Kangaroo gene mapping and sequencing: insights into mammalian genome evolution
Jennifer A. Marshall GravesLa Trobe Institute of Molecular Sciences, La Trobe University, Melbourne, Vic. 3186, Australia and Research School of Biology, Australian National University, Canberra, ACT 2060, Australia. Email: jenny.graves@anu.edu.au
Australian Journal of Zoology 61(1) 4-12 https://doi.org/10.1071/ZO13002
Submitted: 7 January 2013 Accepted: 30 April 2013 Published: 7 June 2013
Journal Compilation © CSIRO Publishing 2013 Open Access CC BY-NC-ND
Abstract
The deep divergence of marsupials and eutherian mammals 160 million years ago provides genetic variation to explore the evolution of DNA sequence, gene arrangement and regulation of gene expression in mammals. Following the pioneering work of Professor Desmond W. Cooper, emerging techniques in cytogenetics and molecular biology have been adapted to characterise the genomes of kangaroos and other marsupials. In particular, genetic and genomic work over four decades has shown that marsupial sex chromosomes differ significantly from the eutherian XY chromosome pair in their size, gene content and activity. These differences can be exploited to deduce how mammalian sex chromosomes, sex determination and epigenetic silencing evolved.
Introduction
‘Why would anyone want to map genes in a kangaroo?’ I asked Des Cooper in exasperation. At Des’ suggestion, I had recently arrived at La Trobe University, fresh from my Ph.D. at Berkeley and with new cell fusion technology ready to explore the control of DNA synthesis in mammals. I was quite dismissive of Australian scientists who worked on the ‘local fauna’, so I was nonplussed at Des’ insistence that I use my somatic cell genetic techniques to map genes on the kangaroo X chromosome. But, like the outcome of many of my arguments with Des, he carried the day, and I have pursued his far-sighted vision ever since. With my colleague Rory Hope (another Adelaide contemporary), I spent the next 10 years developing rodent–marsupial cell hybrids and assigning genes to the X chromosome in several marsupial species. Mapping kangaroo genes took over my life and led to my participation in the Human Gene Mapping workshops that ultimately grew into mammalian (including marsupial) genome projects.
What Des propounded from our earliest interaction (as graduate students in Adelaide in the 1960s) was that marsupial and eutherian (‘placental’) mammals constitute independent experiments in mammal evolution. Marsupials last shared a common ancestor with placental mammals ~160 million years ago (Mya) (Luo et al. 2011). So there has been plenty of time – more than twice the time that humans and mice have been evolving separately – for the two groups of mammals to evolve different genome arrangements, novel genes and different ways of regulating them. Within marsupials there are also closely and distantly related species that can be analysed; for instance, kangaroo species radiated only ~18 Mya from an ancestral macropodid that diverged ~45 Mya from the dasyurid marsupials. Australian marsupials diverged ~70 Mya from American marsupials such as the opossum.
Of particular interest to Des, and to me, was the contributions marsupials could make to our understanding of sex chromosome function and evolution. The X chromosome of placental mammals is extremely conserved in gene content and gene order, and shares the same complex mechanism of epigenetic silencing that renders one X inactive in female somatic cells. The Y chromosome, on the other hand, contains few active genes and is quite variable in gene content, the result of loss of active genes from the Y as it degrades from its original identity with the X. Again, the very ancient divergence of marsupials might mean that the sex chromosomes have different gene content and different means of regulation and could provide clues about how the X and Y evolved in mammals and how they work to determine sex and to regulate activity. This approach proved more productive than either of us could possibly have imagined.
Marsupial chromosomes
With their low diploid number and large size, marsupial chromosomes have provided material for classic studies of chromosome structure, evolution and radiation biology. Even before the introduction of g-banding, careful observations of sizes and arm ratios strongly hinted that marsupial chromosomes are highly conserved across all 21 families, and the occurrence of modes at 2n = 14 and 2n = 22 (Sharman 1973; Hayman and Martin 1974) suggested that one or other might be ancestral. The observation that the 2n = 14 mode was g-band identical (Rofe and Hayman 1985) meant that in Adelaide our money was on the 2n = 14 karyotype, but the presence of telomere sequences at junction points in South American marsupials offered an alternative view that the low number was secondarily derived from centric fusions in an Australian marsupial ancestor (Svartman and Vianna-Morgante 1999).
Marsupial chromosomes show some striking differences in behaviour, displaying low recombination in females rather than males (Bennett et al. 1986; van Oorschot et al. 1992), the opposite pattern from that observed in placental mammals.
Marsupial chromosomes also display some unusual structures. Marsupial centromeres are relatively short and simple, consisting of a few hundred kilobases, rather than the megabases of α repetitive DNA of the mouse centromere. Several types of repeat are interspersed (Carone and O’Neill 2010), and kangaroo centromeres have accumulated an endogenous retroviral sequence KERV, which is transcribed and has actively amplified in several macropodid lineages (Carone and O’Neill 2010; Ferreri et al. 2011). This element may facilitate the frequent Robertsonian fusions observed in macropodid marsupials (Bulazel et al. 2007).
The chromosome ends of one marsupial group are also unusual. Dasyurid telomeres have recently been found to exhibit a parent-specific control of length (Bender et al. 2012). This was first observed in Tasmanian devils as extremely heterogeneous binding of telomere-specific sequences to the two homologues of each chromosome: one haploid set had small telomeres, and the other set had enormous telomeres. Every animal examined showed the same effect, excluding hybridisation between a long- and a short-telomere subpopulation, and other dasyurids exhibit the same phenomenon. In male-derived cells the X had small, and the Y very large, telomeres, suggesting that telomeres are lengthened in the testis and shortened in the ovary. This parent-specific control of telomere length does not conform with either the well established telomerase mechanism (Blackburn et al. 2006), or the ALT method of telomere lengthening (Cesare and Reddel 2010). It is possible that a completely novel mechanism has evolved in this family to counteract stress in males that undergo frenetic mating and die after their first year.
Marsupials share with placental mammals an XX female : XY male system of chromosomal sex determination in which Y determines testis, although some other male phenotypes are independently determined by the X (O et al. 1988). The X chromosome is smaller than the 5% that is highly conserved in placental mammals (Hayman and Martin 1974), in accordance with ‘Ohno’s Law’ (Ohno 1967). The Y is minute in some species. The X and Y do not undergo homologous pairing at male meiosis, but segregate from a proteinaceous plate (Fernandez-Donosa et al. 2010). Fusions between autosomes and sex chromosomes are rather common in marsupials, with several species displaying X1X2Y and XY1Y2 systems resulting from Y-autosome and X-autosome fusions.
In placental mammals, the dosage imbalance of X chromosomes in XX female and XY male is mitigated by an X-inactivation system in which one X chromosome becomes inactive in somatic cells of females (Lyon 1961). This process is random, complete, stable and somatically heritable in the mouse, and constitutes a splendid model system for studying epigenetic silencing. Des and I shared a fascination with marsupial X chromosome inactivation. My honours project was to determine whether in female marsupials as well as female humans and mice, one X showed delayed DNA replication, the cytological hallmark of inactivation. It did, and I set sail for Berkeley thinking that kangaroos are just like humans and mice in this respect (Graves 1967). But while I was away, Des, with colleagues from Macquarie University, showed that the kangaroo X chromosome differs fundamentally from that of placental mammals in that it is always the paternal X that is inactivated, the first demonstration of imprinting in a mammal (Richardson et al. 1971; Sharman 1971). Not only was the process in marsupials imprinted, but it was incomplete and tissue specific (Cooper et al. 1993; Deakin et al. 2009).
In order to use these differences to track how X inactivation evolved and how it works, it became important to know whether the marsupial and placental X chromosomes were monophyletic; hence Des’ interest in kangaroo gene mapping.
Marsupial gene mapping
Des Cooper pioneered the mapping of genes in kangaroo species, discovering allozyme variation (revealed by his beloved starch gel electrophoresis) in several species. He was able to show that human sex-linked enzymes PGK, G6PD and GLA are sex linked also in kangaroos and dasyurid marsupials (Richardson et al. 1971; Cooper et al. 1971). Later the same markers were shown to be sex linked in the Virginia opossum and the grey short-tailed opossum (Samollow et al. 1987), suggesting that the X was completely conserved over all therian mammals, as predicted by Ohno (1967).
But the polymorphisms were scattered over several species and it was not possible to perform dihybrid crosses that would establish a linkage map of the kangaroo X. Des hoped that we might get further using somatic cell hybridisation, which depends on random loss of marsupial chromosomes (and the genes they bear) in hybrid clones. The many years I spent, in collaboration with Rory Hope, obtaining and analysing rodent–marsupial cell hybrids were frustrating in the extreme. Although we could fuse mouse and kangaroo cells and obtain heterokaryons, something awful seemed to happen to the marsupial chromosomes, which were ripped apart or thrown out very early in the hybrid’s life.
We typed hundreds of struggling clones for allozyme markers, using Des’ starch gel electrophoresis skills. We found that they retained the kangaroo form of the selected marker HPRT, but hardly any had other kangaroo markers. Out of some hundreds of hybrids that retained the HPRT, fewer than 10 retained any sign of marsupial chromosomes (Hope and Graves 1978). However, a crucial six hybrids retained a more-or-less intact marsupial X chromosome, and we could show by reversion analysis that three genes (HPRT, PGK-A, G6PD) that were located on the human and mouse X were all gained or lost with the marsupial X (Graves et al. 1979; Dawson and Graves 1986).
Since most hybrids retained only fragments of the X that contained the selected marker, we could use the frequency of retention of unselected markers to propose a gene order on the X; this was the forerunner of radiation hybrids, only our marsupial cells did not need to be irradiated (Dobrovic and Graves 1986).
Our attempts to map autosomal markers by the same strategy were even more frustrating, since most hybrids retained only fragments of the X chromosome, and rarely contained an autosome (Dawson and Graves 1987). Gene mapping over the entire kangaroo genome therefore had to wait for in situ hybridisation. Radioactive in situ hybridisation, using heterologous probes (largely human cDNAs) provided the first detailed description of the marsupial X chromosome (Spencer et al. 1991a). The introduction of fluorescence in situ hybridisation was a dream come true. Now it was possible to screen a marsupial bacterial artificial chromosome (BAC) library for a large-insert clone bearing a particular gene. Aided by trace archive sequence of the tammar, we were able to design overgo probes to conserved genes and very efficiently clone positives (Deakin et al. 2012). From mapping three genes in three years, it became possible to map fifty in a week.
Chromosome painting also became available to us, thanks to the flow sorting of individual marsupial chromosomes by Malcolm Ferguson-Smith’s laboratory in Cambridge, supplemented by microdissection in my laboratory. This made it possible to compare homologous regions in different species. In collaboration with Willem Rens and Malcolm Ferguson-Smith we compared chromosomes between closely related Australian marsupials, and then between Australian and American marsupials (Rens et al. 2001, 2003). Reciprocal painting between species from all the major marsupial families showed that all marsupial karyotypes comprise different arrangements of the same 19 conserved segments. This confirmed the classic observations of Hayman and Martin (1974) that the karyotypes of marsupials, unlike karyotypes of eutherian mammals, are highly conserved.
This high conservation of marsupial chromosomes was always held to be unusual, being very different from the great variety of eutherian chromosome numbers, sizes and morphology. However, in this conservation, marsupials are similar to birds and reptiles, whose karyotypes are strikingly homologous with each other – e,g. chicken, emu and even turtle have chromosomes that are homologous by painting (Graves and Shetty 2001); thus it is the highly variable karyotypes of placental mammals that are unusual, perhaps relating to the acquisition of mobile elements.
The Boden Conference and the Kangaroo Genome Centre
Marsupial (and monotreme) genetics was beginning to excite some international interest when, in 1988, with Des Cooper and Rory Hope, I made a bid for a small conference to gather the few experts in the world to share their knowledge of marsupial breeding, gene mapping and chromosomes. Out of this came a book ‘Mammals from Pouches and Eggs’ (Graves et al. 1990) edited by the three organisers (Fig. 1a, b). Marsupial gene mapping took centre stage when, with Marilyn Renfree and Des Cooper, I initiated a bid for a Centre of Excellence in Kangaroo Genomics in 2003. We were successful in securing limited support, with partner investigators bioinformaticist Terry Speed (Hall Institute) and genomicist Sue Forrest (Australian Genome Research Facility [AGRF]) (Fig. 1c).

|
We set about to complete a map of the entire tammar genome in preparation for sequencing the tammar genome. Since we already had information about X-borne genes, a start was made with the tammar X and Chromosome 5 (Alsop et al. 2005), but this was slow going.
A strategy was devised to efficiently map the entire tammar genome. First we identified blocks of genes shared between human and opossum, then we chose genes to mark each end of each group, pulled out BACs that contained them and mapped only these. A physical map of the tammar genome, containing 548 markers, was rapidly built up. Since we built the map from highly conserved genes with orthologues in all vertebrates, this map could be compared with maps of the opossum, human and even chicken. The comparative map of tammar relative to human shows that the genome has been rearranged at least 120 times; somewhat fewer than between mouse and human.
Combining comparative gene mapping with chromosome painting made it possible to establish detailed homologies between different marsupial species, and also between marsupials, eutherians and even birds. This work confirmed that all marsupials share 19 conserved segments that have undergone simple rearrangements. The arrangement of these segments was almost identical among more than 60 dasyurid species, as well as a family of South American opossums. Comparison with chicken enabled identification of ancestral gene arrangements in 2n = 14 species and 2n = 22 South American species. It was demonstrated that the 2n = 14 karyotype common in Australian marsupials (but not the 2n = 22 karyotype of South American marsupials) shared several gene arrangements with chicken, implying that these gene arrangements were present in the common ancestor of mammals and reptiles (Deakin et al. 2010). This is consistent with a 2n = 14 ancestral marsupial karyotype. The more variable karyotypes of macropodids (kangaroos and wallabies) are easily derived by (largely Robertsonian) fusions between these segments, while the 2n = 22 chromosomes of South American marsupials require several fissions and fusions.
At the same time, Des Cooper’s group in Sydney (initially at Macquarie University, then at the University of New South Wales) had set up crosses and backcrosses between subspecies of tammar that differed in many fixed alleles, and produced the first tammar map based on microsatellites (McKenzie et al. 1997; Zenger et al. 2002). A microsatellite map of the opossum genome was later produced that could be anchored to opossum chromosomes (Samollow et al. 2007). The numbers of markers for the tammar map were greatly expanded by searching for microsatellites within conserved genes which could be mapped by FISH, and this strategy enabled the linkage maps to be anchored to physical chromosomes (Wang et al. 2011a). The physical and linkage maps could be combined into an integrated map of the tammar genome (Wang et al. 2011b).
Kangaroo sex chromosomes
Susumo Ohno predicted long ago that the gene content of the mammalian X chromosome would prove to be identical in all mammals, since rearrangement that disrupted the whole X-inactivation system would be selected against. Early demonstrations that PGK and G6PD were sex linked in several kangaroo species, dasyurids and opossum supported ‘Ohno’s Law’. Somatic cell hybridisation added HPRT to this list, and radioactive in situ hybridisation showed that several genes that lie on the long arm of the human X mapped onto the X in kangaroos (Spencer et al. 1991a) and dasyurids. So far, Ohno’s Law held.
However, Andrew Sinclair in my laboratory discovered that genes on the short arm of the human X chromosome were not, as we had expected, located on the marsupial X, but grouped on Chromosome 5 in tammar and Chromosome 3 in dasyrurids (Sinclair et al. 1987; Spencer et al. 1991b). This was confirmed by chromosome painting: the kangaroo X paint hybridised only to the long arm and pericentric region of the human X (Glas et al. 1999). This constituted the first breach of Ohno’s Law. I happened to be visiting the City of Hope Medical Center in Los Angeles at the time, and took it upon myself to personally inform the great man, whom I found in his office downstairs, composing music from DNA sequence. ‘So?’ he shrugged. A valuable lesson to me that laws are made to be broken.
Our finding could mean either that a piece of the X was lost from a large ancestral X, or that a piece of autosome was added to the X in placental mammals (Graves 1987). Comparison with orthologous genes on the chicken favoured the latter explanation, for the chicken orthologues of genes of the human X lie in two blocks, both autosomal. One (Chicken 4p) represents the region that is shared between the human and marsupial X, the other (Chicken 1) represents the region that is on the X in placentals but is autosomal in marsupials. It was proposed that the marsupial X represents the ancestral mammal X, to which an autosomal region was fused between 160 Mya, when marsupials and placentals diverged, and 105 Mya, when placentals radiated (Graves 1995). A recent study of the elephant X chromosome (Rodriguez Delgado et al. 2009) showed a correspondence between the order of genes on the human and elephant X, but placed the boundary of the conserved and added regions right at the centromere: this suggests that an original Robertsonian fusion occurred, and Afrotheria retain the original centromere; however, the ancestor of other mammals underwent a centric shift (Fig. 2).
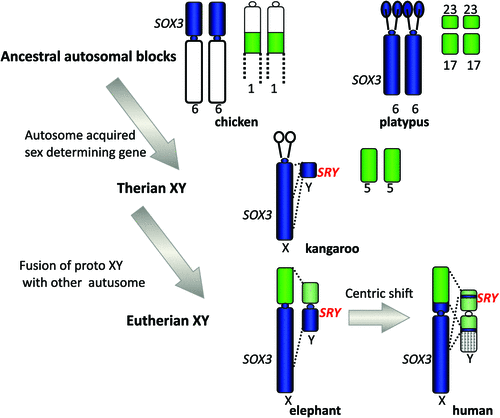
|
We could go back further in time by mapping genes orthologous to those on the marsupial X (equivalent to the ancestral therian X) in birds and reptiles, frogs and fish. They formed autosomal blocks in all of these species (e.g. Nanda et al. 1999), implying that mammal sex chromosomes originated in an autosome some time between the divergence of mammals and reptiles, 310 Mya, and the divergence of marsupials and eutherians, 160 Mya. What was really surprising, however, was our finding that in the basal group of monotreme mammals, genes that are on the therian X are autosomal in platypus and echidna. The complex sex chromosomes of monotremes have homology, instead, to the bird ZW pair. This brings forward the time at which the therian sex chromosomes evolved to only 166–160 Mya (Veyrunes et al. 2008).
The marsupial Y chromosome, like the human Y, is expected to determine testis because XY and XXY embryos develop testes but XX and XO animals do not (even though some other secondary sexual characteristics are independently controlled – see O et al. 1988).
Thus marsupials furnished an important test of the identity of candidate human sex-determining genes: they should be shared by the marsupial Y. The autosomal location in marsupials of the first human candidate sex-determining gene was the first clue that ZFY was the wrong gene (Sinclair et al. 1988), and the trigger for finding the right gene SRY (Sinclair et al. 1990). The presence of SRY on the marsupial Y was an important piece of evidence that it was the right gene (Foster et al. 1992), and the discovery of SOX3 on the X was the first indication that even Y-borne genes with a function in males evolved from genes on the X (Foster and Graves 1994).
The tiny marsupial Y is completely isolated from the X and does not recombine with it; this ‘minimal Y’ is a good model system (Toder et al. 2000). It contains, as well as SRY, orthologues of three other human Y genes (RBMY, UBE1Y, KDMCY), all of which were shown to have X partners on the X from which they obviously diverged (Delbridge et al. 1999). However, all of the other 47 unique protein-coding genes on the human Y originated from the added region of the human X, so are autosomal in marsupials (Waters et al. 2001; Graves 2006).
Surprisingly, the little marsupial Y contains several other genes, all originating from the original therian X chromosome, but lost from the Y in placental mammals. ATRY was the first to be discovered (Pask et al. 2000). Others were found by screening a tammar BAC library with DNA from a microdissected Y (Sankovic et al. 2006). Sequencing Y-borne BACs yielded another seven tammar Y genes, six of which have paralogues on the tammar X chromosome, from which they clearly diverged (Murtagh et al. 2012), consistent with the hypothesis that the Y chromosome evolved by progressive degradation from an original proto–XY pair (Graves 2006).
These characteristics of marsupial sex chromosomes have led to major rethinks about the evolution of mammalian sex chromosome structure and function.
The kangaroo genome
When I was asked by the National Human Genome Research Institute (NHGRI) in 2002 to prepare a proposal (‘White Paper’) to sequence the genome of a model marsupial, I replied that of course it had to be a classic kangaroo. I enlisted Des Cooper and Marilyn Renfree and many other colleagues to prepare the case for sequencing (at the cost, still, of many millions of dollars) our model kangaroo, the tammar wallaby (Graves et al. 2002). However, the decision was to sequence the South American grey short-tailed opossum (because, in the absence of any financial support from Australia, ‘it should be an American marsupial’). The opossum genome was published by a consortium that included 26 Australian marsupial researchers (Mikkelsen et al. 2007).
It was clear that a second marsupial genome was required to enable identification of marsupial-specific and eutherian-specific characteristics. Sue Forrest led a search for funding to commence sequencing the tammar genome. The Victorian State government, supplemented by Applied Biosystems and matched by NHGRI (thanks to Francis Collins) commenced Sanger sequencing in Brisbane (AGRF) and Houston (Baylor Genome Center, directed by Australian Richard Gibbs). The advances in sequencing technology (and the plummeting cost) meant that the 1.5X Sanger sequencing could be supplemented by SOLiD and Illumina sequencing. A hybrid assembly was published in 2011 (Renfree et al. 2011). Since that time, the genome of the Tasmanian devil has been reported (Murchison et al. 2012), and several other marsupials are being sequenced on the recommendation of the Genome 10K Community of Scientists (2009).
Even before the assembly, tammar sequence from trace archives proved important in providing handles to clone tammar genes of interest, for instance HOX genes (Yu et al. 2012), orthologues of imprinted genes (Suzuki et al. 2005; Rapkins et al. 2006) and the flanking markers to prove that XIST does not exist in marsupials (Hore et al. 2007). It greatly facilitated the identification of BACs that define conserved blocks for mapping.
The opossum and tammar sequence established that the size of marsupial genomes, and the size of the non-repetitive fraction, is within the range for eutherian genomes, but the composition of the repeats is very different. There are fewer segmental duplications, and more transposable elements, including more than 500 repeat families (many lineage-specific) especially enriched in LINE elements. The genome has a low %GC, perhaps related to the low recombination.
Like genomes of placental mammals, both marsupial genomes have at least 18 000 protein-coding genes, nearly all of which have orthologues in the human genome. Marsupial-specific genes are largely duplicates and pseudogenes, although a few prove to have homologues in chicken but not human, so represent ancient reptilian genes that were lost in the placental mammal lineage. Many marsupial-specific genes prove to be receptors or transcriptional regulators, and some have characteristics of milk proteins, as might be expected from the complex lactation in marsupials. Many gene families, such as olfactory receptor genes, cytokines, defensins, have been independently amplified in marsupials and eutherians, and may have evolved unique functions: for instance, in adaptation of smell to nocturnal marsupial life (Delbridge et al. 1999) and antimicrobial activity in the pouch to protect the altricial young (Wang et al. 2011c). The arrangement of genes within the MHC locus is different from that of placental mammals, and more similar to that of frog, and tammar is unique in distribution of Class I MHC genes around the genome rather than in a tandem array with other MHC genes (Siddle et al. 2009).
Comparison between orthologous regions of marsupial and eutherian genomes, such as the region containing HOX genes, efficiently identifies exons and conserved non-coding elements. Most of the sequences conserved across this 160-million-year interval proved to be non-coding sequences, many conserved also in birds, suggesting a vital function in all amniotes. This was confirmed by the overlap with functional elements and proximity to developmentally important genes. It is possible to date the origin of these elements by comparing their appearance in birds, marsupials, placentals, and to study both ancient amniote-specific and recently evolved placental- or marsupial-specific elements. The overlap of novel elements with transposable elements suggests that transposable elements furnished the raw material for the evolution of new elements (Mikkelsen et al. 2007; Renfree et al. 2011). The absence of accumulation of LINE1 elements on the X suggests that X chromosome inactivation in marsupials is not regulated in the same way as for the human and mouse X.
Conclusions
Des Cooper’s foresight in exploring the marsupial genome has paid off in ways we could not have foreseen. When I started mapping kangaroo genes at his behest, other scientists told me I was wasting my time; some avowed that marsupials would be so different from human and mouse that comparison would be meaningless, while others warned that the exercise was pointless as they would prove to be exactly the same as our familiar models.
They are neither just the same, nor too different to compare – in fact, they occupy an important middle ground that delivers informative genetic variation on conserved gene arrangements and regulation. The wide array of uses to which this knowledge has been put are summarised in the recent book ‘Marsupial Genetics and Genomics’ (Deakin et al. 2010). Sequence comparisons have proved to be efficient in spotting conserved genes and potential regulatory regions, and comparisons of gene arrangement have opened the way for a comprehensive look at mammalian chromosome evolution. This is particularly evident in the study of the organisation, function and evolution of sex chromosomes, where marsupials have provided a robust picture of the smaller ancestral X and Y, and documented steps in the building up of the complex epigenetic silencing of the X.
References
Alsop, A. E., Miethke, P., Rofe, R., Koina, E., Sankovic, N., Deakin, J., Haines, H., Rapkins, R. W., and Graves, J. A. M. (2005). Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby). Chromosome Research 13, 627–636.| Characterizing the chromosomes of the Australian model marsupial Macropus eugenii (tammar wallaby).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVWiu7vK&md5=31dac2aaa004000a1e4567b8c5a86659CAS | 16170627PubMed |
Bender, H. S., Murchison, E. P., Pickett, H. A., Deakin, J. E., Strong, M. A., Conlan, C., McMillan, D. A., Neumann, A. A., Greider, C. W., Hannon, G. J., Hannon, G. J., Reddel, R. R., and Graves, J. A. M. (2012). Extreme telomere length dimorphism in the Tasmanian devil and related marsupials suggests parental control of telomere length. PLoS ONE 7, e46195.
| Extreme telomere length dimorphism in the Tasmanian devil and related marsupials suggests parental control of telomere length.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVGhtLjM&md5=b5560779046982c83734960f13ffde2eCAS | 23049977PubMed |
Bennett, J. H., Hayman, D. L., and Hope, R. M. (1986). Novel sex differences in linkage values and meiotic chromosome behavior in a marsupial. Nature 323, 59–60.
| Novel sex differences in linkage values and meiotic chromosome behavior in a marsupial.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL28zhtFegsQ%3D%3D&md5=de6c686d6c36edf05bed401cea5fa3fdCAS | 3748181PubMed |
Blackburn, E. H., Greider, C. W., and Szostak, J. W. (2006). Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nature Medicine 12, 1133–1138.
| Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVChsrvM&md5=ab5ebeb452ae397264e04b7512f6ced1CAS | 17024208PubMed |
Bulazel, K. V., Ferreri, G. C., Eldridge, M, and O’Neill, R. J. (2007). Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biology 8, R170.
| Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages.Crossref | GoogleScholarGoogle Scholar | 17708770PubMed |
Carone, D. M., and O’Neill, R. J. (2010). Marsupial centromeres and telomeres: dynamic chromosome domains. In ‘Marsupial Genetics and Genomics’. (Eds J. E., Deakin, P. D. Waters, and J. A. M. Graves.) pp. 55–74. (Springer: Dordrecht, Heidleberg, London, New York.)
Cesare, A. J., and Reddel, R. R. (2010). Alternative lengthening of telomeres: models, mechanisms and implications. Nature Reviews Genetics 11, 319–330.
| Alternative lengthening of telomeres: models, mechanisms and implications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkvVSqs70%3D&md5=c9a9fb1bccca2990d693a0f519b10610CAS | 20351727PubMed |
Cooper, D. W., VandeBerg, J. L., Sharman, G. B., and Poole, W. E. (1971). Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nature: New Biology 230, 155–157.
| 1:CAS:528:DyaE3MXhtlOksb8%3D&md5=90b73d720607d41e4c34ce639ff8451bCAS |
Cooper, D. W., Johnston, P. G., Graves, J. A. M., and Watson, J. M. (1993). X-inactivation in marsupials and monotremes. Seminars in Developmental Biology 4, 117–128.
| X-inactivation in marsupials and monotremes.Crossref | GoogleScholarGoogle Scholar |
Dawson, G. W., and Graves, J. A. M. (1986). Gene mapping in marsupials and monotremes. III. Assignment of four genes to the X chromosome of the wallaroo and the euro (Macropus robustus). Cytogenetics and Cell Genetics 42, 80–84.
| Gene mapping in marsupials and monotremes. III. Assignment of four genes to the X chromosome of the wallaroo and the euro (Macropus robustus).Crossref | GoogleScholarGoogle Scholar |
Dawson, G. W., and Graves, J. A. M. (1987). Gene mapping in marsupials and monotremes. IV. Assignment of PEPA to chromosome 4 of the wallaroo (Macropus robustus). Cytogenetics and Cell Genetics 45, 1–4.
| Gene mapping in marsupials and monotremes. IV. Assignment of PEPA to chromosome 4 of the wallaroo (Macropus robustus).Crossref | GoogleScholarGoogle Scholar |
Deakin, J. E., Chaumeil, J, Hore, T. A., and Graves, J. A. M. (2009). Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Research 17, 671–685.
| Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1WhsLrE&md5=59e555748b77ae2c345647720ebf7712CAS | 19802707PubMed |
Deakin, J. E., Waters, P. D., and Graves, J. A. M. (Eds.) (2010). ‘Marsupial Genetics and Genomics.’ (Springer: Berlin.)
Deakin, J. E., Graves, J. A. M., and Rens, W. R. (2012). The evolution of marsupial and monotreme chromosomes. Cytogenetic and Genome Research 137, 113–129.
| The evolution of marsupial and monotreme chromosomes.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38josFGlug%3D%3D&md5=93e77312ec538c5226d844462750eb47CAS | 22777195PubMed |
Delbridge, M. L., Lingenfelter, P. A., Disteche, C. M., and Graves, J. A. M. (1999). The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nature Genetics 22, 223–224.
| The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXkt1eqsLg%3D&md5=e48f1bce0bdc678b87eb39e9275ff60cCAS | 10391206PubMed |
Dobrovic, A., and Graves, J. A. M. (1986). Gene mapping in marsupials and monotremes. II. Assignments to the X chromosome of dasyurid species. Cytogenetics and Cell Genetics 4, 9–13.
Fernandez-Donosa, R., Berrios, S., Rufas, J. S., and Page, J. (2010). Marsupial sex chromosome behavior during male meiosis. In ‘Marsupial Genetics and Genomics’. (Eds J. E. Deakin, P. D. Waters, and J. A. M. Graves.) pp. 187–204. (Springer: Berlin.)
Ferreri, G. C., Brown, J. D., Obergfell, C., Jue, N., Finn, C. E., O’Neill, M. J., and O’Neill, R. J. (2011). Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere. Journal of Virology 85, 4761–4771.
| Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1Wlt7bE&md5=07c5bb27ca8bbead1c3d7bd911deb03cCAS | 21389136PubMed |
Foster, J. W., and Graves, J. A. M. (1994). An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proceedings of the National Academy of Sciences of the United States of America 91, 1927–1931.
| An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXivFWisr0%3D&md5=95efdcb0c5fd4455f79d186871a55eabCAS | 8127908PubMed |
Foster, J. W., Brennan, F. E., Hampikian, G. K., Goodfellow, P. N., Sinclair, A. H., Lovell-Badge, R., Selwood, L., Renfree, M. B., Cooper, D. W., and Graves, J. A. M. (1992). Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359, 531–533.
| Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXltlamt70%3D&md5=0094a30050b26265663fdbc9fc1a99b8CAS | 1406969PubMed |
Glas, R., Graves, J. A. M., Toder, R., Ferguson-Smith, M. A., and O’Brien, P. C. O. (1999). Cross-species chromosome painting between human and marsupial directly demonstrates the ancient region of the mammalian X. Mammalian Genome 10, 1115–1116.
| Cross-species chromosome painting between human and marsupial directly demonstrates the ancient region of the mammalian X.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXns1Srurg%3D&md5=f647d4e020caf03cc026f8b52250eaeeCAS | 10556436PubMed |
Graves, J. A. M. (1967). DNA synthesis in chromosomes of cultured leucocytes from two marsupial species. Experimental Cell Research 46, 37–57.
| DNA synthesis in chromosomes of cultured leucocytes from two marsupial species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaF2sXktlSnt7c%3D&md5=db7185d0611aa40a14144d4aae726d11CAS |
Graves, J. A. M. (1987). The evolution of mammalian sex chromosomes and dosage compensation – clues from marsupials and monotremes. Trends in Genetics 3, 252–256.
| The evolution of mammalian sex chromosomes and dosage compensation – clues from marsupials and monotremes.Crossref | GoogleScholarGoogle Scholar |
Graves, J. A. M. (1995). The origin and function of the mammalian Y chromosome and Y-borne genes – an evolving understanding. BioEssays 17, 311–320.
| The origin and function of the mammalian Y chromosome and Y-borne genes – an evolving understanding.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2M3mtVehsA%3D%3D&md5=8a1683356bc88bd40dd7987419627492CAS |
Graves, J. A. M. (2006). Sex chromosome specialization and degeneration in mammals. Cell 124, 901–914.
| Sex chromosome specialization and degeneration in mammals.Crossref | GoogleScholarGoogle Scholar |
Graves, J. A. M., and Shetty, S (2001). Sex from W to Z – evolution of vertebrate sex chromosomes and sex determining genes. Journal of Experimental Zoology 281, 472–481.
| Sex from W to Z – evolution of vertebrate sex chromosomes and sex determining genes.Crossref | GoogleScholarGoogle Scholar |
Graves, J. A. M., Chew, G. K., Cooper, D. W., and Johnston, P. G. (1979). Marsupial–mouse cell hybrids containing fragments of the marsupial X chromosome. Somatic Cell Genetics 5, 481–489.
| Marsupial–mouse cell hybrids containing fragments of the marsupial X chromosome.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL3c%2Fjs1Kkug%3D%3D&md5=f89534f71652f26a015b3e3364310ff9CAS |
Graves, J. A. M., Hope, R. M., and Cooper, D. W. (Eds.) (1990). ‘Mammals from Pouches and Eggs: Genetics, Breeding and Evolution of Marsupials and Monotremes.’ (CSIRO Publishing: Melbourne.)
Graves, J. A. M., Wakefield, M. J., Renfree, M. B., Cooper, D. W., Speed, T., Lindblad-Toh, K., Lander, E. S., and Wilson, R. K. (2002). Proposal to sequence the genome of the model marsupial Macropus eugenii (tammar wallaby). Available at: http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/WallabySEQ.pdf
Genome 10K Community of Scientists (2009). A proposal to obtain whole genome sequence for 10,000 vertebrate species The Journal of Heredity 100, 675–680.
| 19656817PubMed |
Hayman, D. L., and Martin, P. G. (1974). ‘Animal Cytogenetics. Mammalia I: Monotremata and Marsupialia.’ (Gebruder Borntraeger: Berlin.)
Hope, R. M., and Graves, J. A. M. (1978). Fusion and hybridization of marsupial and eutherian cells. VI. Hybridization. The Australian Journal of Biological Sciences 31, 527–543.
| 1:STN:280:DyaE1M7os12ltg%3D%3D&md5=0a87c28f1200b164b47c0cbf1e28a17fCAS |
Hore, T. A., Koina, E., Wakefield, M. J., and Graves, J. A. M. (2007). The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Research 15, 147–161.
| The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXisVaqtLo%3D&md5=6d5bd34975f1476b8d30da18440631c5CAS | 17333539PubMed |
Luo, Z. X., Yuan, C. X., Meng, Q. J., and Ji, Q. (2011). A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476, 442–445.
| A Jurassic eutherian mammal and divergence of marsupials and placentals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVOnu7jO&md5=8dde1241df0bb8ca9305345db8ca6bc7CAS | 21866158PubMed |
Lyon, M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373.
| Gene action in the X-chromosome of the mouse (Mus musculus L.).Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaF3c%2FosVGksw%3D%3D&md5=6f59bc6a91cfdd5b941820c76437d6b7CAS | 13764598PubMed |
McKenzie, L. M., Collet, C., and Cooper, D. W. (1997). Use of a subspecies cross for efficient development of a linkage map for a marsupial mammal, the tammar wallaby (Macropus eugenii). The Journal of Heredity 88, 398–400.
| Use of a subspecies cross for efficient development of a linkage map for a marsupial mammal, the tammar wallaby (Macropus eugenii).Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK1c%2FhvVSkuw%3D%3D&md5=079b0e3cc5798fad54f24124676438c0CAS | 9378916PubMed |
Mikkelsen, T. S., Wakefield, M. J., Aken, B., Amemiya, C. T., Chang, J. L., et al (2007). Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177.
| Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXltVGrsLw%3D&md5=e57e19dec553edd426b7ae91cedf8b04CAS | 17495919PubMed |
Murchison, E. P., Schulz-Trieglaff, O. B., Ning, Z., Alexandrov, L. B., Bauer, M. J., et al (2012). Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148, 780–791.
| Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjtV2qsLs%3D&md5=c351ff9ccc5822f55e87625f19742ce1CAS | 22341448PubMed |
Murtagh, V. J., O’Meally, D., Sankovic, N., Delbridge, M. L., Kuroki, Y., Boore, J. L., Toyoda, A., Jordan, K. S., Pask, A. J., Renfree, M. B., Fujiyama, A., Graves, J. A. M., and Waters, P. D. (2012). Evolutionary history of novel genes on the tammar wallaby Y chromosome: implications for sex chromosome evolution. Genome Research 22, 498–507.
| Evolutionary history of novel genes on the tammar wallaby Y chromosome: implications for sex chromosome evolution.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XjsV2lsb0%3D&md5=9adc435fae6aa1dd5dad7f209ee18987CAS | 22128133PubMed |
Nanda, I., Shan, Z., Schartl, M., Burt, D. W., Koehler, M., Nothwang, H., Grützner, F., Paton, I. R., Windsor, D., Dunn, I., Engel, W., Staeheli, P., Mizuno, S., Haaf, T., and Schmid, M. (1999). 300 million years of conserved synteny between chicken Z and human chromosome 9. Nature Genetics 21, 258–259.
| 300 million years of conserved synteny between chicken Z and human chromosome 9.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXitVCit7k%3D&md5=15950bbb3ac858cfe8ef8b46ccbd3694CAS | 10080173PubMed |
O, W. S., Short, R.V., Renfree, M. B., and Shaw, G (1988). Primary genetic control of somatic sexual differentiation in a mammal. Nature 331, 716–717.
| Primary genetic control of somatic sexual differentiation in a mammal.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL1c7ktlWmtA%3D%3D&md5=b4fd4d36f2035712846f82858bffdba3CAS | 3344046PubMed |
Ohno, S. (1967). ‘Sex Chromosomes and Sex-Linked Genes.’ (Springer-Verlag: New York.)
Pask, A. J., Renfree, M. B., and Graves, J. A. M. (2000). The human sex-reversing ATRX gene has a homologue on the marsupial Y chromosome, ATRY: implications for the evolution of mammalian sex determination. Proceedings of the National Academy of Sciences of the United States of America 97, 13198–13202.
| The human sex-reversing ATRX gene has a homologue on the marsupial Y chromosome, ATRY: implications for the evolution of mammalian sex determination.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXosVKqurY%3D&md5=22fe3d769999ec2b65d897be4636c56cCAS |
Rapkins, R. W., Hore, T., Smithwick, M., Ager, E., Pask, A., Renfree, M. B., Kohn, M., Hameister, H., Nicholls, R. D., Deakin, J. E., and Graves, J. A. M. (2006). Recent assembly and initiating of imprinting in the Prader-Willi/Angelman imprinted domain. PLOS Genetics 2, e182.
| Recent assembly and initiating of imprinting in the Prader-Willi/Angelman imprinted domain.Crossref | GoogleScholarGoogle Scholar | 17069464PubMed |
Renfree, M. B., Papenfuss, A. T., Deakin, J. E., Lindsay, J., Heider, T., et al (2011). Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biology 12, R81.
| Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development.Crossref | GoogleScholarGoogle Scholar | 21854559PubMed |
Rens, W., O’Brien, P. C. M., Yang, F., Solanky, N., Perelman, P., Graphodatsky, A. S., Ferguson, M. W. J., Svartman, M., De Leo, A. A., Graves, J. A. M., and Ferguson-Smith, M. A. (2001). Karyotype relationships between distantly related marsupials from South America and Australia. Chromosome Research 9, 301–308.
| Karyotype relationships between distantly related marsupials from South America and Australia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXksFCitbk%3D&md5=b2f7e8397d378d3518afad48f067c6ddCAS | 11419794PubMed |
Rens, W., O’Brien, P. C. M., Fairclough, H., Harman, L., Graves, J. A. M., and Ferguson-Smith, M. A. (2003). Reversal and convergence in marsupial chromosome evolution. Cytogenetic and Genome Research 102, 282–290.
| Reversal and convergence in marsupial chromosome evolution.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD2c%2FotVKrug%3D%3D&md5=5260a2258a838e6a5cf4d56ceb26a3c8CAS | 14970718PubMed |
Richardson, B. J., Czuppon, A. B., and Sharman, G. B. (1971). Inheritance of glucose-6-phosphate dehydrogenase variation in kangaroos. Nature: New Biology 31, 154–155.
Rodriguez Delgado, C. L., Waters, P. D., Gilbert, C., Robinson, T. J., and Graves, J. A. M. (2009). Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Research 17, 917–926.
| Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtl2ksL3I&md5=7c7855f8d837689a906595e7f06d0613CAS |
Rofe, R., and Hayman, D. L. (1985). G-banding evidence for a conserved complement in the Marsupialia. Cytogenetics and Cell Genetics 39, 40–50.
| G-banding evidence for a conserved complement in the Marsupialia.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL2M7ltlCgtA%3D%3D&md5=abc33dbdcdd3e1d425711147488b59a1CAS | 3979118PubMed |
Samollow, P. B., Ford, A. L., and VandeBerg, J. L. (1987). X-linked gene expression in the Virginia opossum: differences between the paternally derived Gpd and Pgk-A loci. Genetics 115, 185–189.
| 1:CAS:528:DyaL2sXhtFOis7o%3D&md5=977964e9a14a700f5153e4d0fd3e3e48CAS | 3557111PubMed |
Samollow, P. B., Gouin, N., Miethke, P., Mahaney, S. M., Kenney, M., VandeBerg, J. L., Graves, J. A. M., and Kammerer, C. M. (2007). A microsatellite-based, physically anchored linkage map for the gray, short-tailed opossum (Monodelphis domestica). Chromosome Research 15, 269–281.
| 1:CAS:528:DC%2BD2sXmsV2jsbk%3D&md5=711ba593fdf304fd6eae1e5b78ce7b1fCAS | 17333535PubMed |
Sankovic, N., Delbridge, M. L., Gruetzner, F., Ferguson-Smith, M., O’Brien, P. C. M., and Graves, J. A. M. (2006). Creation of a highly enriched marsupial Y chromosome specific BAC sub-library using isolated Y chromosomes. Chromosome Research 14, 657–664.
| Creation of a highly enriched marsupial Y chromosome specific BAC sub-library using isolated Y chromosomes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XptlSmtbw%3D&md5=e07cf90a43ab234d73002a410e97b4b3CAS | 16964572PubMed |
Sharman, G. B. (1971). Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230, 231–232.
| Late DNA replication in the paternally derived X chromosome of female kangaroos.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaE3M7js1GhtA%3D%3D&md5=d091f87a97d8a72b1e97f544388bc140CAS | 4926712PubMed |
Sharman, G. B. (1973). The chromosomes of non-eutherian mammals. In ‘Cytotaxonomy and Vertebrate Evolution’. (Eds A. N. Chiarelli, and E. Capanna.) pp. 485–530.(Academic Press: New York.)
Siddle, H. V., Deakin, J. E., Coggill, P., Hart, E., Cheng, Y., Wong, E. S. W., Harrow, J, Beck, S, and Belov, K (2009). MHC-linked and un-linked class I genes in the wallaby. BMC Genomics 10, 310.
| MHC-linked and un-linked class I genes in the wallaby.Crossref | GoogleScholarGoogle Scholar | 19602235PubMed |
Sinclair, A. H., Wrigley, J. M., and Graves, J. A. M. (1987). Autosomal assignment of OTC in marsupials and monotremes: implications for the evolution of sex chromosomes. Genetical Research 50, 131–136.
| Autosomal assignment of OTC in marsupials and monotremes: implications for the evolution of sex chromosomes.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL1c%2FosVCitQ%3D%3D&md5=56c512bbea1e94e9cd56eddffdaeaf74CAS | 3692164PubMed |
Sinclair, A. H., Foster, J. W., Spencer, J. A., Page, D. C., Palmer, M., Goodfellow, P. N., and Graves, J. A. M. (1988). Sequences homologous to ZFY, a candidate human sex-determining gene, are autosomal in marsupials. Nature 336, 780–783.
| Sequences homologous to ZFY, a candidate human sex-determining gene, are autosomal in marsupials.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXovFOjtw%3D%3D&md5=86589c8724dbf6e4840c4119ebae8c3cCAS | 3144651PubMed |
Sinclair, A. H., Berta, P., Palmer, M. S., Hawkins, J. R., Griffiths, B. L., Smith, M. J., Foster, J. W., Frischauf, A. M., Lovell-Badge, R., and Goodfellow, P. N. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244.
| A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3cXkvFaltrg%3D&md5=831a7da62b4645709111e53a5ce31a9aCAS | 1695712PubMed |
Spencer, J. A., Watson, J. M., and Graves, J. A. M. (1991a). The X chromosome of marsupials shares a highly conserved region with eutherians. Genomics 9, 598–604.
| The X chromosome of marsupials shares a highly conserved region with eutherians.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXksVeis78%3D&md5=6818f7cc197ddb47a2961d98371ed312CAS | 2037290PubMed |
Spencer, J. A., Sinclair, A. H., Watson, J. M., and Graves, J. A. M. (1991b). Genes on the short arm of the human X chromosome are not shared with the marsupial X. Genomics 11, 339–345.
| Genes on the short arm of the human X chromosome are not shared with the marsupial X.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38Xlt1Kmug%3D%3D&md5=896c6422b462ea10450911e7965658a2CAS | 1769650PubMed |
Suzuki, S., Renfree, M. B., Pask, A. J., Shaw, G., Kobayashi, S., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2005). Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mechanisms of Development 122, 213–222.
| Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkvFensw%3D%3D&md5=c61f09dfa40f7979ea67b87195853d84CAS | 15652708PubMed |
Svartman, M., and Vianna-Morgante, A. M. (1999). Comparative genome analysis in American marsupials: chromosome banding and in-situ hybridization. Chromosome Research 7, 267–275.
| Comparative genome analysis in American marsupials: chromosome banding and in-situ hybridization.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXltV2jtrs%3D&md5=489c773f6b35828a19e592de3e031583CAS | 10461872PubMed |
Toder, R., Wakefield, M., and Graves, J. A. M. (2000). The minimal mammalian Y chromosome – the marsupial Y as a model system. Cytogenetics and Cell Genetics 91, 285–292.
| The minimal mammalian Y chromosome – the marsupial Y as a model system.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3M7otFyltw%3D%3D&md5=edbfd28bdbdd57c434f3a59241b67a2bCAS | 11173870PubMed |
van Oorschot, R. A., Porter, P. A., Kammerer, C. M., and VandeBerg, J. L. (1992). Severely reduced recombination in females of the South American marsupial Monodelphis domestica. Cytogenetics and Cell Genetics 60, 64–67.
| Severely reduced recombination in females of the South American marsupial Monodelphis domestica.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK383lvVSquw%3D%3D&md5=0428db9bbad4a1201c14c1b1e909bb23CAS | 1582262PubMed |
Veyrunes, F., Waters, P. D., Miethke, P., Rens, W., McMillan, D., Alsop, A. E., Grützner, F., Deakin, J. E., Whittington, C. M., Schatzkamer, K., Kremitzky, C. L., Graves, T., Ferguson-Smith, M. A., Warren, W., and Graves, J. A. M. (2008). Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Research 18, 965–973.
| Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnsVygsbY%3D&md5=b05983bdd248703a2c41473236036e9dCAS | 18463302PubMed |
Wang, C., Webley, L., Wei, K.-J., Wakefield, M. J., Patel, H. R., Deakin, J. E., Alsop, A., Graves, J. A. M., Cooper, D. W., Nicholas, F. W., and Zenger, K. R. (2011a). A second-generation anchored linkage map of the tammar wallaby (Macropus eugenii). BMC Genetics 12, 72.
| A second-generation anchored linkage map of the tammar wallaby (Macropus eugenii).Crossref | GoogleScholarGoogle Scholar | 21854616PubMed |
Wang, C., Deakin, J. E., Rens, W., Zenger, K. L., Belov, K., Graves, J. A. M., and Nicholas, F. W. (2011b). An integrated tammar wallaby map and its use in creating a virtual tammar wallaby genome map. BMC Genomics 12, 422.
| An integrated tammar wallaby map and its use in creating a virtual tammar wallaby genome map.Crossref | GoogleScholarGoogle Scholar | 21854555PubMed |
Wang, J., Wong, E. S. W., Whitley, J. C., Li, J., Stringer, J. M., Short, K. R., Renfree, M. B., Belov, K, and Cocks, B. G. (2011c). Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS ONE 6, e24030.
| Ancient antimicrobial peptides kill antibiotic-resistant pathogens: Australian mammals provide new options.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1Wrt77N&md5=40e56e01e6cf0fb76f8e4854b145e63eCAS | 21912615PubMed |
Waters, P., Duffy, B., Frost, C. J., Delbridge, M. L., and Graves, J. A. M. (2001). The human Y chromosome derives largely from a single autosomal region added 80–130 MYA. Cytogenetics and Cell Genetics 92, 74–79.
| The human Y chromosome derives largely from a single autosomal region added 80–130 MYA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXjs12gsb8%3D&md5=8fb3f6d1fd30ccd78cea802a6e003f1dCAS | 11306800PubMed |
Yu, H., Lindsay, J., Feng, Z. P., Frankenberg, S., Hu, Y., et al (2012). Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics 13, 251.
| Evolution of coding and non-coding genes in HOX clusters of a marsupial.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXot1Wmt7c%3D&md5=4553427d8be1175e2b4931befae24896CAS | 22708672PubMed |
Zenger, K. R., McKenzie, L. M., and Cooper, D. W. (2002). The first comprehensive genetic linkage map of a marsupial: the tammar wallaby (Macropus eugenii). Genetics 162, 321–323.
| 1:CAS:528:DC%2BD38Xotl2gsbk%3D&md5=eedc7bec66484671bd37a50fdc21f2d1CAS | 12242243PubMed |


