (van Gils et al. 2014) using MAXENT 3.3.3k (Phillips et al. 2006) to build SDMs of the two study species, with the aim of estimating their full distribution and identifying areas to target future surveys (Raxworthy et al. 2003; Bourg et al. 2005). A presence-only method was selected because, before this study, presence-only data were collected for this species, and because it is often difficult to distinguish true absences from failed detection in cryptic and localised species. Furthermore, because these species have small and highly localised distributions, the number of maximum possible unique presence points is low, and MAXENT is better suited than other widely used presence-only algorithms to deal with small sample sizes (Hernandez et al. 2006).
The preliminary environmental dataset included 28 variables, described in detail in Table S1 (Supplementary Material). The environmental layers included: accuCLIM01 to accuCLIM07 from Storlie et al. (2013), which are an improved version of the temperature BioClim layers for the Australian Wet Tropics region; Bio08 to Bio19 from the BioClim dataset (Hutchinson et al. 2009); surfaces of elevation (DEM), slope, aspect, distance to stream and foliage projected cover (FPC) for the Australian Wet Tropics region from Storlie et al. (2013); surfaces of broad vegetation groups (BVG) for Queensland at 1 : 2 million and 1 : 5 million scale (Neldner et al. 2017); and surfaces of static rainforest refugia and preclearing rainforest cover (RFP) for the Australian Wet Tropics region (Graham et al. 2010).
BVG layers were obtained in shapefile format from the Queensland Government Data portal and converted to raster using ArcGIS 10.1 (ESRI 2012), while all other layers were directly obtained in raster format by the eResearch group at James Cook University at a resolution of 250 m. All layers were clipped to the same extent using the ‘raster’ package (Hijmans et al. 2015) within R (R Development Core Team 2013). Pearson’s correlation coefficient was estimated for combinations of all 28 variables using the LayerStats function from the package ‘raster’ (Hijmans et al. 2015). The occurrence dataset for each species was produced using previously known records from CJH’s personal database and records collected during Round 1 of field surveys in this study. Presence records were then overlaid onto environmental grids and one record only per grid cell was retained.
Because recent studies have shown that using MAXENT with default parameters can result in poorly performing models (Shcheglovitova and Anderson 2013; Radosavljevic and Anderson 2014), and that species-specific tuning can increase robustness to sampling bias (Anderson and Gonzalez 2011), all models were fine-tuned using the R package ‘ENMeval’ (Muscarella et al. 2014). Four feature class combinations (Linear, L; Linear and Quadratic, LQ; Linear, Quadratic and Hinge, LQH; Hinge, H) and regularisation multipliers from 0.5 to 5, with intervals of 0.25, were tested. The function ENMevaluate calculates several evaluation metrics: area under the curve (AUC), minimum training presence omission rate (ORmin), 10% training omission rate (OR10) (Peterson 2011), and the Akaike Information Criterion corrected for small samples sizes (AICc) (Burnham and Anderson 2004; Warren and Seifert 2011). AUC has been widely adopted to evaluate the discriminatory ability of SDMs, but recent studies have shown its limitations, especially for presence-only methods used to predict potential distributions (Jiménez‐Valverde 2012). Thus, the best model was selected using AICc, as Warren and Seifert (2011) found AICc to be the best-performing metric for small sample sizes. All SDMs adopted 10 000 pseudoabsence points, selected at random from a 75-km buffer around occurrence points. The extent of the buffer was chosen following VanDerWal et al. (2009a) on the extent for pseudoabsence selection in the Australian Wet Tropics region. Targeted choice of pseudoabsences to correct for sampling bias was not implemented, as no common sampling bias could be identified for all presence points. Sampling was conducted in different environments (i.e. hilltops and mountaintops, rainforest gullies, rocky outcrops) and at different distances from roads (<1 km to 10 km from roads).
Due to the small number of unique occurrence points available for each species, parsimony in the number of predictors was necessary to avoid overfitting the model to the available records, which would reduce the model’s ability to identify novel areas of suitable habitat (Anderson and Gonzalez 2011). Following the hierarchical approach from van Gils et al. (2014) we created three models for each species: a ‘Climatic’, a ‘Non-Climatic’, and an ‘All’ (overall combined) model. To do so, the preliminary set of predictor variables was divided into two categories, climatic and non-climatic, and four subcategories: temperature, precipitation, topography and vegetation (Table S1). Model parameters were fine-tuned for each subcategory of variables using ENMeval, and predictors with a permutation importance of zero in the model with the lowest AICc were removed from successive steps. The remaining predictors were then joined according to their category, thus merging precipitation with temperature predictors (Climatic), and topographic with vegetation predictors (Non-Climatic). Parameters for Climatic and Non-Climatic models were tuned with ENMeval, and predictors with a permutation importance of zero removed. This created the Climatic and Non-Climatic models. Finally, to create the ‘All’ model, all remaining predictors were combined, the best-performing parameters estimated with ENMeval, and predictors with a permutation importance of zero removed. ENMeval was run once more to fine tune the final model with the most parsimonious number of predictors.
Permutation importance was adopted instead of percentage contribution to select variables because the latter is heuristically defined and depends on the path taken by MAXENT to produce the model, while the former depends only on the final model (Phillips et al. 2006). For the Climatic, Non-Climatic and All models, predictors with a Pearson’s correlation coefficient higher than 0.7 and with the lower permutation importance value of the correlated pair were removed, and parameters tuned once more without the removed predictor using ENMeval. Predictor and parameter selection was conducted separately for the two study species. This modelling approach was repeated on a reduced dataset including only occurrence points available before this study.
Field surveys – Round 2
Follow-up surveys were conducted between September 2016 and April 2018, to survey additional areas predicted as suitable by the SDMs. These surveys targeted areas identified as potential suitable habitat in the SDMs that had previously received no, or limited, survey effort for these species (e.g. central Paluma Range, Cape Cleveland, Mt Stuart, central Hervey Range, and other areas around The Pinnacles). Methodology was otherwise as for the first round of surveys.
Genetic sequencing and phylogenetic analysis
A small tissue sample was taken from the tip of the tail, from up to four individuals per site, and preserved in 100% ethanol. Genomic DNA was extracted using a modified version of the Salting Out method (Miller et al. 1988). The entire mitochondrial NADH dehydrogenase subunit 2 (ND2) gene was sequenced using published primers (Table 1) and GoTaq Flexi® reagents (Promega, Madison, WI, USA). For each 25 μL of PCR reaction, reagent concentrations and volumes were as follows: Colorless Flexi Buffer 1X, GoTaq® DNA polymerase 0.125 u, MgCl2 1.5 mm, each dNTP 0.2 mm, forward primer 0.4 mm, reverse primer 0.4 mm and ~50 ng of genomic DNA. The following PCR profile was adopted for both species: denaturation at 94°C for 1 min, then 30 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 1 min and final elongation at 72°C for 3 min. PCR products were then cleaned with Sephadex® and sequenced in both directions at Macrogen Inc. (Seoul, Korea). All sequences have been deposited in GenBank (MH719328–MH719357).

|
Sequences were edited in Geneious 9 (http://www.geneious.com: Kearse et al. 2012) and aligned using ClustalW with standard parameters (Thompson et al. 2002). The alignment was conducted separately for P. gulbaru and P. amnicola and visually inspected. The two alignments were then jointly aligned with a Saltuarius cornutus sequence from GenBank (JF807328), with S. cornutus being a basal outgroup to both species (Hoskin et al. 2003). Uncorrected p-distances were calculated in MEGA7 (Tamura et al. 2013).
PartitionFinder was adopted to select the best-fit model of nucleotide evolution for phylogenetic analysis (Lanfear et al. 2012). For this purpose, ND2 was partitioned into its codon positions, a necessary step for accurate phylogenetic analysis (Lanfear et al. 2012). The predefined partitioning scheme comprised three partitions, representing the three codon positions of ND2. A user search was implemented, with linked branch lengths. The Bayesian Information Criterion was adopted to select the best-fit model following Aho et al. (2014). PartitionFinder was run twice, constraining evaluated models to those available in RAxML (Stamatakis 2014) and MrBayes (Ronquist et al. 2012) respectively.
Phylogenetic analysis was carried out using maximum likelihood (ML) and Bayesian inference (BI) methods. ML analysis was performed with RAxML 8 (Stamatakis 2014), using the best-fit model inferred with PartitionFinder (GTR+G). The autoMRE function to find the needed number of bootstrap replicates was adopted, as this function was estimated to perform the best by Pattengale et al. (2010). BI analysis was carried out with MrBayes 3.2.6 (Ronquist et al. 2012). The best-fit models identified by PartitionFinder were specified for each partition: HKY+G, HKY, and HKY respectively for the first, second and third codon positions of ND2. BI analysis was run for 5 × 106 generations, sampling every 1000 generations, with four chains and a temperature of 0.2. The default burn-in fraction of 25% was retained, and the consensus tree produced from the remaining trees. Stationary log-likelihood (lnL) scores and an average standard deviation of split frequencies lower than 0.01 confirmed that enough generations were being applied (Ronquist et al. 2012).
Results
Field surveys – Round 1
Prior to this study, P. gulbaru was known only from south Patterson’s Gorge and north Patterson’s Gorge (blue symbols in Fig. 1b). In the first round of surveys we found the species at these localities, as well as at an additional three localities: Bluewater Range, Hervey Range, and The Pinnacles (red symbols in Fig. 1b). The new localities were up to 10 km north (Bluewater) and 17 km south-east (The Pinnacles) of the previously known range, greatly increasing the known distribution of P. gulbaru. P. gulbaru was found to be highly localised at each of these localities. Assessment of habitat from satellite imagery (Google Earth) and on foot in the field show that the habitat required by these species, consisting of rainforest and piled rocky boulders, is currently fragmented. This is particularly the case for the southern localities (Hervey Range and The Pinnacles), which are clearly separated from other localities by unsuitable dry habitat. Despite being highly localised, P. gulbaru was generally present at high density in suitable habitat at each site. The exception was The Pinnacles, where only two individuals were found during three thorough surveys of the locality (totalling ~23 person-hours of survey effort).
Prior to this study, P. amnicola was known only from two areas on Mt Elliot: Alligator Creek and Western Boulders Fig. 1c). We found the species at these two localities, as well as at an additional locality on Mt Elliot (Cockatoo Creek). We also discovered a population of P. amnicola on the adjacent mountain, Saddle Mountain (Fig. 1c). New localities for P. amnicola were relatively close to previously known ones (~1–4 km) (Fig. 1c) but the population discovered on Saddle Mountain represents the first record of P. amnicola outside of Mt Elliot. P. amnicola was found to be restricted to rocky habitat at each of these localities and was generally at high densities within this habitat. The exception was the Western Boulders locality, where the species occurs at low density on exposed boulder-field with little associated rainforest.
Species distribution modelling
In total, 14 unique occurrence points were used to model the distribution of P. gulbaru, and 13 points for P. amnicola (Fig. 1b, c). Occurrence points for P. gulbaru did not include the Mt Halifax site and one of the Pinnacles sites, as these were discovered in Round 2 of the surveys with the aid of the SDMs (see below). The preliminary environmental dataset included 28 variables, described in detail in Table S1. Following the stepwise hierarchical selection of variables outlined in the Methods, three final models were produced for each species: Climatic, Non-Climatic, and All. Models for P. gulbaru retained 4–6 variables, while models for P. amnicola retained 2–4 variables (Table 2).
For P. gulbaru, variables included in the Climatic model were mean diurnal range (AC02), mean temperature of coldest quarter (BC11), precipitation of wettest month (BC13) and precipitation seasonality (BC15), while for P. amnicola they were temperature seasonality (AC04) and precipitation of wettest month (BC13). The Non-Climatic model for P. gulbaru included DEM, slope, distance to stream (SDist), preclearing rainforest (RFP), broad vegetation categories (BVG5) and foliage projected cover (FPC), while for P. amnicola the Non-Climatic model included aspect, slope, BVG5 and FPC. Finally, the All model retained BC13, BC15, DEM and FPC for P. gulbaru, and AC04, BC13, slope and aspect for P. amnicola.
The six final models had full AUC scores from 0.95 (e.g. P. amnicola, Climatic) to 0.99 (e.g. P. gulbaru, All). Regularisation multipliers and feature classes selected with ENMeval matched standard parameters by Phillips and Dudik (2008) for the All model for both species and for the Climatic model for P. amnicola, whereas they differed for the remaining three models, having higher regularisation multipliers and the addition of hinge features for the P. gulbaru climatic model (see Table 2).
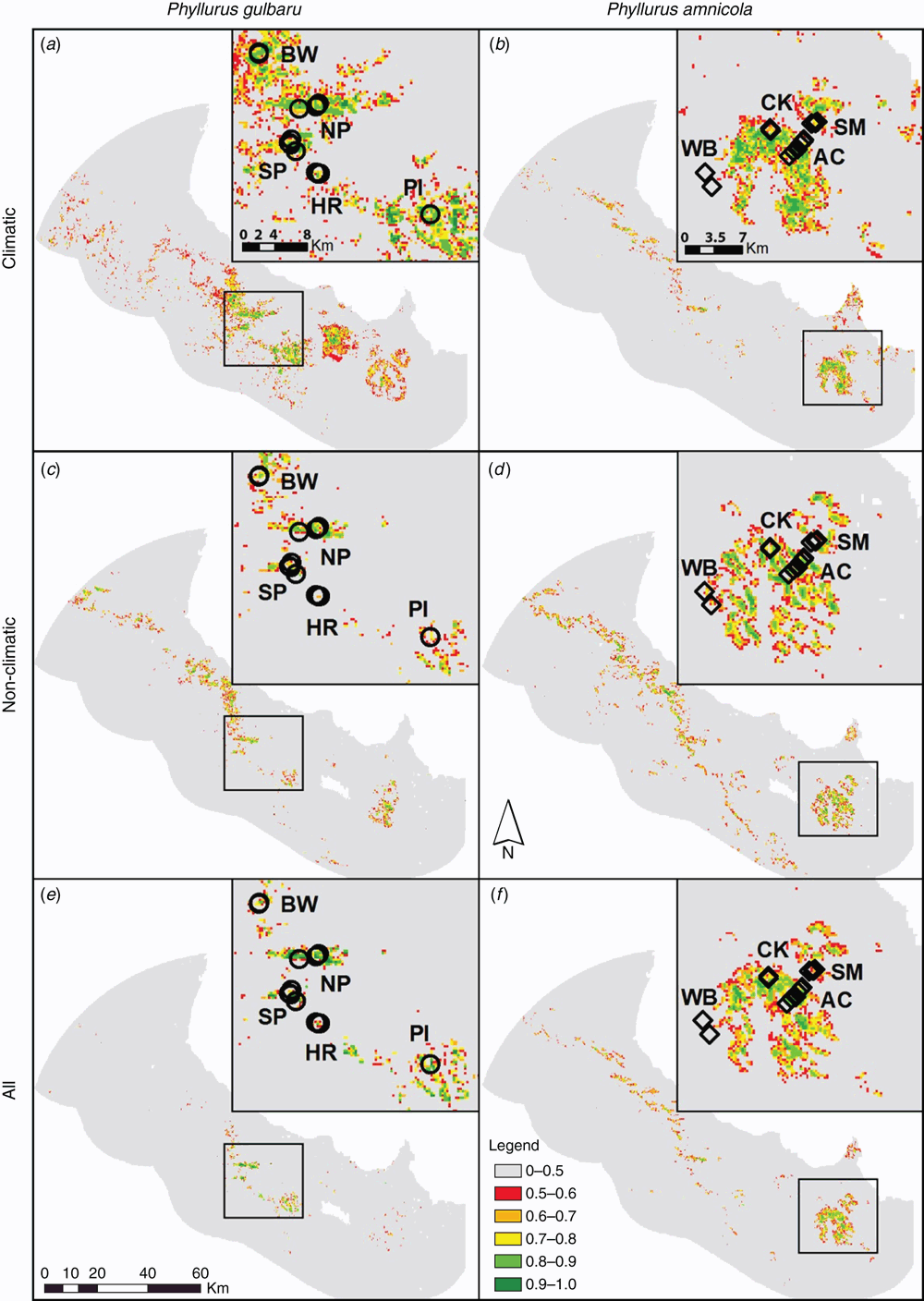
For the three final models for each species, the logistic output was reclassified into suitability categories for ease of interpretation (see Fig. 2). All models for P. gulbaru show logistic values above 0.5 for localities where the species was detected in the first round of surveys (i.e. the training records) (Fig. 2a, c, e). The Climatic model (Fig. 2a) also identifies Mt Elliot and Mt Stuart as areas with predicted probability of presence >0.5, while the Non-Climatic model (Fig. 2c) identifies Mt Elliot and the eastern slopes of the Paluma Range as suitable areas for the species. The All model for P. gulbaru has a limited area of predicted suitability, primarily focussed around the known locations (Fig. 2e).
For P. amnicola the three models are less variable, and they all predict four main areas as suitable for this species: Mt Elliot, Saddle Mountain, Cape Cleveland, and the eastern slopes of the Paluma Range (Fig. 2b, d, f). Of the three P. amnicola models, only the Non-Climatic model (Fig. 2d) has values of predicted probability of presence >0.5 where the Western Boulders occurrence points are found.
SDMs obtained with the reduced dataset including only occurrence records available prior to this study identified similar areas with a predicted probability of presence >0.5 for P. amnicola. In contrast, for P. gulbaru these models included predicted suitable areas well outside the current known range of the species, with the Climatic model in particular lacking resolution (Fig. S1, Supplementary Material).
Field surveys – Round 2
Follow-up surveys were based on the SDMs, targeting areas identified as moderate to high predicted occurrence that had previously received no, or limited, survey effort for these species. Targeted areas included the central Paluma Range (north of known P. gulbaru records) for P. gulbaru, additional areas around The Pinnacles for P. gulbaru, Cape Cleveland for P. amnicola, and Mt Stuart for either species (see Fig. 3). The complete survey data (Round 1, Round 2, and all unsuccessful surveys for each species) are shown in Fig. 3. The follow-up surveys were successful in finding P. gulbaru at two additional sites. The first was in the central Paluma Range, at Mt Halifax. This location had probability of presence >0.5 only for the Climatic and Non-Climatic models (Fig. 2a, c), and it fell outside of predicted areas in the All SDM for P. gulbaru (Fig. 3). The second was an additional site in The Pinnacles area. This location had probability of presence >0.6 for all three models (Fig. 3). No other locations were found for either species.
Genetic population structure
The final nucleotide alignment comprised 1041 base pairs. The sequencing products included in the analyses had unambiguous single peaks and the ND2 region for all individuals translated into amino acids. For all P. amnicola samples, a three-base-pair deletion was present in ND2 at position 953–955 of the alignment, resulting in a loss of one amino acid with no changes to other amino acids. Frequencies of bases A, C, G, T within ND2 were 0.3, 0.34, 0.11 and 0.25 respectively. The bias against G matched mitochondrial base frequency patterns in other vertebrates (Kocher et al. 1989). The BI and ML analysis consensus trees had nearly identical topologies, so the BI consensus tree is presented here with BI posterior probabilities and ML bootstrap values (see Fig. 1d).
Within P. gulbaru, sampling localities showed monophyly with generally high support values (Fig. 1d). The Pinnacles locality was highly divergent (uncorrected p-distance = 6.5%) from all other localities of P. gulbaru, with maximum support values for both ML and BI (Table 3; Fig. 1d). Interestingly, despite the geographical proximity among the remaining localities, sequences from each were monophyletic. However, sequence divergence among these localities was low (uncorrected p-distance = 0.6%) (Table 3; Fig. 1d). The P. amnicola sequences from Mt Elliot and Saddle Mountain were each monophyletic, and mean uncorrected p-distance between Saddle Mountain and Mt Elliot (Alligator Creek, Cockatoo Creek, Western Boulders) samples was 2.9% (Table 3; Fig. 1d). Population structuring on Mt Elliot was limited, with low divergence of the Western Boulders samples but no clear structure among the Alligator Creek and Cockatoo Creek samples (Table 3; Fig. 1d). The mean uncorrected p-distance between P. amnicola localities at Mt Elliot was 0.7% (Table 3).
Discussion
The rainforest fragments along the east coast of Queensland are characterised by highly localised reptile and frog species (Moritz et al. 2005; Rosauer et al. 2017). These narrow endemics are biogeographically interesting because they provide information on rainforest stability, lineage persistence, and speciation through time (Moritz et al. 2005, 2009; Hoskin et al. 2011). These species are also of conservation interest due to their small distributions and increasing concern over threats such as altered burning practices (e.g. Hoskin et al. 2003) and climate change (e.g. Williams et al. 2003; Fordham et al. 2016). A primary consideration with these narrowly restricted endemics is resolving just how narrow their distributions are, and whether population genetic structuring is present. This information is central to decisions on management, such as species conservation listing, because range size and population subdivision are two primary criteria in species listing (e.g. IUCN 2012).
Queensland rainforest herpetofauna is typical of many groups globally in that many species have received little survey attention since their original description, so their distribution and population structuring remain poorly resolved. This means that many would be considered Data Deficient in threatened species listing, which inhibits conservation recognition and formulation of management actions. Further, poor distribution knowledge limits our ability to assess potential threats because it is hard to estimate, infer, or project the level of threat from factors such as fire regime changes, climate change or invasive species when only part of a species’ distribution is known. Although microendemics, by definition, have small ranges, resolving their fine-scale distribution may have significant conservation outcomes, both in terms of determining threatened species status and devising management actions. We used field surveys, species distribution modelling, and phylogenetics to better resolve the distribution and population genetic structure of two representative rainforest reptiles.
The first round of field surveys was based on expert opinion. This resulted in the discovery of multiple previously unknown populations of P. gulbaru and P. amnicola (Fig. 3). Expert opinion was based on considerable prior field survey experience for leaf-tailed geckos, and on close scrutiny of topography, vegetation, and substrate on Google Earth. These surveys targeted previously unsurveyed areas around the known distributions that appeared to have significant amounts of suitable habitat buffered by topography (i.e. gullies, slopes, aspect) or rock to enable persistence over millennia (i.e. ‘lithorefugia’: Couper and Hoskin 2008). Round 1 of surveys greatly increased the number of known sites, particularly for P. gulbaru. This increase in number and spread of occurrence records for P. gulbaru (Fig. 1b) helped improve the resolution of SDMs (Fig. 2 versus Fig. S1). The increase in occurrence records made little difference for the P. amnicola SDMs (Fig. 2 versus Fig. S1), most likely because the new records were generally close to the records available before this study (Fig. 1c), and sampled similar environmental conditions.
All three final SDMs for each species (i.e. Climatic, Non-Climatic and All models) were then used to target additional survey efforts (Round 2). For both species, the SDMs predicted into the range of the other species (see Figs 2 and 3). In particular, the P. amnicola SDM predicted north through the distribution of P. gulbaru. This overprediction is interesting because it suggests ecological similarity between these two species. This similarity would probably result in competition if the species co-occurred, and this likely explains (along with dispersal limitations) why none of the nine Phyllurus species occur sympatrically (Hoskin et al. 2003). The ‘Climatic’ and ‘Non-Climatic’ SDMs for P. gulbaru suggested that small areas of suitable habitat exist just to the north of the known sites in the southern Paluma Range (Fig. 2e). A follow-up survey in this area indeed found a localised population at Mt Halifax (Fig. 3). It is likely that other small populations occur in this area. Surveys to the north of Mt Halifax in the central and northern Paluma Range failed to find any Phyllurus geckos (Fig. 3) but did find the northern leaf-tailed gecko (Saltuarius cornutus) at most sites. The ‘All’ SDM for P. amnicola identified Cape Cleveland and Mt Stuart as areas with high probability of presence, but Round 2 of surveys at these sites failed to find any Phyllurus geckos (see Figs 2f and 3). The habitat at Cape Cleveland, which was surveyed multiple times, appeared highly suitable. The absence of leaf-tailed geckos on Cape Cleveland probably reflects severe contraction or loss of rainforest habitat at this site historically, such as during the last glacial maxima when rainforest distribution in the Wet Tropics was much reduced (VanDerWal et al. 2009b).
We believe we have now resolved the breadth of distribution of both species. More undiscovered populations are likely present in between known ones (i.e. in the areas of high suitability in Fig. 3), but negative surveys around the current known sites suggest that we now know the main extent of both species’ distributions. Ground observations during field surveys, visual assessment of satellite imagery, and SDMs show that gaps of unsuitable habitat are currently present between known populations. In some cases these appear to be narrow gaps of rainforest lacking suitable rocky substrate, whereas in other cases the barriers are more substantial, lacking rainforest altogether. For P. gulbaru, the Pinnacles and Hervey Range populations are separated from other populations by barriers of unsuitable dry forest habitat 12 km and 3 km wide, respectively (Fig. 1). P. amnicola populations on Mt Elliot fall within an extensive interconnected patch of suitable habitat, but a narrow gap in suitable habitat (<2 km) exists between Mt Elliot and the Saddle Mountain population (Fig. 2). Although these gaps may not sound particularly large, they are probably significant barriers to current and historic gene flow because these Phyllurus geckos are restricted to rocky rainforest habitat and likely have very limited dispersal capabilities away from suitable microhabitat.
Phylogenetics can provide insights into population genetic structuring, and thus past and present isolation. In general, the genetic data supported the presence of barriers to gene flow in both species. All P. gulbaru sites were monophyletic, but genetic divergence was generally low (Fig. 1; Table 3). The exception was the Pinnacles population, which has an average divergence of ~6.5% from all other P. gulbaru populations (Fig. 1; Table 3). This level of divergence is on par with that between recognised subspecies of Phyllurus ossa (Couper and Hoskin 2013). This suggests that the Pinnacles population is currently isolated and has probably been isolated for at least hundreds of thousands of years, based on coarse molecular clock estimates for protein coding mtDNA genes (e.g. Moritz et al. 2000; Borsboom et al. 2010). There was very little genetic structuring between P. amnicola sites on Mt Elliot (up to 12 km straight-line apart), but ~2.9% divergence between Mt Elliot and Saddle Mountain sites (Fig. 1; Table 3). This suggests that the gap in suitable habitat between these two mountains, estimated currently at ~2 km straight-line, has been in place for a considerable period. Although we have only used mtDNA sequence divergence in this study, mtDNA divergence is generally correlated with nDNA divergence when assessing historical population structuring (e.g. Singhal et al. 2018). Genome-wide nuclear markers such as single nucleotide polymorphisms (SNPs) would be required to resolve patterns of population connectivity in more recent times (Gagnaire et al. 2015).
Species distribution modelling of narrow endemics
This study shows how Species Distribution Models (SDMs) can provide useful information for the conservation of microendemics. As covered above, for both species, models generated from a moderate number of unique occurrence localities gave informative estimates of the range of both species, identified additional areas to survey, found additional populations, identified likely gaps in suitable habitat (in congruence with genetic structuring and field surveys), and gave insights into ecological similarity between the two species.
The SDMs for both species also show areas of modelled habitat where the species do not occur, which reveals limitations in the modelling. As covered above, reasons why the species are restricted to a subset of predicted locations include habitat barriers and dispersal limitations, aspects of microhabitat suitability that the models do not include (e.g. depth of layered rock or rock type), and competition. Competition is important in determining distributions but is complex and not easily incorporated into SDMs. Ecological similarity between Phyllurus geckos is discussed above but competition with Saltuarius leaf-tailed geckos may also play a role in shaping distributions. S. cornutus is common throughout the Wet Tropics, with the most southerly records being at the northern P. gulbaru sites (Mt Halifax, Bluewater Range, Patterson’s Gorge). We found S. cornutus and P. gulbaru in close proximity at two of these sites but S. cornutus was at low density and typically on trees (versus P. gulbaru on rocks). However, S. cornutus is often found on rocks elsewhere in the Wet Tropics and may be a reason why P. gulbaru is not found further north on the Paluma Range (where S. cornutus is common) and, more broadly, why Phyllurus geckos are absent from the Wet Tropics.
It is worth noting that the addition of a modest number of occurrence points by the targeted surveys in this study substantially improved the resolution of the Climatic SDM, particularly for P. gulbaru (Fig. 2 versus Fig. S1). However, it is important to reiterate that deriving ecologically sensible models for microendemic species needs to account for the inherently small number of occurrence points. SDMs produced from small numbers of unique occurrences must be interpreted with caution because of their tendency to overfit and their higher sensitivity to sampling bias. Tuning of regularisation multipliers and feature classes can reduce these effects, and should thus become common practice (Anderson and Gonzalez 2011; Shcheglovitova and Anderson 2013; Radosavljevic and Anderson 2014). As Galante et al. (2018) recently showed, tuning of parameters and model selection can reduce the effect of overfitting and sampling bias by selecting fewer parameters and providing more general and ecologically sensible models. In particular, information criteria such as AICc seem to perform well for model selection in data-poor species (Warren and Seifert 2011; Galante et al. 2018).
Potential threats to P. gulbaru and P. amnicola
With the distributions largely resolved, a better assessment of potential threats and their likely influence is possible. Although these species appear to have persisted in these localised areas for millions of years (Couper et al. 2000; Hoskin et al. 2003), novel human-induced threats are present. Changes to fire regimes has been highlighted as a potential threat to rainforest areas in the Townsville region and other parts of tropical Australia (e.g. Russell-Smith and Stanton 2002; Hoskin et al. 2003). Fire is a natural part of this region but the frequency, intensity, and timing of fires has changed in the last century due to burning for cattle grazing, fire management through high-frequency burning, late dry-season burning, and the introduction of highly flammable grasses. In particular, guinea grass (Megathyrsus maximus) and molasses grass (Melinis minutiflora) flourish on the interface between rainforest and open woodland in this region, creating high fuel loads around rainforest patches (Russell-Smith and Stanton 2002). These higher fuel loads, in conjunction with inappropriate fire regimes, particularly late dry-season burning, may be incrementally reducing rainforest area and connectivity in this region (Hoskin et al. 2003). Increased extreme events such as prolonged drought and heatwaves are projected as climate change progresses (Mann et al. 2017), potentially increasing impacts on fire-sensitive habitats in this region.
Poaching represents another significant threat. P. amnicola individuals have been poached from Mt Elliot in recent years, as revealed by their possession and sale on the internet in the USA and Germany. Australian leaf-tailed geckos (Phyllurus, Saltuarius and Orraya) are highly desired by collectors due to their unusual appearance and their rarity in the wild and in collections. Field surveys conducted in 2013 detected a substantial drop in population density of P. amnicola along a section of Alligator Creek (CJH, pers. obs.) before poaching was confirmed by the online advertisement of individuals for sale by a collector in the USA. The poacher apparently obtained specific site details from a post on the internet that outlined access to the site and where to find the geckos, and probably also from the original description paper. This reinforces the push to keep site details for rare and desirable species secret or vague, even in description papers (Lindenmayer and Scheele 2017).
An additional potential threat to both species is competition from the invasive gecko Hemidactylus frenatus (Hoskin 2011). This species is invading natural habitats in the Townsville region, and occurs as high-density breeding populations in some forest habitats of the Townsville region (Barnett et al. 2017). H. frenatus has not yet been detected at any Phyllurus sites but will presumably spread to some of these with time. Continued monitoring is required to assess the degree to which they will invade these sites and the potential competitive impacts on Phyllurus.
Management recommendations
We assess management for each species by first identifying evolutionarily significant units (ESUs) within each species, then identifying potential management actions for threats outlined above, and finally assessing whether the current conservation status is appropriate based on the results herein. Crandall et al. (2000) argued that the concept of ESUs should incorporate both genetic and phenotypic data so that management preserves adaptive diversity across the range of a species. Their criteria for determining ESUs involves assessing historic and current gene flow (e.g. mtDNA divergence for historic genetic flow; divergence at microsatellites or SNPs for current gene flow) and assessing divergence in phenotypic traits (e.g. morphology, habitat, physiology) (Crandall et al. 2000). In determining ESUs in P. gulbaru and P. amnicola we reject historic gene flow where we find at least moderate mtDNA divergence. We did not assess current gene flow, genetically, but did assess the potential for current gene flow during field surveys and modelling. We reject current gene flow where the field surveys and SDMs suggest substantial current habitat barriers. In terms of phenotypic divergence, we do not present morphological data in this paper but summarise differences in morphology that have been identified between populations (Hoskin, unpubl. data).
Phyllurus gulbaru is more widespread than previously thought; however, it still has a localised and highly fragmented distribution, with each population restricted to a small area of suitable habitat (Fig. 1b). The Pinnacles population is an ESU that should be managed separately from other populations. This population is highly divergent for mtDNA (6.5% uncorrected p-distance) and currently isolated by a substantial habitat barrier (Figs 1, 2). It is also morphologically distinct in terms of body proportions, tail shape, and aspects of scalation (Hoskin, unpubl. data). These genetic and phenotypic differences are of a similar or greater magnitude than seen between subspecies of P. ossa (Couper and Hoskin 2013) and the Pinnacles population is being described as a subspecies (Hoskin, unpubl. data). The remaining populations of P. gulbaru should be considered a series of isolated populations within a single ESU. The mtDNA monophyly of each population, albeit with minimal divergence (Fig. 1), suggests at least recent isolation, as supported by the highly localised nature of suitable habitat observed during field surveys. This is particularly the case for the Hervey Range population, which is the most divergent and isolated in that ESU. However, we have not detected morphological differences among these populations (Hoskin, unpubl. data). Although all one ESU, these populations still need to be managed separately because each occupies an isolated area of habitat that could be subject to different threats. Understanding connectivity within this ESU, through more detailed population genetic analyses, may be useful for future management decisions.
Fire is a threat to the more southerly populations, particularly South Patterson’s Gorge, Hervey Range and The Pinnacles (Fig. 1b). All of these sites consist of small areas of rocky, relatively dry, rainforest habitat surrounded by open, fire-prone vegetation (including high fuel loads of guinea grass). Late dry-season fires, which occur regularly in this area, have incrementally reduced the rainforest of at least two of these sites over the last 15 years (CJH, pers. obs.). Active fire management is required in these areas to protect the rainforest fragments and facilitate expansion into adjacent rocky habitat near P. gulbaru sites. This could involve reducing fire frequency and conducting controlled burning early in the dry season to protect these areas from uncontrolled fires later in the dry season. Poaching is another potential threat and site details should be limited to protected area managers only.
Phyllurus gulbaru should retain its current listing of Endangered (under both IUCN 2012 and Queensland NCA 1992). For example, it fits the IUCN (2012) Endangered listing (B1, B2, a, b) due to its small extent of occurrence and area of occupancy, severely fragmented populations, and observed and projected reduction in area of suitable habitat from inappropriate burning at southern sites. At the federal level, the species is currently listed as Critically Endangered (EPBC Act 1999). Given the increase in distributional area and number of locations/populations in this study, P. gulbaru no longer fulfils a Critically Endangered listing and we suggest that this be reviewed to consider an Endangered EPBC listing. Description of The Pinnacles population as a subspecies (Hoskin, unpubl. data) would then require individual assessment of conservation status. A research priority will be estimating population size at The Pinnacles, where only four individuals have been observed at two sites over five targeted surveys.
Phyllurus amnicola has a highly localised distribution, being restricted to Mt Elliot and Saddle Mountain. The species is widespread and probably fairly connected across Mt Elliot, but appears to be more localised on Saddle Mountain. The Mt Elliot and Saddle Mountain populations should be managed independently as separate ESUs. This is due to mtDNA divergence (2.9% uncorrected p-distance), suggesting historical isolation, and the observed probable barrier of unsuitable habitat between the two mountains, suggesting no current gene flow. Additionally, the Saddle Mountain adults are ~25% smaller (for snout–vent length) than those at all sites on Mt Elliot (Hoskin, unpubl. data). The sites on Mt Elliot should be managed as a set of patchy, but at least partially connected, subpopulations, with the aim of maintaining connectivity and gene flow.
Fire appears to be less of a threat to P. amnicola because the sites on Mt Elliot and Saddle Mountain are generally moister and more buffered by extensive rocky areas than P. gulbaru sites. Nonetheless, efforts to reduce fire incursion of rainforest are required for P. amnicola and other highly localised rainforest endemics of Mt Elliot (e.g. the Mt Elliot nursery-frog (Cophixalus mcdonaldi), the Mt Elliot sunskink (Lampropholis elliotensis), the Mt Elliot crayfish (Euastacus bindal), and the Mt Elliot assassin spider (Austrarchaea hoskini). As for P. gulbaru sites, management efforts for P. amnicola could involve reducing fire frequency, limiting late dry-season burning, and strategic burning to protect specific areas. As mentioned above, illegal collection of P. amnicola is known to have occurred. Stopping poaching is difficult but should involve limiting site information to protected area managers only and installing hidden cameras in P. amnicola habitat to monitor visitation.
Phyllurus amnicola fits the IUCN (2012) Endangered criteria in terms of extent of occurrence (B1), area of occupancy (B2), and small number of locations (Mt Elliot and Saddle Mountain) (B2a), but fulfilling a B category listing is dependent on an observed, estimated or projected decline in habitat area or number of mature individuals (B2b) (IUCN 2012). Poaching is a known threat but whether the observed reduction in adults at one site due to poaching has significantly impacted the species, or whether poaching is a continuing threat, is unknown. Fire is another potential threat but currently fire does not appear to be impacting the known sites. A research priority is monitoring whether H. frenatus will invade P. amnicola habitat and assessing potential competitive impacts on Phyllurus. We recommend that P. amnicola remain unlisted, pending further research on potential threats.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This research was conducted under Queensland Environment and Heritage Protection permits (WISP16444615, WITK16444715, WITK13653913) and James Cook University Animal Ethics (A2225). This research did not receive any specific funding. We thank Stephen Zozaya, Anders Zimny, Leah Carr, Jenny Cocciardi, Jaimie Hopkins and Tobias Baumgartner for help during field surveys; Diana Pazmino and Jaimie Hopkins for help in the laboratory; and Collin Storlie for help in obtaining environmental layers and modelling. We also thank local protected area managers Kenneth McMahon, Leigh Benson and Marty McLaughlin for site access information and general discussions.
References
Aho, K., Derryberry, D., and Peterson, T. (2014). Model selection for ecologists: the worldviews of AIC and BIC. Ecology 95, 631–636.| Model selection for ecologists: the worldviews of AIC and BIC.Crossref | GoogleScholarGoogle Scholar |
Anderson, R. P., and Gonzalez, I. (2011). Species-specific tuning increases robustness to sampling bias in models of species distributions: an implementation with Maxent. Ecological Modelling 222, 2796–2811.
| Species-specific tuning increases robustness to sampling bias in models of species distributions: an implementation with Maxent.Crossref | GoogleScholarGoogle Scholar |
Auliya, M., Altherr, S., Ariano-Sanchez, D., Baard, E. H., Brown, C., Brown, R. M., Cantu, J., Gentile, G., Gildenhuys, P., Henningheim, E., Hintzmann, J., Kanari, K., Krvavac, M., Lettink, M., Lippert, J., Luiselli, L., Nilson, G., Nguyen, T. Q., Nijman, V., Parham, J. F., Pasachnik, S. A., Pedrono, M., Rauhaus, A., Cordova, D. R., Sanchez, M., Schepp, U., van Schingen, M., Schneeweiss, N., Segniagbeto, G. H., Somaweera, R., Sy, E. Y., Turkozan, O., Vinke, S., Vinke, T., Vyas, R., Williamson, S., and Ziegler, T. (2016). Trade in live reptiles, its impact on wild populations, and the role of the European market. Biological Conservation 204, 103–119.
| Trade in live reptiles, its impact on wild populations, and the role of the European market.Crossref | GoogleScholarGoogle Scholar |
Barnett, L. K., Phillips, B. L., and Hoskin, C. J. (2017). Going feral: time and propagule pressure determine range expansion of Asian house geckos into natural environments. Austral Ecology 42, 165–175.
| Going feral: time and propagule pressure determine range expansion of Asian house geckos into natural environments.Crossref | GoogleScholarGoogle Scholar |
Bateman, B. L., VanDerWal, J., Williams, S. E., and Johnson, C. N. (2012). Biotic interactions influence the projected distribution of a specialist mammal under climate change. Diversity & Distributions 18, 861–872.
| Biotic interactions influence the projected distribution of a specialist mammal under climate change.Crossref | GoogleScholarGoogle Scholar |
Bell, R. C., Parra, J. L., Tonione, M., Hoskin, C. J., Mackenzie, J. B., Williams, S. E., and Moritz, C. (2010). Patterns of persistence and isolation indicate resilience to climate change in montane rainforest lizards. Molecular Ecology 19, 2531–2544.
Borsboom, A. C., Couper, P. J., Amey, A., and Hoskin, C. J. (2010). Distribution and population genetic structure of the critically endangered skink Nangura spinosa, and the implications for management. Australian Journal of Zoology 58, 369–375.
| Distribution and population genetic structure of the critically endangered skink Nangura spinosa, and the implications for management.Crossref | GoogleScholarGoogle Scholar |
Bourg, N. A., McShea, W. J., and Gill, D. E. (2005). Putting a CART before the search: successful habitat prediction for a rare forest herb. Ecology 86, 2793–2804.
| Putting a CART before the search: successful habitat prediction for a rare forest herb.Crossref | GoogleScholarGoogle Scholar |
Burnham, K. P., and Anderson, D. R. (2004). Multimodel inference understanding AIC and BIC in model selection. Sociological Methods & Research 33, 261–304.
| Multimodel inference understanding AIC and BIC in model selection.Crossref | GoogleScholarGoogle Scholar |
Couper, P., and Hoskin, C. (2008). Litho-refugia: the importance of rock landscapes for the long-term persistence of Australian rainforest fauna. Australian Zoologist 34, 554–560.
| Litho-refugia: the importance of rock landscapes for the long-term persistence of Australian rainforest fauna.Crossref | GoogleScholarGoogle Scholar |
Couper, P., and Hoskin, C. J. (2013). Two new subspecies of the leaf-tailed gecko Phyllurus ossa (Lacertilia: Carphodactylidae) from mid-eastern Queensland, Australia. Zootaxa 3664, 537–553.
| Two new subspecies of the leaf-tailed gecko Phyllurus ossa (Lacertilia: Carphodactylidae) from mid-eastern Queensland, Australia.Crossref | GoogleScholarGoogle Scholar |
Couper, P. J., Schneider, C. J., Hoskin, C. J., and Covacevich, J. A. (2000). Australian leaf-tailed geckos: phylogeny, a new genus, two new species and other new data. Memoirs of the Queensland Museum 45, 253–266.
Crandall, K. A., Bininda-Emonds, O. R., Mace, G. M., and Wayne, R. K. (2000). Considering evolutionary processes in conservation biology. Trends in Ecology & Evolution 15, 290–295.
Cunningham, H. R., Rissler, L. J., Buckley, L. B., and Urban, M. C. (2016). Abiotic and biotic constraints across reptile and amphibian ranges. Ecography 39, 1–8.
| Abiotic and biotic constraints across reptile and amphibian ranges.Crossref | GoogleScholarGoogle Scholar |
Curtis, L. K. (Ed.) (2012). ‘Queensland’s Threatened Animals.’ (CSIRO Publishing: Melbourne.)
ESRI (2012). ‘ArcGIS 10.1.’ (ESRI: Redlands, CA.)
Fordham, D. A., Brook, B. W., Hoskin, C. J., Pressey, R. L., VanDerWal, J., and Williams, S. E. (2016). Extinction debt from climate change for frogs in the wet tropics. Biology Letters 12, 20160236.
| Extinction debt from climate change for frogs in the wet tropics.Crossref | GoogleScholarGoogle Scholar |
Gagnaire, P. A., Broquet, T., Aurelle, D., Viard, F., Souissi, A., Bonhomme, F., Arnaud-Haondm, S., and Bierne, N. (2015). Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evolutionary Applications 8, 769–786.
| Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era.Crossref | GoogleScholarGoogle Scholar |
Galante, P. J., Alade, B., Muscarella, R., Jansa, S. A., Goodman, S. M., and Anderson, R. P. (2018). The challenge of modeling niches and distributions for data‐poor species: a comprehensive approach to model complexity. Ecography 41, 726–736.
| The challenge of modeling niches and distributions for data‐poor species: a comprehensive approach to model complexity.Crossref | GoogleScholarGoogle Scholar |
Gilpin, M. E., and Soule, M. E. (1986). Minimum viable populations: processes of species extinction. In ‘Conservation Biology: the Science of Scarcity and Diversity’. (Ed. M. E. Soule.) pp. 19–34. (Sinauer Associates: Sunderland, MA.)
Graham, C. H., VanDerWal, J., Phillips, S. J., Moritz, C., and Williams, S. E. (2010). Dynamic refugia and species persistence: tracking spatial shifts in habitat through time. Ecography 33, 1062–1069.
| Dynamic refugia and species persistence: tracking spatial shifts in habitat through time.Crossref | GoogleScholarGoogle Scholar |
Harvey, M. S., Rix, M. G., Framenau, V. W., Hamilton, Z. R., Johnson, M. S., Teale, R. J., Humphreys, G., and Humphreys, W. F. (2011). Protecting the innocent: studying short-range endemic taxa enhances conservation outcomes. Invertebrate Systematics 25, 1–10.
| Protecting the innocent: studying short-range endemic taxa enhances conservation outcomes.Crossref | GoogleScholarGoogle Scholar |
Hernandez, P. A., Graham, C. H., Master, L. L., and Albert, D. L. (2006). The effect of sample size and species characteristics on performance of different species distribution modelling methods. Ecography 29, 773–785.
| The effect of sample size and species characteristics on performance of different species distribution modelling methods.Crossref | GoogleScholarGoogle Scholar |
Hijmans, R. J., van Etten, J., Cheng, J., Mattiuzzi, M., Sumner, M., Greenberg, J. A., Lamigueiro, O. P., Bevan, A., Racine, E. B., Shortridge, A. (2015). ‘Raster’. R package version 2.6-7.
Hoskin, C. J. (2004). Australian microhylid frogs (Cophixalus and Austrochaperina): phylogeny, taxonomy, calls, distributions and breeding biology. Australian Journal of Zoology 52, 237–269.
| Australian microhylid frogs (Cophixalus and Austrochaperina): phylogeny, taxonomy, calls, distributions and breeding biology.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J. (2011). The invasion and potential impact of the Asian house gecko (Hemidactylus frenatus) in Australia. Austral Ecology 36, 240–251.
| The invasion and potential impact of the Asian house gecko (Hemidactylus frenatus) in Australia.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J. (2014). A new skink (Scincidae: Carlia) from the rainforest uplands of Cape Melville, north-east Australia. Zootaxa 3869, 224–236.
| A new skink (Scincidae: Carlia) from the rainforest uplands of Cape Melville, north-east Australia.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J., and Couper, P. J. (2013). A spectacular new leaf-tailed gecko (Carphodactylidae: Saltuarius) from the Melville Range, north-east Australia. Zootaxa 3717, 543–558.
| A spectacular new leaf-tailed gecko (Carphodactylidae: Saltuarius) from the Melville Range, north-east Australia.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J., and Couper, P. J. (2014). Two new skinks (Scincidae: Glaphyromorphus) from rainforest habitats in north-eastern Australia. Zootaxa 3869, 1–16.
| Two new skinks (Scincidae: Glaphyromorphus) from rainforest habitats in north-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J., Couper, P. J., and Schneider, C. J. (2003). A new species of Phyllurus (Lacertilia: Gekkonidae) and a revised phylogeny and key for the Australian leaf-tailed geckos. Australian Journal of Zoology 51, 153–164.
| A new species of Phyllurus (Lacertilia: Gekkonidae) and a revised phylogeny and key for the Australian leaf-tailed geckos.Crossref | GoogleScholarGoogle Scholar |
Hoskin, C. J., Tonione, M., Higgie, M., MacKenzie, J. B., Williams, S. E., VanDerWal, J., and Moritz, C. (2011). Persistence in peripheral refugia promotes phenotypic divergence and speciation in a rainforest frog. American Naturalist 178, 561–578.
| Persistence in peripheral refugia promotes phenotypic divergence and speciation in a rainforest frog.Crossref | GoogleScholarGoogle Scholar |
Hugall, A., Moritz, C., Moussalli, A., and Stanisic, J. (2002). Reconciling paleodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875). Proceedings of the National Academy of Sciences of the United States of America 99, 6112–6117.
| Reconciling paleodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875).Crossref | GoogleScholarGoogle Scholar |
Hutchinson, M., Xu, T., Houlder, D., Nix, H., and McMahon, J. (2009). ANUCLIM 6.0 User’s Guide. Fenner School of Environment and Society, Australian National University, Canberra.
IUCN (2012). ‘IUCN Red List Categories and Criteria: Version 3.1.’ (IUCN: Gland, Switzerland.)
Jiménez‐Valverde, A. (2012). Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Global Ecology and Biogeography 21, 498–507.
| Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling.Crossref | GoogleScholarGoogle Scholar |
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., and Drummond, A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Pääbo, S., Villablanca, F. X., and Wilson, A. C. (1989). Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences of the United States of America 86, 6196–6200.
| Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers.Crossref | GoogleScholarGoogle Scholar |
Lanfear, R., Calcott, B., Ho, S. Y., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29, 1695–1701.
| PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |
Laurance, W. F. (1997). A distributional survey and habitat model for the endangered northern bettong Bettongia tropica in tropical Queensland. Biological Conservation 82, 47–60.
| A distributional survey and habitat model for the endangered northern bettong Bettongia tropica in tropical Queensland.Crossref | GoogleScholarGoogle Scholar |
Lindenmayer, D., and Scheele, B. (2017). Do not publish. Science 356, 800–801.
| Do not publish.Crossref | GoogleScholarGoogle Scholar |
MacArthur, R. H., and Wilson, E. O. (1967). ‘The Theory of Island Biogeography.’ (Princeton University Press: Princeton, NJ.)
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Molecular Biology and Evolution 14, 91–104.
| Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome.Crossref | GoogleScholarGoogle Scholar |
Mann, M. E., Rahmstorf, S., Kornhuber, K., Steinman, B. A., Miller, S. K., and Coumou, D. (2017). Influence of anthropogenic climate change on planetary wave resonance and extreme weather events. Scientific Reports 7, 45242.
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 16, 1215.
| A simple salting out procedure for extracting DNA from human nucleated cells.Crossref | GoogleScholarGoogle Scholar |
Moritz, C., Patton, J. L., Schneider, C., and Smith, T. B. (2000). Diversification of rainforest faunas: an integrated molecular approach. Annual Review of Ecology and Systematics 31, 533–563.
| Diversification of rainforest faunas: an integrated molecular approach.Crossref | GoogleScholarGoogle Scholar |
Moritz, C., Hoskin, C., Graham, C. H., Hugall, A., Moussalli, A., and Purvis, A. (2005). Historical biogeography, diversity and conservation of Australia’s tropical rainforest herpetofauna. In ‘Phylogeny and Conservation’. (Eds A. Purvis, J. L. Gittleman, and T. Brooks.) pp. 243–264. (Cambridge University Press: New York.)
Moritz, C., Hoskin, C. J., MacKenzie, J. B., Phillips, B. L., Tonione, M., Silva, N., VanDerWal, J., Williams, S. E., and Graham, C. H. (2009). Identification and dynamics of a cryptic suture zone in tropical rainforest. Proceedings of the Royal Society B: Biological Sciences 276, 1235–1244.
| Identification and dynamics of a cryptic suture zone in tropical rainforest.Crossref | GoogleScholarGoogle Scholar |
Murray, P., and de Jong, C. (2017). Range extension for the leaf-tailed gecko Phyllurus gulbaru in north Queensland. Memoirs of the Queensland Museum 60, 176.
Muscarella, R., Galante, P. J., Soley‐Guardia, M., Boria, R. A., Kass, J. M., Uriarte, M., and Anderson, R. P. (2014). ENMeval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods in Ecology and Evolution 5, 1198–1205.
| ENMeval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models.Crossref | GoogleScholarGoogle Scholar |
Neldner, V. J., Niehus, R. E., Wilson, B. A., McDonald, W. J. F., Ford, A. J., and Accad, A. (2017). The vegetation of Queensland. Descriptions of broad vegetation groups. Version 3.0. Queensland Herbarium, Department of Science, Information Technology and Innovation.
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R., Moret, B. M., and Stamatakis, A. (2010). How many bootstrap replicates are necessary? Journal of Computational Biology 17, 337–354.
| How many bootstrap replicates are necessary?Crossref | GoogleScholarGoogle Scholar |
Peterson, A. T. (2011). ‘Ecological Niches and Geographic Distributions.’ (Princeton University Press: Princeton, NJ.)
Phillips, S. J., and Dudik, M (2008). Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175.
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modelling of species geographic distributions. Ecological Modelling 190, 231–259.
| Maximum entropy modelling of species geographic distributions.Crossref | GoogleScholarGoogle Scholar |
Pope, L. (2000). Population structure of the northern bettong. Ph.D. Thesis, University of Queensland, Brisbane.
R Development Core Team (2013). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.r-project.org.
Radosavljevic, A., and Anderson, R. P. (2014). Making better Maxent models of species distributions: complexity, overfitting and evaluation. Journal of Biogeography 41, 629–643.
| Making better Maxent models of species distributions: complexity, overfitting and evaluation.Crossref | GoogleScholarGoogle Scholar |
Raxworthy, C. J., Martinez-Meyer, E., Horning, N., Nussbaum, R. A., Schneider, G. E., Ortega-Huerta, M. A., and Peterson, A. T. (2003). Predicting distributions of known and unknown reptile species in Madagascar. Nature 426, 837–841.
| Predicting distributions of known and unknown reptile species in Madagascar.Crossref | GoogleScholarGoogle Scholar |
Read, K., Keogh, J. S., Scott, I. A., Roberts, J. D., and Doughty, P. (2001). Molecular phylogeny of the Australian frog genera Crinia, Geocrinia, and allied taxa (Anura: Myobatrachidae). Molecular Phylogenetics and Evolution 21, 294–308.
| Molecular phylogeny of the Australian frog genera Crinia, Geocrinia, and allied taxa (Anura: Myobatrachidae).Crossref | GoogleScholarGoogle Scholar |
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A., and Huelsenbeck, J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
| MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Crossref | GoogleScholarGoogle Scholar |
Rosauer, D. F., Catullo, R. A., VanDerWal, J, Moussalli, A, Hoskin, C. J., and Moritz, C (2017). Correction: lineage range estimation method reveals fine-scale endemism linked to Pleistocene stability in Australian rainforest herpetofauna. PLoS One 12, e0169726.
Russell-Smith, J., and Stanton, P. (2002). Fire regimes and fire management of rainforest communities across northern Australia. In ‘Flammable Australia: the Fire Regimes and Biodiversity of a Continent’. (Eds R. A. Bradstock, J. Williams, and A. M. Gill.) pp. 329–350. (Cambridge University Press: Cambridge.)
Shcheglovitova, M., and Anderson, R. P. (2013). Estimating optimal complexity for ecological niche models: a jack-knife approach for species with small sample sizes. Ecological Modelling 269, 9–17.
| Estimating optimal complexity for ecological niche models: a jack-knife approach for species with small sample sizes.Crossref | GoogleScholarGoogle Scholar |
Singhal, S., Hoskin, C. J., Couper, P., Potter, S., and Moritz, C. (2018). A framework for resolving cryptic species: a case study from the lizards of the Australian Wet Tropics. Systematic Biology , .
| A framework for resolving cryptic species: a case study from the lizards of the Australian Wet Tropics.Crossref | GoogleScholarGoogle Scholar |
Sistrom, M. J., Hutchinson, M. N., Hutchinson, R. G., and Donnellan, S. C. (2009). Molecular phylogeny of Australian Gehyra (Squamata: Gekkonidae) and taxonomic revision of Gehyra variegata in south-eastern Australia. Zootaxa 2277, 14–32.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
| RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Crossref | GoogleScholarGoogle Scholar |
Storlie, C. J., Phillips, B. L., VanDerWal, J. J., and Williams, S. E. (2013). Improved spatial estimates of climate predict patchier species distributions. Diversity & Distributions 19, 1106–1113.
| Improved spatial estimates of climate predict patchier species distributions.Crossref | GoogleScholarGoogle Scholar |
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729.
| MEGA6: molecular evolutionary genetics analysis version 6.0.Crossref | GoogleScholarGoogle Scholar |
Terborgh, J., and Winter, B. (1980). Some causes of extinction. In ‘Conservation Biology: an Evolutionary–Ecological Perspective’. (Eds M. E. Soulé, and B. A. Wilcox.) pp. 119–133. (Sinauer Associates: Sunderland, MA.)
Thompson, J. D., Gibson, T., and Higgins, D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics 00, 2–3.
van Gils, H., Westinga, E., Carafa, M., Antonucci, A., and Ciaschetti, G. (2014). Where the bears roam in Majella National Park, Italy. Journal for Nature Conservation 22, 23–34.
| Where the bears roam in Majella National Park, Italy.Crossref | GoogleScholarGoogle Scholar |
VanDerWal, J., Shoo, L. P., Graham, C., and Williams, S. E. (2009a). Selecting pseudo-absence data for presence-only distribution modelling: how far should you stray from what you know? Ecological Modelling 220, 589–594.
| Selecting pseudo-absence data for presence-only distribution modelling: how far should you stray from what you know?Crossref | GoogleScholarGoogle Scholar |
VanDerWal, J., Shoo, L. P., and Williams, S. E. (2009b). New approaches to understanding late Quaternary climate fluctuations and refugial dynamics in Australian wet tropical rain forests. Journal of Biogeography 36, 291–301.
| New approaches to understanding late Quaternary climate fluctuations and refugial dynamics in Australian wet tropical rain forests.Crossref | GoogleScholarGoogle Scholar |
Vernes, K., and Pope, L. C. (2006). Capture success and population density of the northern bettong Bettongia tropica in northeastern Queensland. Australian Mammalogy 28, 87–92.
| Capture success and population density of the northern bettong Bettongia tropica in northeastern Queensland.Crossref | GoogleScholarGoogle Scholar |
Warren, D. L., and Seifert, S. N. (2011). Ecological niche modelling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications 21, 335–342.
| Ecological niche modelling in Maxent: the importance of model complexity and the performance of model selection criteria.Crossref | GoogleScholarGoogle Scholar |
Williams, S. E., Bolitho, E. E., and Fox, S. (2003). Climate change in Australian tropical rainforests: an impending environmental catastrophe. Proceedings. Biological Sciences 270, 1887–1892.
| Climate change in Australian tropical rainforests: an impending environmental catastrophe.Crossref | GoogleScholarGoogle Scholar |
Wilson, S., and Swan, G. (2013). ‘Complete Guide to Reptiles of Australia.’ 4th edn. (New Holland Publishers: Sydney.)