Historical persistence and isolation by distance of Mirbelia viminalis (Fabaceae) across the Hamersley Range of the Pilbara bioregion
Melissa A. Millar A * , Rachel M. Binks
A * , Rachel M. Binks  A , Sarah-Louise Tapper A , Bronwyn M. Macdonald A , Shelley L. McArthur A , Margaret Hankinson A , David J. Coates
A , Sarah-Louise Tapper A , Bronwyn M. Macdonald A , Shelley L. McArthur A , Margaret Hankinson A , David J. Coates  A , Stephen van Leeuwen
A , Stephen van Leeuwen  A B and Margaret Byrne
A B and Margaret Byrne  A
A
A Department of Biodiversity, Conservation and Attractions, Biodiversity and Conservation Science, Locked Bag 104, Bentley Delivery Centre, Bentley, WA 6983, Australia.
B Present address: School of Molecular and Life Sciences, Curtin University, GPO Box U1987, Perth, WA 6845, Australia.
Australian Journal of Botany 70(5) 358-371 https://doi.org/10.1071/BT22014
Submitted: 7 February 2022 Accepted: 25 July 2022 Published: 26 August 2022
© 2022 The Department of Biodiversity, Conservation and Attractions, WA. Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Empirical studies of intraspecific genetic diversity and population structure can inform the evolutionary and demographic history of individual species and of landscapes at the bioregional level.
Aims: We aimed to assess intraspecific genetic variation at macroevolutionary and microevolutionary temporal scales for Mirbelia viminalis, a key species present on the Hamersley Range in the ancient and highly diverse landscape of the Pilbara bioregion of northwest Western Australia.
Methods: We sampled extant populations and assessed diversity and structure using sequences (chloroplast DNA, 1759 base pairs) and microsatellite markers (nuclear DNA, 15 loci) data.
Key results: Significant phylogeographic structure and a lack of historical demographic signals of population contraction or expansion suggest historical population persistence. Moderate chloroplast haplotype diversity (h = 15) and moderate divergence among extant haplotypes indicates a degree of historical connectivity via seed dispersal across central populations on the Hamersley Range. Levels of nuclear genetic diversity were low to moderate (allelic richness = 3.554, expected heterozygosity = 0.489, observed heterozygosity = 0.462) and depauperate compared to another member of the Mirbelia genus present further south in the Midwest region. Nuclear diversity revealed a strong signal of isolation by distance with localised admixture among populations and some contemporary genetic clustering along a north-west to south-east transect of the Hamersley Range.
Conclusions: Low nuclear genetic diversity may be related to recent reductions in population size for M. viminalis. Historical population persistence with few barriers to dispersal other than geographic distance may be common for members of the Fabaceae across the Hamersley Range.
Keywords: arid landscape, demography, dispersal, Fabaceae, genetic connectivity, genetic diversity, genetic structure, haplotypes, phylogeography.
Introduction
Empirical studies of intraspecific patterns of genetic structure and genetic diversity can provide valuable insights into the evolutionary history and the historical and contemporary demographic processes of species and the landscapes they occupy. Phylogeographic studies reveal genetic structure and diversity that has evolved over macroevolutionary temporal scales (Hewitt 2004). Such studies can indicate historical demographic patterns of population persistence, contraction and/or expansion, which in plant species, are driven by seed dispersal and can be tracked by analysis of the chloroplast genome. They can also aid in the identification of landscape features that have acted as potential historical refugia, or barriers to dispersal at the bioregeographical scale (Hewitt 2004; Hickerson et al. 2010; Keppel et al. 2012). Contemporary patterns of genetic structure, genetic diversity, and demography are a result of more recent or microevolutionary patterns of gene flow driven largely by pollen dispersal and revealed by the nuclear genome (Ennos et al. 1999). Whilst intraspecific patterns of genetic structure and genetic diversity, and population demographic history may vary over time, in general, species with more limited capability to maintain genetic connectivity via seed and pollen dispersal are likely to experience greater genetic drift and develop patterns of genetic structure among populations. They may also display reduced levels of diversity at both macro and micro evolutionary temporal and spatial scales (Ellstrand and Elam 1993).
Studies that assess genetic structure, genetic diversity, and demographic history at both the macroevolutionary and microevolutionary scale have been historically underrepresented in the Southern Hemisphere (Beheregaray 2008; Hickerson et al. 2010), and in the Australian landscape in general (Beheregaray 2008). Species specific studies have been informative in some environments, such as in the biodiverse southwest of Western Australia (e.g. Bradbury et al. 2016, 2019; Sampson et al. 2018; Nistelberger et al. 2021), yet studies remain limited in more arid regions of the state. The Interim Biogeographic Regionalisation for Australia (IBRA, Thackway and Cresswell 1995) Pilbara bioregion of Western Australia is a biodiverse, highly structured, and ancient landscape (Cracraft 1991; Pepper et al. 2013). The Pilbara bioregion extends from the offshore islands in the west, transitioning across alluvial coastal plains and tablelands, granitic plains, ranges, and uplands of the Pilbara Craton, through low lying alluvial flats and river systems, to the sandy deserts of the vast central Australian arid zone in the east, and the arid rangelands to the south (McKenzie et al. 2009; Pepper et al. 2013). The region is predominantly arid and is characterised by extreme summer temperatures and tropical summer cyclonic rainfall events (Leighton 2004). The landscape is dominated by two topographically complex ranges that provide a high degree of spatial complexity, and microclimatic variability. The iron-rich sedimentary Hamersley Range (elevation up to 1249 m) comprises a series of mountainous ranges, ridges, eroded gorges, and hills, with an elevated plateau dominated by Acacia shrubland and woodlands over spinifex hummock grassland. The Chichester Range (elevation up to 367 m) to the north, comprises a basaltic, granitic/greenstone escarpment and tableland with jagged peaks, gorges, rolling hills, and tree lined water courses, with spinifex hummock grassland (Triodia spp.), and Eucalyptus leucophloia Brooker steppe. The Fortescue Valley forms a topographical barrier between the two Ranges and supports an extensive salt marsh, unique samphire heath (Tecticornia spp.), shrub, bunch and tussock grass (Poaceae), and mulga (Acacia aneura sen. lat. Benth) woodlands (Beard 1975; McKenzie et al. 2009). The region is recognised as a significant centre of species endemism across flora and fauna biomes for the Australian continent (Cracraft 1991; Unmack 2001; Eberhard et al. 2005; Ladiges et al. 2006; Gibson and McKenzie 2009; McKenzie and Bullen 2009; McKenzie et al. 2009; Burbidge et al. 2010; Durrant et al. 2010; Guthrie et al. 2010; Doughty et al. 2011), and there is growing interest in understanding the historical and contemporary patterns of genetic structure and diversity and the demographic histories of the flora of this complex, arid bioregion (Byrne et al. 2016, 2017; Levy et al. 2016; Nistelberger et al. 2020; Millar et al. 2022).
The Fabaceae family contains the Mirbelia genus, consisting of 26 species endemic to Australia (Barrett et al. 2021). Little is known of the evolutionary history or population dynamics of Mirbelia, with genetic structure, diversity and demographic history being investigated to date in only one other species Mirbelia sp. Bursarioides T.R. Lally 760 (Millar et al. 2016). Mirbelia viminalis (Benth) C. A. Gardner, is the only member of the genus present in the Pilbara. It has a widespread range (>600 km, Moran and Hopper 1987) being patchily distributed throughout much of the northern half of the continent (Fig. 1), although within the Pilbara it has a regional distribution (150–600 km, Moran and Hopper 1987) centred on the Hamersley Range. Most M. viminalis populations occur in skeletal red-brown clayey sand over sandstone, on low rises, upper slopes, and crests of haematite ridges, and in shrubland or open shrub mallee on the Hamersley Range, with some populations to the north-west off the main Hamersley Range, but still at elevation. The Hamersley Range is rich in iron ore, with extensive existing or proposed footprints of mining operations making M. viminalis a key species for use in the ecological restoration of mined habitat and a focus for conservation management activities that would benefit from knowledge of historical and contemporary patterns of genetic structure, genetic diversity, and demographic history. We use both chloroplast (cpDNA) sequencing data and nuclear microsatellite (nDNA) data to assess phylogeographic and contemporary patterns of genetic structure and levels of genetic diversity for Pilbara populations of M. viminalis. We conduct tests for demographic scenarios of historical population persistence or population expansion and tests for recent contraction in population size. We assess pollen to seed dispersal ratios to provide a context for historical and contemporary pattens of genetic connectivity and assess the correlation between genetic diversity and environmental or landscape variables that may indicate the presence of environmental refugia.
|
|
Materials and methods
Study species
M. viminalis, also known as yellow broom or leafless mirbelia, is an erect to spreading shrub, 0.4–2 m high. The small shrubs are spiny and many branched, with leaves that are reduced to scales. Small yellow flowers appear from March to October. The small seed forms in small, flat, non-fleshy pods and is released on dehiscence (Sweedman and Brand 2006). No specific pollinators have been recorded for M. viminalis, although other Mirbelia are considered to be primarily bee pollinated. The mating system is likely to be self-compatible but preferentially outcrossed, as is common for other Fabaceae (Stone et al. 2003).
Sample collection and DNA extraction
Distributional information was determined from specimens lodged at the Western Australian Herbarium (Perth) through the FloraBase website (http://florabase.dpaw.wa.gov.au). Collections of vegetative material were made from 24 plants in each of 19 populations (total of 456 plants) that spanned the species’ distribution in the Pilbara bioregion (Fig. 1, Table 1). Field identification was straightforward and voucher specimens were collected at each sampled population and lodged with the Western Australian Herbarium. Material was freeze-dried before DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987), adding 1% polyvinylpyrrolidone (PVP) to the extraction buffer.
Chloroplast sequencing
Non-coding chloroplast regions known to show variability in Australian plant species and widely used for barcoding were tested with DNA amplification and analysis following Byrne and Hankinson (2012). Raw polymerase chain reaction (PCR) products were sequenced using forward and reverse primers by the Australian Genome Research Limited Facility (AGRF, Perth, Australia). The regions selected for full analysis (ndhA, trnV and atpF) were chosen based on sequence quality and nucleotide diversity and were sequenced for eight individuals randomly selected from each population (total of 152 samples). Sequence reactions were carried out following Byrne and Hankinson (2012) with thermocycling conditions according to Shaw et al. (2007) with a Serapure method to purify PCR products (Rohland and Reich 2012; Faircloth and Glenn 2014). Sequence data were aligned using CLUSTAL W v2.1 (Thompson et al. 1994) and corrected manually where necessary using BioEdit v7.2 (Hall 1999). Variable sites were scored as a single mutation and indels coded as single binary states regardless of length. Indels arising from mononucleotide repeats were removed. These sequence data have been submitted to the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database under accession numbers OM389460–389483.
Nuclear microsatellite library construction, primer, and locus assessment
Nuclear microsatellite markers were developed specifically through partial genome sequencing on a 454 platform at the AGRF using the methods of Gardner et al. (2011). The resulting microsatellite libraries were sorted for functionality and 80 primers were tested for polymorphism across multiple populations. Fifteen reliable microsatellite loci were optimised. Microsatellite amplification was performed in 7.25 μL multiplexed reactions containing 3.5 μL QIAGEN Multiplex PCR Master Mix (Qiagen, Venlo, Netherlands), 0.75 μL primer mix, 2 μL sterile distilled water and 1 μL (10 ng) of template DNA. Cycling conditions were the same for all loci: 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 60°C for 90 s and 72°C for 60 s, followed by a final extension of 60°C for 30 min. Forward primers contained a fluorescent colour of the G5 label set (FAM, VIC, NED or PET). Diluted PCR product (1 μL) was added to 12 μL of GeneScan™ LIZ®500(-250) size Standard (Applied Biosystems, Waltham, USA)/formamide, and PCR products were visualised on a Biosystems 3730 Sequencer (Applied Biosystems). Genotypes were scored for 23 (one individual from SOL failed to amplify well) or 24 plants from each of 19 sites (total of 455 samples) using Genemapper™ v3.7 (Applied Biosystems) software with amplification and fragment analysis repeated once for any individuals that failed to amplify or produce scorable bands in the first instance. Allele bins were manually assigned, and alleles automatically scored and manually checked. Sequence data of microsatellite containing regions have been submitted to the GenBank database under accession numbers MW138757–MW138771 (See Supplementary material Table S1 for further detail of primers and loci).
Individual loci were tested for departure from Hardy–Weinberg equilibrium (HWE) and locus pairs tested for linkage disequilibrium (LD) over all populations, using exact tests as implemented in GENEPOP v4.0 (Raymond and Rousset 1995). Presence and frequency of null alleles was assessed with the FREENA software program (Chapuis and Estoup 2007).
Data analysis
Chloroplast
The number of haplotypes (H) and the number of private haplotypes (Hp) in each population were counted. Estimates of nucleotide and haplotype diversity including the total number of variable sites (S), total number of haplotypes (h), haplotype diversity (Hd), and nucleotide diversity (π), were obtained using DNASP v6.12.03 (Librado and Rozas 2009). Measures of phylogenetic diversity within (HS), and total diversity within and among populations (HT), as well as coefficients of phylogenetic differentiation for unordered (GST) and ordered alleles (NST), were calculated with PERMUT v2.0 following Pons and Petit (1996). Phylogeographic structure was evaluated by assessing whether NST was significantly greater than GST using the U test for 1000 permutations in PERMUT. Parameters of neutrality tests that can indicate historical changes in population size (Tajima’s D, Fu’s Fs statistic and Ramos-Onsins and Rozas’ R2 test) were obtained with significance of values assessed using a null distribution of 10 000 coalescent based simulations using DNASP. Tajima’s D considers the genetic diversity and the number of variable sites in a sequence to test for demographic range expansions although significant values of D can also be due to bottlenecks, selective effects, or heterogeneity of mutation rates. Fs uses information on the distribution of haplotypes as a test for demographic expansion and is more sensitive to population growth than Tajima’s D. R2 contrasts the number of singletons and the mean number of differences to also test for recent population expansion.
Maximum parsimony phylogenetic networks (Polzin and Daneshmand 2003) were constructed, and the shortest, least complex trees produced using Median-Joining (MJ) network algorithm for multi-state data (Bandelt et al. 1999), as implemented in NETWORK v5.0.1.1 (Fluxus Technology Ltd, Sudbury, UK). Character states had equal weighting, ε was set to 0 and MJ networks were verified using the Reduced Median (RM) algorithm. No star contraction pre-processing was applied. Haplotypes present in each population were mapped across the species’ distribution.
Nuclear
Estimates of nuclear diversity (allelic richness NR), the proportion of polymorphic loci (%P), observed (Ho) and expected (He) heterozygosity and the Fixation index (FIS) were calculated using MSA v4.05 (Dieringer and Schlötterer 2003). The number of private alleles (NAp) was calculated using GenAlEx v6.5 (Peakall and Smouse 2012).
Global measures of genetic divergence, Wright’s measure of allelic fixation (FST, Wright 1943) and Jost’s measure of allelic identity (DST, Jost 2008), were estimated with P-values calculated on 999 permutations using GenAlEx. Pairwise among-population measures of FST were estimated. The global divergence measure of allelic size (RST, Slatkin 1995) that considers microsatellite allele size under a strict stepwise mutational model (SMM) was also estimated. Phylogeographic pattern within populations was tested for by permuting allele sizes among alleles within populations (Hardy et al. 2003), and phylogeographic pattern among populations was tested by permuting alleles at population locations among populations using 1000 permutations conducted in SPAGeDi v1.7 (Hardy and Vekemans 2002). The effect of mutation in relation to drift was also tested by comparing global RST to global FST.
Hierarchical partitioning of population genetic structure was characterised using the Analysis of Molecular Variance (AMOVA) method conducted on FST values at the population level. Principal Coordinates Analysis (PCoA) was conducted based on pairwise individual genetic distances using a standardised data set for all populations. Mantel tests were used to test for the association of genetic differentiation (linearised FST (FST/1 − FST)) with log geographic distance among populations. Significance tests were conducted using 999 permutations of sample site locations. This analysis was conducted in GenAlEx.
We also used model-based clustering methods to determine a potentially optimal number of biologically meaningful genetic clusters. Genetic structure was investigated using the Bayesian assignment approach in STRUCTURE v2.3.2.1 (Pritchard et al. 2000). Analysis used the admixture ancestry model with the assumption of correlated allele frequencies amongst samples given the likelihood of high population similarity due to gene flow (Falush et al. 2003). A burn-in period of 50 000 was applied with 100 000 Markov chain Monte Carlo (MCMC) replications to assess K values ranging from K = 1 to K = the number of populations (19) plus one (20), with 10 iterations of each K value. The potentially optimal K value was determined by assessing maxima with limited deviation for the heuristic model likelihood statistics LK and ΔK (Evanno et al. 2005) estimated with STRUCTURE HARVESTER v.0.6.93 (Earl and vonHoldt 2012). Mean permuted proportion of membership (Q) values for all individuals were graphed and mean Q values for populations were mapped.
It can often be difficult to describe sets of individuals that form continuous clines or discrete clusters using clustering models. Models such as those used in STRUCTURE, assume underlying frequencies are identical for all individuals assigned to clusters determined by optimising HWE (Pritchard et al. 2000). This can often result in the overestimation of the discreteness of clusters and underestimation of admixture among them. For species that show a continuous pattern of differentiation, such as that produced by geographic isolation by distance (IBD) (Wright 1943; Novembre and Stephens 2008; Frantz et al. 2009), models that incorporate distinct spatial geographical data provide more biologically meaningful estimates of population structure and admixture. Like STRUCTURE, the tess3r v1.1.0 package (Caye et al. 2016) implemented in R (R Core Team 2019) is based on optimising HWE to compute the likelihood of a given clustering solution, but also incorporates geographically constrained non-negative matrix factorisation that models continuous geographic variation through space. We assessed K values ranging from K = 1 to K = the number of populations plus one (20), assessed the optimal number of clusters via minima in cross-validation scores, and obtained matrices of individual and population assignments for the optimal K number of clusters in tess3r. Mean permuted proportion of membership (Q) values for all individuals were graphed and mean Q values for populations were mapped.
The ratio of pollen to seed dispersal was calculated following Ennos (1994) where Pollen: Seed = [A(1 + FIS) − 2C]/C, where A = (1/FST) − 1 and where C = (1/GST) − 1. Correlation between levels of genetic diversity and environmental or landscape variables that may suggest the presence of environmental refugia was assessed using regression analysis. Genetic diversity parameters included H, NR, Ho and He and environmental and landscape variables included aridity index, annual mean precipitation, annual mean evaporation, annual mean temperature, elevation, and topographical relief (variation in relief within 5 km of sampling location) for each population, available from Atlas of Living Australia (www.ala.org.au). Bounded genetic diversity measures Ho and He were arcsine transformed prior to analysis.
Populations that have experienced a recent reduction of their effective population size exhibit a correlative reduction of the allele numbers and expected heterozygosities with allelic diversity reduced faster than the expected heterozygosity (Piry et al. 1999), providing an explicit test for population size reductions. This has been demonstrated only for loci evolving under the Infinite Allele Model (IAM) (Maruyama and Fuerst 1985). However, choosing both a proportion of Step wise Mutation Model (SMM) in a Two Phase Model (TPM) with a variance of the geometric distribution for TPM = 0.36 corresponds to sensible parameter values for most microsatellites (Piry et al. 1999). We analysed populations for recent size reductions using the IAM and TPM in the BOTTLENECK v1.2.02 program (Piry et al. 1999).
Results
Chloroplast DNA
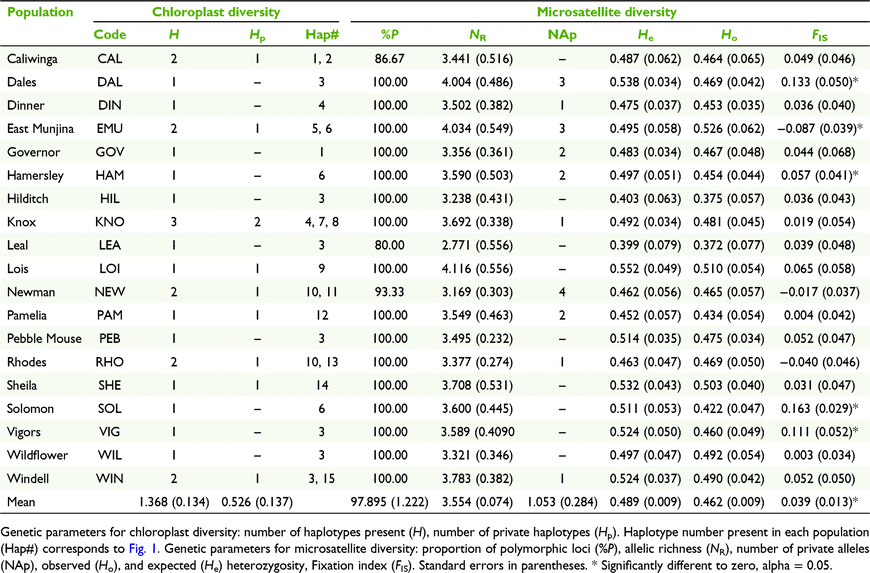
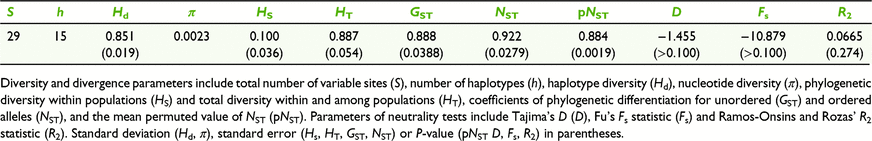
For cpDNA, 1759 sites were scored. There were 29 variable sites with 23 parsimony-informative sites, leading to the identification of 15 haplotypes (Table 2). Mean haplotype diversity (H), the mean number of private haplotypes (Hp) per population (Table 1) and overall haplotype diversity (Hd, Table 2) were moderate. Sharing of haplotypes among populations was common (Fig. 1), although private haplotypes were detected in nine populations located across most of the sampled range (Fig. 1). Distinct haplotypes in the same population were on average more closely related than distinct haplotypes from different populations indicating significant phylogeographic structure (observed NST > mean permuted NST [pNST], P = 0.05, Table 2). All neutrality tests for population expansion were non-significant, suggesting historically stable population sizes (Table 2).
The phylogenetic network was structured with some closely related haplotypes and others with varying degrees of divergence (Fig. 1). The distribution of haplotypes showed three general clusters in the northwestern (purple–green), central (blue), and southeastern (yellow–orange) areas of the Hamersley Range. The most common haplotype (H3) was present across seven sampled populations (DAL, HIL, PEB, VIG, WIL, WIN) in the central area of the distribution, as well as in LEA to the north-west off the Hamersley Range (Fig. 1). The least divergent haplotypes (H1–H9 and H14, purple-blue-green) occurred in the north-west and centre of the sampled population distribution, and the most divergent haplotypes (H10–H14) located in south-eastern populations (RHO, PAM, NEW, yellow-orange), (Fig. 1).
Nuclear DNA
All loci conformed to HWE (P < 0.001) and no pairs of loci showed evidence of LD (P < 0.001). Estimated null allele frequencies were low for all loci across all populations (≤0.048 ± 0.013). Nuclear microsatellite diversity was low to moderate (Table 1). Observed heterozygosity was lower than expected heterozygosity at most populations but not significantly so. Although Fixation indices varied among populations, mean values were low, and few were significantly different to zero (Table 1).
Global genetic divergence among populations was moderate; genetic fixation FST = 0.139, P < 0.001 and allelic differentiation DST = 0.155 P < 0.001. Pairwise population measures of genetic fixation were visualised as a heatmap (Fig. 2a) and indicated the greatest levels of divergence between the most north-westerly population LEA and the south-easterly populations PAM, NEW and RHO. Pairwise population divergence was generally high between these peripheral populations (LEA, PAM, NEW and RHO) and more centrally located populations. Pairwise values of divergence between more central populations were generally lower but showed significant variation. Global estimates of genetic divergence without the most divergent and geographically disjunct peripheral populations LEA, PAM, NEW and RHO were somewhat lower, but still moderate (FST = 0.102 P < 0.001 and DST = 0.113 P < 0.001). The mutation rate in nuclear loci was not sufficient to affect phylogeographic structure within populations (observed RST = 0.145 > mean permuted RST = 0.127, P = 0.228), or among populations globally (observed RST = 0.129 ≥ mean permuted RST = 0.129, P = 1.000), although phylogeographic structure was associated with geographic distance (slope RST/1 − RST = 0.072, P = 0.000, equivalent to a Mantel test). Global RST was greater than FST but not significantly greater (RST = 0.145, s.e. = 0.020; FST = 0.139, s.e. = 0.016) suggesting that overall, mutation in microsatellite regions is negligible in relation to genetic drift.
|
|
AMOVA revealed 86% of genetic diversity partitioned within populations and 14% among populations. The first two axes of the PCoA explained 19.4% of the total variation and showed some separation of the south-easterly populations PAM, NEW and RHO from other populations (Fig. 2b). There was a significant positive correlation between geographic distance and genetic differentiation indicating a significant effect of IBD across the sampled region (Rxy = 0.678, R2 = 0.459, P < 0.01).
Assessment of the ad hoc model likelihood statistic L(K) from STRUCTURE analyses showed a slowing but continual increase up to K = 10, with a reasonable maximum and low standard deviation for K = 5 (Fig. 3a). The ΔK statistic provided strong evidence for a maximum and an optimal number of clusters K = 3 (Fig. 3b). Mean Q values for all individuals and populations (Fig. 3c) indicated three genetic clusters running along a north-west to south-east transect of the Hamersley Range with a small amount of admixture between them (Fig. 3d). In a geographic context, Cluster 1 included (population Q values > 0.5) populations to the north-west (CAL, HAM, LEA, LOI, SHE, SOL), Cluster 2 included more central populations (DAL, DIN, EMU, GOV, HIL, KNO, PEB, VIG, WIL and WIN) and Cluster 3 included the three most south-easterly populations (NEW, PAM and RHO).
|
|
Assessment of cross-validation scores from tess3r analyses showed an initial minimum at K = 5 (Fig. 4a). Mean Q values for all individuals (Fig. 4b) and populations indicate five genetic clusters running along the north-west to south-east transect along the Hamersley Range (Fig. 4c). Mean Q values for individuals and populations using a model that incorporates distinct spatial geographical data revealed far greater amounts of admixture than by Bayesian analysis (Fig. 4c), with some grouping of admixed populations into the three geographic areas. The more disjunct populations showed high membership to a cluster, LEA to cluster 1 and the south-eastern populations to cluster 5. The other populations tended to have high levels of admixture; north-western populations generally had admixture of clusters 1 and 2, and the central populations had admixture of clusters 3 and 4.
|
|
The pollen to seed dispersal ratio was high (49:1). There was no correlation of environmental or landscape variables with measures of genetic diversity (all P ≥ 0.304). However, populations located on the Hamersley Range had greater measures of genetic diversity, than the one population located off the Hamersley Range (LEA). Twelve populations (CAL, GOV, HAM, LEA, LOI, NEW, PEB, RHO, SHE, SOL, VIG, WIL) showed evidence of having undergone recent reductions in population size or genetic bottlenecks under the IAM (one tailed He excess Wilcoxon test P < 0.05), and four showed evidence under the TPM (LEA, LOI, SHE, WIL).
Discussion
Our analysis of patterns of historical phylogeographic and contemporary population genetic diversity and structure in M. viminalis showed historical persistence and contemporary connectivity associated with three general groups across the regional distribution in the Hamersley Range. The level of contemporary genetic diversity was lower than may be expected given the species widespread range and diversity estimates for a highly comparable study on another Mirbelia with the same life history traits. Low diversity in the nuclear genome may be related to recent reductions in size for Pilbara populations of M. viminalis. Pollen dispersal appears currently sufficient to maintain genetic admixture among neighbouring populations but becomes increasingly limited with geographic distance.
Historical persistence in the Pilbara
A signal of phylogeographic structure and a lack of demographic signal for historical expansion for extant populations suggests historical persistence for Pilbara populations of M. viminalis over a macroevolutionary time scale. Significant phylogeographic structure indicates some limitation to historical genetic connectivity among M. viminalis populations across the Hamersley Range. For example, the presence of private and divergent haplotypes in the three most south-easterly populations suggests that these populations have been more isolated from the rest of the distribution, despite a lack of obvious topographical or other environmental barriers. Limitations to gene flow via seed dispersal may be expected for geographically disjunct populations of Mirbelia and other species where unspecialised primary seed dispersal by gravity and perhaps secondarily by ants is likely to result in short distance dispersal of only a few metres (Hughes and Westoby 1992; Gómez and Espadaler 1998). Shallow divergence and the presence of seven haplotypes (H1, H2, H3, H4, H6, H9, H14) shared in multiple populations indicates greater historical genetic connectivity across the rest of the sampled populations, especially in the central area with more geographically continuous populations. Historical genetic connectivity may also be maintained among more geographically distant populations in this landscape via longer distance seed dispersal events. Such events may be regularly facilitated by meteorological phenomena (Nathan et al. 2008) including willy willies or dust devils, and the strong wind gusts and sometimes-extensive flooding and surface water flows associated with tropical depressions and cyclonic activity that are common across the region (Leighton 2004). In addition, dispersal may be facilitated by endozoochory and exozoochory by migratory animal vectors (Nathan et al. 2008) such as emus and kangaroos (Calviño-Cancela et al. 2006, 2008; Nathan 2006) and historically by the now locally extinct bandicoots, bettongs, other small macropods, and larger rodents (Nistelberger et al. 2020).
It has been suggested that upland areas with high topographical complexity are more mesic or thermally buffered compared to surrounding lowland areas and may be important refugial areas (Trew and Maclean 2021). The habitat heterogeneity of the topographically complex Pilbara ranges may have provided microrefugia over historical periods of climate change promoting population persistence and the maintenance of population size, and genetic diversity (Byrne et al. 2008; Keppel et al. 2012). There is evidence of the mesic ranges acting as refugia in E. leucophloia (Byrne et al. 2017), Callitris columellaris F. Muell. and Acacia ancistrocarpa Maiden and Blakely where populations had greater genetic connectivity and larger effective population sizes on the range (Levy et al. 2016). In M. viminalis, the species distribution itself implies that extant populations in the Pilbara have been restricted to topographically complex elevated areas for some time. This is supported by the lower nuclear genetic diversity in the LEA population compared to populations on the range, although there was no correlation of nuclear diversity with environmental or landscape variables to indicate microrefugia. Greater sampling of M. viminalis throughout its wider range is required to further elucidate the species’ demographic history.
Limited contemporary genetic diversity
Overall, extant Pilbara populations of M. viminalis maintained low to moderate levels of contemporary genetic diversity. This may seem inconsistent with population genetic theory that predicts that plant species with widespread ranges maintain greater diversity than species with restricted range sizes. However, although M. viminalis has a very widespread range on the Australian continent, populations within the Pilbara bioregion are almost exclusively restricted to the uplands of the Hamersley Range and can be considered to have a regional range. In this case and given the evidence for contemporary reductions in population size, more moderate levels of genetic diversity may be expected. It is not clear what may have driven reductions in populations’ size, although vast areas of the Pilbara are highly degraded due to unsustainable commercial cattle grazing and resulting erosion, as well as feral animal grazing, weeds, and altered fire regimes (Department of Primary Industries and Regional Development 2020). Populations of M. viminalis were surprisingly genetically depauperate compared to another member of the Mirbelia genus; Mirbelia sp. Bursarioides, which has an even more restricted endemic regional range (160 km) in the semi-arid Avon, wheatbelt, Murchison and Yalgoo IBRA region of Western Australia with populations that have also experienced recent changes in population size (Millar et al. 2016). Whilst the number of chloroplast haplotypes were similar (h = 15 and 16 respectively), nuclear diversity for the Pilbara populations of M. viminalis was considerably lower than that observed for Mirbelia sp. Bursarioides (NA = 8.63, Ho = 0.723, Millar et al. 2016). It would be interesting to assess genetic diversity across the full geographic range of M. viminalis to determine whether the lower levels of genetic diversity detected in this study are reflective of the species as a whole or are associated more specifically with the Hamersley Range.
Intrapopulation genetic diversity may become reduced in the presence of increased levels of selfing or mating among related individuals for predominantly outcrossed species. Many Fabaceae are self-compatible but preferentially outcrossed (Bernhardt et al. 1984; Moran et al. 1989; Muona et al. 1991; Broadhurst et al. 2008; Millar et al. 2008, 2019; Ng et al. 2009) and, although no studies have been conducted, mating systems of Mirbelia are expected to be similarly largely outcrossed. We found no evidence of elevated levels of selfing or bi-parental inbreeding for M. viminalis in this study. Alternatively, low genetic diversity in Pilbara populations of M. viminalis may be a result of recent reductions in effective population sizes limiting the quantity of pollen available for dispersal, as indicated for a number of populations by bottleneck analyses. In contrast, most populations of Mirbelia sp. Bursarioides showed evidence for recent expansion in size (Millar et al. 2016).
Isolation by geographic distance
Despite the maintenance of only low to moderate measures of contemporary genetic diversity within populations, a signal of IBD and moderate estimates of population genetic differentiation imply a stepping model of genetic connectivity via contemporary gene flow, at least among neighbouring populations of M. viminalis in the Pilbara. A signal of IBD along the north-west to south-east transect of the Hamersley Range suggests that connectivity among extant populations is predominantly limited by spatial geographic distance and not by any distinct barriers relating to currently obvious ecological, environmental, or landscape features. The elevated degree of divergence in the nuclear genome among south-easterly populations PAM, NEW and RHO reflects that observed in the chloroplast genome, although barriers that may be affecting this macro and microevolutionary divergence are unclear. The influence of IBD is reflected in the strongest pairwise genetic structure occurring among the most geographically distal populations (LEA and NEW, 307 km apart) with a clear pattern of admixture among the five genetic clusters identified by tess3r, with some structure into three groups similar to the three genetic clusters identified by STRUCTURE analysis, of the north-western, central and south-eastern populations. The Bayesian model of clustering used by STUCTURE is known to overestimate the discreteness of clusters compared with models that incorporate spatial data. The tess3r analysis probably provides a more biologically meaningful interpretation of genetic structure, with better estimates of admixture in the context of a known pattern of IBD (Meirmans 2012; Caye et al. 2016).
Neutral nuclear DNA variation reflects both pollen and seed dispersal; and estimates of ratios of pollen to seed dispersal from a range of plants generally indicate that genetic connectivity via contemporary gene flow is driven largely by pollen dispersal (Ennos 1994). The relative contributions of pollen and seed dispersal to overall genetic connectivity do vary considerably for flora of the Pilbara with previous estimates ranging from 3:1 for Acacia pruinocarpa Tindale (Nistelberger et al. 2020) to 44:1 for A. ancistrocarpa (unpubl. data but see Levy et al. 2016). Interestingly, the pollen to seed dispersal ratio for M. viminalis is the highest recorded so far in this landscape, a result that might be unexpected given the likely primary pollen dispersers are introduced and native bees (Scaccabarozzi et al. 2020). Combined with a signal of IBD, moderate estimates of population genetic differentiation, and high contemporary genetic admixture among populations, the pollen to seed dispersal ratio suggests genetic connectivity is largely maintained via pollen dispersal for M. viminalis. It would be informative to conduct direct studies to confirm mating systems and methods of both pollen and seed dispersal to investigate relative contributions of pollen and seed dispersal in flora of the Pilbara. This may inform on the low levels of contemporary genetic diversity detected despite the presence of genetic admixture among populations.
Conclusions
Phylogeographic diversity and structure was consistent with historical population persistence for M. viminalis in the Pilbara bioregion, with some connectivity maintained via seed dispersal across the Hamersley Range, as seen for a number of plant species, including other Fabaceae, present in the Pilbara. Contemporary genetic connectivity maintained largely via pollen dispersal appears to be sufficient to maintain genetic admixture especially among neighbouring populations but becomes increasingly limited with geographic distance. The maintenance of low levels of contemporary genetic diversity may be an inherent trait of the species or associated with recent reductions in population size.
Supplementary material
Supplementary material is available online.
Data availability
cpDNA sequence data (accession numbers OM389460–389483) and nuclear microsatellite primer sequence data (accession numbers MW138757–MW138771) generated for this study have been deposited in the GenBank database http://www.ncbi.nlm.nih.gov/genbank/. Haplotype and genotype data has been deposited in the Mendeley Data V1, database https://data.mendeley.com/, Doi:10.17632/xg7chf69wk.1.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This work was funded by Rio Tinto (http://www.riotinto.com/) and BHP (https://www.bhp.com/our-businesses/minerals-australia/western-australia-iron-ore/).
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48.| Median-joining networks for inferring intraspecific phylogenies.Crossref | GoogleScholarGoogle Scholar |
Barrett RL, Clugston JAR, Cook LG, Crisp MD, Jobson PC, Lepschi BJ, Renner MAM, Weston PH (2021) Understanding diversity and systematics in Australian Fabaceae tribe Mirbelieae. Diversity 13, 391
| Understanding diversity and systematics in Australian Fabaceae tribe Mirbelieae.Crossref | GoogleScholarGoogle Scholar |
Beard JS (1975) ‘Vegetation survey of Western Australia 1: 1 000 000 vegetation series. Sheet 5, Pilbara.’ (The University of Western Australia: Perth, WA)
Beheregaray LB (2008) Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Molecular Ecology 17, 3754–3774.
| Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere.Crossref | GoogleScholarGoogle Scholar |
Bernhardt P, Kenrick J, Knox RB (1984) Pollination ecology and the breeding system of Acacia retinoides (Leguminosae: Mimosoideae). Annals of the Missouri Botanical Garden 71, 17–29.
| Pollination ecology and the breeding system of Acacia retinoides (Leguminosae: Mimosoideae).Crossref | GoogleScholarGoogle Scholar |
Bradbury D, Tapper S-L, Coates D, McArthur S, Hankinson M, Byrne M (2016) The role of fire and a long-lived soil seed bank in maintaining persistence, genetic diversity and connectivity in a fire-prone landscape. Journal of Biogeography 43, 70–84.
| The role of fire and a long-lived soil seed bank in maintaining persistence, genetic diversity and connectivity in a fire-prone landscape.Crossref | GoogleScholarGoogle Scholar |
Bradbury D, Binks RM, Coates DJ, Byrne M (2019) Conservation genomics of range disjunction in a global biodiversity hotspot: a case study of Banksia biterax (Proteaceae) in southwestern Australia. Biological Journal of the Linnean Society 127, 390–406.
| Conservation genomics of range disjunction in a global biodiversity hotspot: a case study of Banksia biterax (Proteaceae) in southwestern Australia.Crossref | GoogleScholarGoogle Scholar |
Broadhurst LM, Young AG, Forrester R (2008) Genetic and demographic responses of fragmented Acacia dealbata (Mimosaceae) populations in southeastern Australia. Biological Conservation 141, 2843–2856.
| Genetic and demographic responses of fragmented Acacia dealbata (Mimosaceae) populations in southeastern Australia.Crossref | GoogleScholarGoogle Scholar |
Burbidge AH, Johnstone RE, Pearson DJ (2010) Birds in a vast arid upland: avian biogeographical patterns in the Pilbara region of Western Australia. Records of the Western Australian Museum Supplement 78, 247–270.
| Birds in a vast arid upland: avian biogeographical patterns in the Pilbara region of Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Hankinson M (2012) Testing the variability of chloroplast sequences for plant phylogeography. Australian Journal of Botany 60, 569–574.
| Testing the variability of chloroplast sequences for plant phylogeography.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Yeates DK, Joseph L, et al. (2008) Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Molecular Ecology 17, 4398–4417.
| Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Coates D, Macdonald B, Hankinson M, McArthur S, van Leeuwen S (2016) High nuclear genetic differentiation, but low chloroplast diversity in a rare species, Aluta quadrata (Myrtaceae), with a disjunct distribution in the Pilbara, Western Australia. Australian Journal of Botany 64, 687–695.
Byrne M, Millar MA, Coates DJ, MacDonald BM, McArthur SM, Zhou M, van Leeuwen S (2017) Refining expectations for environmental characteristics of refugia: two ranges of differing elevation and topographical complexity are mesic refugia in an arid landscape. Journal of Biogeography 44, 2539–2550.
| Refining expectations for environmental characteristics of refugia: two ranges of differing elevation and topographical complexity are mesic refugia in an arid landscape.Crossref | GoogleScholarGoogle Scholar |
Calviño-Cancela M, Dunn RR, van Etten EJB, Lamont BB (2006) Emus as non-standard seed dispersers and their potential for long-distance dispersal. Ecography 29, 632–640.
| Emus as non-standard seed dispersers and their potential for long-distance dispersal.Crossref | GoogleScholarGoogle Scholar |
Calviño-Cancela M, He T, Lamont BB (2008) Distribution of myrmecochorous species over the landscape and their potential long-distance dispersal by emus and kangaroos. Diversity and Distributions 14, 11–17.
| Distribution of myrmecochorous species over the landscape and their potential long-distance dispersal by emus and kangaroos.Crossref | GoogleScholarGoogle Scholar |
Caye K, Deist TM, Martins H, Michel O, François O (2016) TESS3: fast inference of spatial population structure and genome scans for selection. Molecular Ecology Resources 16, 540–548.
| TESS3: fast inference of spatial population structure and genome scans for selection.Crossref | GoogleScholarGoogle Scholar |
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24, 621–631.
| Microsatellite null alleles and estimation of population differentiation.Crossref | GoogleScholarGoogle Scholar |
Cracraft J (1991) Patterns of diversification within continental biotas: heirarchical congruence among the areas of endemism of Australian vertebrates. Australian Systematic Botany 4, 211–227.
| Patterns of diversification within continental biotas: heirarchical congruence among the areas of endemism of Australian vertebrates.Crossref | GoogleScholarGoogle Scholar |
Department of Primary Industries and Regional Development (2020) ‘Status of the Western Australian pastoral rangelands 2020: condition, trend and risk.’ (DPIRD, Western Australian Government: Perth, WA)
Dieringer D, Schlötterer C (2003) MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes 3, 167–169.
| MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets.Crossref | GoogleScholarGoogle Scholar |
Doughty P, Rolfe JK, Burbidge AH, Pearson DJ, Kendrick PG (2011) Herpetological assemblages of the Pilbara biogeographic region, Western Australia: ecological associations, biogeographic patterns and conservation. Records of the Western Australian Museum Supplement 78, 315–350.
| Herpetological assemblages of the Pilbara biogeographic region, Western Australia: ecological associations, biogeographic patterns and conservation.Crossref | GoogleScholarGoogle Scholar |
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15.
Durrant BJ, Harvey MS, Framenau VW, Ott R, Waldock JM (2010) Patterns in the composition of ground-dwelling spider communities in the Pilbara bioregion, Western Australia. Records of the Western Australian Museum Supplement 78, 185–204.
| Patterns in the composition of ground-dwelling spider communities in the Pilbara bioregion, Western Australia.Crossref | GoogleScholarGoogle Scholar |
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4, 359–361.
| STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method.Crossref | GoogleScholarGoogle Scholar |
Eberhard SM, Halse SA, Humphreys WF (2005) Stygofauna in the Pilbara region, north-west Western Australia: a review. Journal of the Royal Society of Western Australia 88, 167–176.
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24, 217–242.
| Population genetic consequences of small population size: implications for plant conservation.Crossref | GoogleScholarGoogle Scholar |
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72, 250–259.
| Estimating the relative rates of pollen and seed migration among plant populations.Crossref | GoogleScholarGoogle Scholar |
Ennos RA, Sinclair WT, Hu X-S, Langdon A (1999) Using organelle markers to elucidate the history, ecology and evolution of plant populations. In ‘Molecular systematics and plant evolution’. (Eds PM Hollingsworth, RM Bateman, RJ Gornall) pp. 1–99. (Taylor and Francis: London, UK)
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study.Crossref | GoogleScholarGoogle Scholar |
Faircloth BC, Glenn TC (2014) ‘Protocol: preparation of an AMPure XP substitute (AKA Serapure).’
| Crossref |
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
| Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies.Crossref | GoogleScholarGoogle Scholar |
Frantz AC, Cellina S, Krier A, Schley L, Burke T (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? Journal of Applied Ecology 46, 493–505.
| Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance?Crossref | GoogleScholarGoogle Scholar |
Gardner MG, Fitch AJ, Bertozzi T, Lowe AJ (2011) Rise of the machines – recommendations for ecologists when using next generation sequencing for microsatellite development. Molecular Ecology Resources 11, 1093–1101.
| Rise of the machines – recommendations for ecologists when using next generation sequencing for microsatellite development.Crossref | GoogleScholarGoogle Scholar |
Gibson LA, McKenzie NL (2009) Environmental associations of small ground-dwelling mammals in the Pilbara region, Western Australia. Records of the Western Australian Museum Supplement 78, 91–122.
| Environmental associations of small ground-dwelling mammals in the Pilbara region, Western Australia.Crossref | GoogleScholarGoogle Scholar |
Gómez C, Espadaler X (1998) Myrmecochorous dispersal distances: a world survey. Journal of Biogeography 25, 573–580.
| Myrmecochorous dispersal distances: a world survey.Crossref | GoogleScholarGoogle Scholar |
Guthrie NA, Weir T, Will K (2010) Localised and regional patterns in ground-dwelling beetle assemblages in a semi-tropical arid zone environment. Records of the Western Australian Museum Supplement 78, 169–184.
| Localised and regional patterns in ground-dwelling beetle assemblages in a semi-tropical arid zone environment.Crossref | GoogleScholarGoogle Scholar |
Hall TA (1999) BioEdit: a user friendly sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98.
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2, 618–620.
| SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels.Crossref | GoogleScholarGoogle Scholar |
Hardy OJ, Charbonnel N, Fréville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163, 1467–1482.
| Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation.Crossref | GoogleScholarGoogle Scholar |
Hewitt GM (2004) The structure of biodiversity – insights from molecular phylogeography. Frontiers in Zoology 1, 4
| The structure of biodiversity – insights from molecular phylogeography.Crossref | GoogleScholarGoogle Scholar |
Hickerson MJ, Carstens BC, Cavender-Bares J, Crandall KA, Graham CH, Johnson JB, Rissler L, Victoriano PF, Yoder AD (2010) Phylogeography’s past, present, and future: 10 years after Avise, 2000. Molecular Phylogenetics and Evolution 54, 291–301.
| Phylogeography’s past, present, and future: 10 years after Avise, 2000.Crossref | GoogleScholarGoogle Scholar |
Hughes L, Westoby M (1992) Fate of seeds adapted for dispersal by ants in Australian sclerophyll vegetation. Ecology 73, 1285–1299.
| Fate of seeds adapted for dispersal by ants in Australian sclerophyll vegetation.Crossref | GoogleScholarGoogle Scholar |
Jost L (2008) GST and its relatives do not measure differentiation. Molecular Ecology 17, 4015–4026.
| GST and its relatives do not measure differentiation.Crossref | GoogleScholarGoogle Scholar |
Keppel G, van Niel KP, Wardell-Johnson GW, Yates CJ, Byrne M, Mucina L, Schut AGT, Hopper SD, Franklin SE (2012) Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecology and Biogeography 21, 393–404.
| Refugia: identifying and understanding safe havens for biodiversity under climate change.Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Ariati SR, Murphy DJ (2006) Biogeography of the Acacia victoriae, pyrifolia and murrayana species groups in arid Australia. Journal of Arid Environments 66, 462–476.
| Biogeography of the Acacia victoriae, pyrifolia and murrayana species groups in arid Australia.Crossref | GoogleScholarGoogle Scholar |
Leighton K (2004) Climate. In ‘An inventory and condition survey of the Pilbara region, Western Australia’. Vol. No. 92. (Eds AME van Vreeswyk, AL Payne, KA Leighton, P Hennig) (Western Australian Department of Agriculture: Perth, WA, Australia)
Levy E, Byrne M, Coates DJ, Macdonald BM, McArthur S, van Leeuwen S (2016) Contrasting influences of geographic range and distribution of populations on patterns of genetic diversity in two sympatric Pilbara Acacias. PLoS ONE 11, e0163995
| Contrasting influences of geographic range and distribution of populations on patterns of genetic diversity in two sympatric Pilbara Acacias.Crossref | GoogleScholarGoogle Scholar |
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
| DnaSP v5: a software for comprehensive analysis of DNA polymorphism data.Crossref | GoogleScholarGoogle Scholar |
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequlibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111, 675–689.
| Population bottlenecks and nonequlibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck.Crossref | GoogleScholarGoogle Scholar |
McKenzie NL, Bullen RD (2009) The echolocation calls, habitat relationships, foraging niches and communities of Pilbara microbats. Records of the Western Australian Museum Supplement 78, 123–155.
| The echolocation calls, habitat relationships, foraging niches and communities of Pilbara microbats.Crossref | GoogleScholarGoogle Scholar |
McKenzie NL, van Leeuwen S, Pinder AM (2009) Introduction to the Pilbara biodiversity survey, 2002–2007. Records of the Western Australian Museum Supplement 78, 3–89.
| Introduction to the Pilbara biodiversity survey, 2002–2007.Crossref | GoogleScholarGoogle Scholar |
Meirmans PG (2012) The trouble with isolation by distance. Molecular Ecology 21, 2839–2846.
| The trouble with isolation by distance.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Byrne M, Nuberg I, Sedgley M (2008) High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites. Tree Genetics & Genomes 4, 367–377.
| High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Byrne M, Coates DJ, Roberts JD (2016) Contrasting diversity and demographic signals in sympatric narrow-range endemic shrubs of the south-west Western Australian semi-arid zone. Biological Journal of the Linnean Society 118, 315–329.
| Contrasting diversity and demographic signals in sympatric narrow-range endemic shrubs of the south-west Western Australian semi-arid zone.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Coates DJ, Byrne M, Krauss SL, Jonson J, Hopper SD (2019) Assessment of genetic diversity and mating system of Acacia cyclops restoration and remnant populations. Restoration Ecology 27, 1327–1338.
| Assessment of genetic diversity and mating system of Acacia cyclops restoration and remnant populations.Crossref | GoogleScholarGoogle Scholar |
Millar MA, Binks RM, Tapper S-L, MacDonald BM, McArthur SL, Hankinson M, Coates DJ, van Leeuwen S, Byrne M (2022) Limited phylogeographic and genetic connectivity in Acacia species of low stature in an arid landscape. Ecology and Evolution 12, e9052
| Limited phylogeographic and genetic connectivity in Acacia species of low stature in an arid landscape.Crossref | GoogleScholarGoogle Scholar |
Moran G, Hopper S (1987) Conservation of the genetic resources of rare and widespread eucalypts in remnant vegetation. In ‘Nature conservation: the role of native vegetation’. (Eds D Saunders, G Arnold, A Burbidge, A Hopkins) pp. 151–162. (Surry Beatty & Sons: -Chipping Norton, NSW)
Moran GF, Muona O, Bell JC (1989) Breeding systems and genetic diversity in Acacia auriculiformis and A. crassicarpa. Biotropica 21, 250–256.
| Breeding systems and genetic diversity in Acacia auriculiformis and A. crassicarpa.Crossref | GoogleScholarGoogle Scholar |
Muona O, Moran GF, Bell JC (1991) Hierarchical patterns of correlated mating in Acacia melanoxylon. Genetics 127, 619–626.
| Hierarchical patterns of correlated mating in Acacia melanoxylon.Crossref | GoogleScholarGoogle Scholar |
Nathan R (2006) Long-distance dispersal of plants. Science 313, 786–788.
| Long-distance dispersal of plants.Crossref | GoogleScholarGoogle Scholar |
Nathan R, Schurr FM, Spiegel O, Steinitz O, Trakhtenbrot A, Tsoar A (2008) Mechanisms of long-distance seed dispersal. Trends in Ecology & Evolution 23, 638–647.
| Mechanisms of long-distance seed dispersal.Crossref | GoogleScholarGoogle Scholar |
Ng C-H, Lee S-L, Ng KK-S, Muhammad N, Ratnam W (2009) Mating system and seed variation of Acacia hybrid (A. mangium × A. auriculiformis). Journal of Genetics 88, 25–31.
| Mating system and seed variation of Acacia hybrid (A. mangium × A. auriculiformis).Crossref | GoogleScholarGoogle Scholar |
Nistelberger HM, Binks RM, van Leeuwen S, Coates DJ, McArthur SL, Macdonald BM, Hankinson M, Byrne M (2020) Extensive genetic connectivity and historical persistence are features of two widespread tree species in the ancient Pilbara region of Western Australia. Genes 11, 863
| Extensive genetic connectivity and historical persistence are features of two widespread tree species in the ancient Pilbara region of Western Australia.Crossref | GoogleScholarGoogle Scholar |
Nistelberger HM, Tapper S-L, Coates DJ, McArthur SL, Byrne M (2021) As old as the hills: Pliocene palaeogeographical processes influence patterns of genetic structure in the widespread, common shrub Banksia sessilis. Ecology and Evolution 11, 1069–1082.
| As old as the hills: Pliocene palaeogeographical processes influence patterns of genetic structure in the widespread, common shrub Banksia sessilis.Crossref | GoogleScholarGoogle Scholar |
Novembre J, Stephens M (2008) Interpreting principal component analyses of spatial population genetic variation. Nature Genetics 40, 646–649.
| Interpreting principal component analyses of spatial population genetic variation.Crossref | GoogleScholarGoogle Scholar |
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539.
| GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update.Crossref | GoogleScholarGoogle Scholar |
Pepper M, Doughty P, Keogh JS (2013) Geodiversity and endemism in the iconic Australian Pilbara region: a review of landscape evolution and biotic response in an ancient refugium. Journal of Biogeography 40, 1225–1239.
| Geodiversity and endemism in the iconic Australian Pilbara region: a review of landscape evolution and biotic response in an ancient refugium.Crossref | GoogleScholarGoogle Scholar |
Piry S, Luikart G, Cornuet J-M (1999) Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity 90, 502–503.
| Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data.Crossref | GoogleScholarGoogle Scholar |
Polzin T, Daneshmand SV (2003) On Steiner trees and minimum spanning trees in hypergraphs. Operations Research Letters 31, 12–20.
| On Steiner trees and minimum spanning trees in hypergraphs.Crossref | GoogleScholarGoogle Scholar |
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245.
| Measuring and testing genetic differentiation with ordered versus unordered alleles.Crossref | GoogleScholarGoogle Scholar |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
| Inference of population structure using multilocus genotype data.Crossref | GoogleScholarGoogle Scholar |
Raymond M, Rousset F (1995) GENEPOP (version 3.4): population genetics software for exact tests and ecumenicism. Journal of Heredity 86, 248–249.
| GENEPOP (version 3.4): population genetics software for exact tests and ecumenicism.Crossref | GoogleScholarGoogle Scholar |
R Core Team (2019) R: a language and environment for statistical computing. (Foundation for Statistical Computing: Vienna, Austria) Available at http://www.R-project.org/
Rohland N, Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research 22, 939–946.
| Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture.Crossref | GoogleScholarGoogle Scholar |
Sampson J, Tapper S, Coates D, Hankinson M, Mcarthur S, Byrne M (2018) Persistence with episodic range expansion from the early Pleistocene: the distribution of genetic variation in the forest tree Corymbia calophylla (Myrtaceae) in south-western Australia. Biological Journal of the Linnean Society 123, 545–560.
| Persistence with episodic range expansion from the early Pleistocene: the distribution of genetic variation in the forest tree Corymbia calophylla (Myrtaceae) in south-western Australia.Crossref | GoogleScholarGoogle Scholar |
Scaccabarozzi D, Dixon KW, Tomlinson S, Milne L, Bohman B, Phillips RD, Cozzolino S (2020) Pronounced differences in visitation by potential pollinators to co-occurring species of Fabaceae in the Southwest Australian biodiversity hotspot. Botanical Journal of the Linnean Society 194, 308–325.
| Pronounced differences in visitation by potential pollinators to co-occurring species of Fabaceae in the Southwest Australian biodiversity hotspot.Crossref | GoogleScholarGoogle Scholar |
Shaw J, Lickey E, Schilling E, Small R (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hair III. American Journal of Botany 94, 275–288.
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139, 457–462.
| A measure of population subdivision based on microsatellite allele frequencies.Crossref | GoogleScholarGoogle Scholar |
Stone GN, Raine NE, Prescott M, Willmer PG (2003) Pollination ecology of acacias (Fabaceae, Mimosoideae). Australian Systematic Botany 16, 103–118.
| Pollination ecology of acacias (Fabaceae, Mimosoideae).Crossref | GoogleScholarGoogle Scholar |
Sweedman L, Brand G (2006) Collection guidelines for common Australian families and genera. In ‘Australian seeds: a guide to their collection, identification and biology’. (Eds L Sweedman, D Merritt) p. 191. (CSIRO Publishing: Melbourne, Vic., Australia)
Thackway R, Cresswell ID (1995) ‘An interim biogeographic regionalisation for Australia: a framework for setting priorities in the National Reserves System Cooperative Program, Version 4.0.’ (Autralian Nature Conservation Agency: Canberra, ACT)
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680.
| CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.Crossref | GoogleScholarGoogle Scholar |
Trew BT, Maclean IMD (2021) Vulnerability of global biodiversity hotspots to climate change. Global Ecology and Biogeography 30, 768–783.
| Vulnerability of global biodiversity hotspots to climate change.Crossref | GoogleScholarGoogle Scholar |
Unmack PJ (2001) Biogeography of Australian freshwater fishes. Journal of Biogeography 28, 1053–1089.
| Biogeography of Australian freshwater fishes.Crossref | GoogleScholarGoogle Scholar |
Wright S (1943) Isolation by distance. Genetics 28, 114–138.
| Isolation by distance.Crossref | GoogleScholarGoogle Scholar |