Selection of transcripts related to low-temperature tolerance using RNA sequencing from F2 plants between japonica and indica rice (Oryza sativa L.) cultivars
Akari Fukuda A E , Tatsuro Hirose B , Yoichi Hashida B , Naohiro Aoki
A E , Tatsuro Hirose B , Yoichi Hashida B , Naohiro Aoki  C and Atsushi J. Nagano D
C and Atsushi J. Nagano D
A Institute of Crop Science, National Agriculture and Food Research Organization, Tsukuba, Ibaraki, Japan.
B Faculty of Agriculture, Takasaki University of Health and Welfare, Takasaki, Gunma, Japan.
C Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.
D Faculty of Agriculture, Ryukoku University, Otsu, Shiga, Japan.
E Corresponding author. Email: akfukuda@affrc.go.jp
Functional Plant Biology 48(10) 984-993 https://doi.org/10.1071/FP21088
Submitted: 23 March 2021 Accepted: 21 May 2021 Published: 11 June 2021
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
At low temperatures (18°C), seedlings of an indica rice (Oryza sativa L.) cultivar Kasalath showed symptoms of chlorosis, although the leaves of a japonica cultivar Arroz da Terra remained green. In this study, transcripts related to the chlorophyll content of rice seedlings grown at 18°C were investigated using RNA-sequencing (RNA-Seq) data for F2 crosses between cultivars Arroz da Terra and Kasalath, as well as their parental cultivars. Differential expression analysis revealed that gene ontology terms related to ‘photosynthesis’ were significantly enriched in lowly expressed genes at 18°C than at 25°C in Kasalath. However, the gene ontology terms related to ‘response to stress’ were significantly enriched in highly expressed genes at 18°C than at 25°C in Kasalath. When the F2 plants were grown at 18°C, their chlorophyll contents varied. Transcripts with expression levels related to chlorophyll content were statistically selected using RNA-Seq data from 21 F2 plants. In regression models, frequently selected genes included four photosynthetic and two stress-responsive genes. The expression values of four photosynthetic and two stress-responsive genes in high-frequency selected genes were significantly correlated with chlorophyll content not only in plants analysed using RNA-Seq but also in 95 F2 plants.
Keywords: cold tolerance, low temperature, LASSO, POX22.3, RNA sequencing (RNA-Seq), TIFY11C, transcriptome, rice, Oryza sativa L.
Introduction
Low temperatures have negative effects on plants including reduced growth rates and increased leaf chlorosis (Huner et al. 1993; Yoshida et al. 1996; Mackill and Lei 1997; Fukuda and Terao 2015; Yamori et al. 2011; Bai et al. 2021). During the direct seeding of rice (Oryza sativa L.), low temperature can cause serious problems, including increased seedling mortality. Therefore, elucidation of the genetic factors affecting low-temperature tolerance is important in rice breeding to improve direct seeding efficacy. Two major rice cultivars, indica and japonica, are known to have differences in their tolerance to low temperatures. The cultivar indica originated in the tropics and is more sensitive to low-temperature stress than the cultivar japonica, which is adapted to temperate conditions (Mackill and Lei 1997; Ohsumi et al. 2012; Fukuda and Terao 2015). indica cultivars develop symptoms of chlorosis at low temperatures (below 18°C). Low chlorophyll contents in leaves have also been observed, which reduce seedling growth rates. Generally, indica cultivars have larger heads and are useful for improving yield capacity (Nagata et al. 2002; Ando et al. 2008; Terao et al. 2010) but their sensitivity to low temperatures at the seedling stage limits their utilisation in temperate regions. Thus, improvement of the low-temperature tolerance of indica-based cultivars is required. Previous studies have examined the quantitative trait loci (QTLs) involved in cold-induced leaf chlorosis (Zhang et al. 2014; Fukuda and Terao 2015) and have found that multiple genes with weak effects control chlorophyll content under cold conditions. However, QTL analysis is laborious and time-consuming, and it is difficult to identify the candidate genes associated with multiple weak QTLs.
An alternate method for elucidating which physiological mechanism is involved in low-temperature tolerance is transcriptome analysis. This measures gene expression levels in a genome-wide manner and reveals genes related to many endogenous and environmental factors (Nagano et al. 2012; Kudapa et al. 2013; Yogendra and Kushalappa 2016; Onaga et al. 2017; Li et al. 2019; Han et al. 2020). RNA sequencing (RNA-Seq) analyses have been performed on cold-tolerant and cold-sensitive rice seedling varieties following short periods of severe cold stress (2–13°C) (Shen et al. 2014; Zhang et al. 2016; da Maia et al. 2017; Buti et al. 2018; Pradhan et al. 2019), and the results of these analyses have suggested that there are transcriptional differences among cultivars under cold stress conditions. However, indica cultivars display symptoms of chlorosis even at medium-low temperatures (18°C). Therefore, it is necessary to analyse gene expression under medium to low temperature stress (hereafter referred to as ‘low temperature’) at 18°C. This temperature is commonly experienced during direct seeding.
In this study, we aimed to determine the transcripts related to the chlorophyll content at low temperatures using a statistical approach. In general, transcriptome analyses require the comparison of two opposite groups to detect transcripts with different expression levels. However, most naturally derived organism traits are continuous and cannot be separated into two opposite groups. A statistical approach was previously used to select transcripts affecting the gradated traits using RNA-Seq data from rice seedlings. This is considered a useful tool for identifying transcripts related to initial seedling growth rates (Fukuda et al. 2018). In this study, we used this statistical approach to select transcripts affecting the chlorophyll contents of F2 plants grown under low-temperature conditions. The results showed that this statistical method effectively selected the genes related to chlorophyll content using a relatively small sample size of 21 F2 plants.
Materials and methods
Plant materials, growth conditions and experimental design
For the parental cultivars, five seedlings each of japonica (Arroz da Terra) and indica (Kasalath) were contained in each biological replicate. There were four biological replicates per treatment. Seeds were sterilised in water at 60°C for 10 min and imbibed in water at 25°C or 18°C for 2 days in an incubator. Then, one seed was sown in one compartment (1.6 cm diameter by 2.5 cm depth, with a distance of 1.9 cm between compartments) within the seedling pots filled with commercial nursery soil (‘Honens nursery soil No.1’, Honen Agri Co. Niigata, Japan) to a depth of 5 mm. Seedling pots were placed in an LH-240 (Nippon Medical and Chemical Instruments, Osaka, Japan) chamber under a 12-h artificial light and 12-h dark cycle. Light was emitted from a neutral white fluorescent lamp (52–77 μmol m–2 s–1 of photosynthetically active radiation). After 7 days of growth at 25°C or 21 days of growth at 18°C, seedlings that reached the second leaf stages were used to measure chlorophyll content and for RNA-Seq. Because low temperatures delayed the seedling growth, it took more time to reach the same growth stage under 18°C (21 days) than 25°C (7 days). Furthermore, 95 F2 individuals derived from a cross between Arroz da Terra and Kasalath were grown at 18°C, as described above, and used in the experiments. There was one biological replicate per individual.
Analysis of the chlorophyll contents
Chlorophyll content was determined using a digital chlorophyll meter (soil and plant analyzer development meter 502Plus, Konica Minolta Inc. Osaka, Japan), which provides a non-invasive method for estimating leaf chlorophyll content by measuring the light absorption of specific spectral bands in living leaves (Kumagai et al. 2009; Takai et al. 2010). Soil plant analysis development (SPAD) values were measured in the upper, middle and base sections of the second leaves of each seedling, and the mean value of the three leaf sections was considered the phenotypical value of SPAD.
RNA isolation
Second-leaf stage seedlings from the parental cultivars, Arroz da Terra and Kasalath, and the F2 plants were used for RNA-Seq analysis. After the seeds and roots of the seedlings were removed, fresh seedlings were frozen in liquid nitrogen and stored at –80°C until analysis. Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. For the parental cultivars, Arroz da Terra and Kasalath, total RNA was extracted from five seedlings per biological replicate. For the F2 individuals, total RNA was extracted from one individual seedling.
RNA-Seq analysis
RNA samples were used to produce sequencing libraries according to the Lasy-Seq method (ver. 1.1) (Kamitani et al. 2019). These were sequenced as 150-bp paired-end reads using an Illumina Hiseq × platform (Illumina Inc., San Diego, CA, USA). Forward-read files of the resulting Fastq files were deposited into the DNA Data Bank of Japan Sequence Read Archive (DRA; accession number DRA011043) (Kodama et al. 2012). RNA-Seq analysis was performed using the CLC Genomics Workbench ver. 11 (Qiagen, Hilden, Germany). After quality trimming, the raw reads were mapped to the Oryza sativa-Nipponbare-Reference-IRGSP-1.0 genome assembly and gene set (Ensemble plants release 25) (Kersey et al. 2016) using the following parameters: a mismatch cost of two, an insertion/deletion cost of three, a length fraction of 0.5, a similarity fraction of 0.8, and a maximum number of hits for a read of 10. Read numbers that were successfully mapped to the Os-Nipponbare-Reference-IRGSP-1.0 genome are listed (see Table S1, available as Supplementary Material to this paper). Transcript expression was calculated as reads per kilobase per million (RPKM) based on the number of uniquely mapped reads that overlapped exon regions.
Differential expression analysis
Differential expression analysis was performed using the RNA-Seq analysis module in CLC Genomics Workbench ver. 11 (Qiagen). Genes with a false discovery rate (FDR) < 0.05 and an absolute log2 fold change (FC) of at least one were considered differentially expressed. Gene ontology (GO) analysis was conducted on the agriGO database (Du et al. 2010) using singular enrichment analysis with an FDR < 0.05 and the National Center for Biotechnology Information (NCBI) ID.
Gene selection frequency
To determine which transcripts were correlated with chlorophyll content and the selection frequencies for the explanatory variables, we performed a statistical analysis method described by Fukuda et al. (2018). Previously, the transcripts related to initial growth rates of rice seedlings were statistically selected using 22 RNA-Seq samples (Fukuda et al. 2018). In this study, 21 F2 RNA-Seq samples were used for the statistical analysis, which was a similar sample size to the previous study. We used 30 614 genes whose average RPKM value was above 0.01 in the RNA-Seq analysis data obtained from the 21 F2 individuals. Gene expression values were log2 transformed after adding 0.01 to each RPKM value. Explanatory variables were determined using an L1-regularised linear regression model constructed using LASSO (Tibshirani 1996). To assess gene selection frequencies, we repeatedly evaluated explanatory variables using subsets of the transcriptome. Among the randomly selected 3061 genes (10% of the transcriptome) used as input variables for LASSO, eight presented non-zero coefficients in LASSO, and therefore, these were designated as explanatory variables. Subset selection and explanatory variable determination steps were repeated 10 000 times. Gene selection frequency was defined as the ratio of trials where the gene was eligible for use as an explanatory variable to the number of subsets that included that gene. The analysis was conducted using R version 3.33 software (R Core Team 2015) and glmnet package version 3.0–1 (Friedman et al. 2010).
Quantitative real-time PCR
Total RNA was extracted from seedling samples from the 95 F2 individuals, as described above. One microgram of each RNA sample was used to synthesise cDNA using the PrimeScript RT reagent Kit with a gDNA Eraser (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s instructions. The resulting cDNA was used for PCR amplification in a Thermal Cycler Dice Real Time System III with a TB Green Premix Ex TaqII (Takara Bio Inc.). One biological replicate and three technical replicates were conducted in relation to each F2 RNA sample. The primers used for quantitative real-time PCR are listed (see Table S2). The relative gene expression values for each target gene were calculated using the rice polyubiquitin gene (UBI1, The Rice Annotation Project Database (RAP-DB) (Ohyanagi et al. 2006) ID: Os06 g0681400) as a reference (Wang et al. 2000).
Statistical analyses
Significant differences in SPAD values were evaluated by analysis of variance (ANOVA) using R ver. 3.33 software (R Core Team 2015), with a significance level of P < 0.05. Pearson’s product-moment correlations between expression values and SPAD were also calculated using R software (R Core Team 2015), with a significance level of P < 0.05.
Results
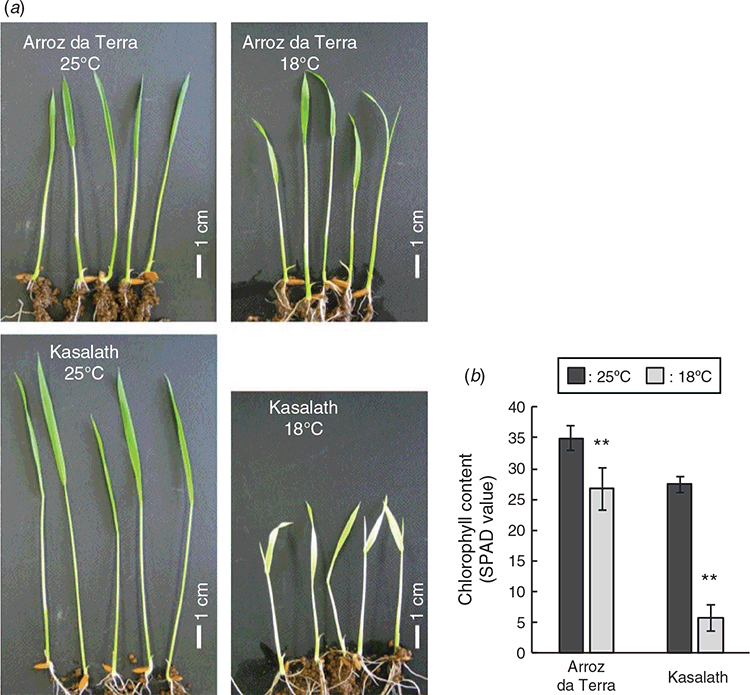
Chlorophyll content of second leaves grown at 25°C or 18°C
To investigate differences in low-temperature tolerance among cultivars, the seedlings of a japonica (cv. Arroz da Terra) and indica (cv. Kasalath) were grown at 25°C or 18°C. When grown at 18°C, leaves from Kasalath showed symptoms of chlorosis, although the leaves of Arroz da Terra remained green (Fig. 1). The mean SPAD value of second leaves from Arroz da Terra was 26.7 in the low-temperature treatment (18°C), which was 76% of the value (35.0) calculated for the same cultivar grown in the high-temperature treatment (25°C) (P < 0.001). However, the SPAD value of Kasalath leaves grown at 18°C was 5.8, which was 21% of the value (27.5) calculated for the same cultivar grown at 25°C (P < 0.001). Comparing the cultivars, the SPAD value calculated for Kasalath was 22% of the SPAD value for Arroz da Terra grown at 18°C (P < 0.001). SPAD values for Kasalath were lower than those for Arroz da Terra even at 25°C (P < 0.001), although this difference was small (79%).
Differentially expressed genes depending on temperature
RNA-Seq analysis was performed on seedling tissues from Arroz da Terra and Kasalath grown at 25°C or 18°C. Approximately 1.9–14.8 million reads per sample were obtained, and 83.7–87.6% of the clean reads were successfully mapped to the Oryza sativa-Nipponbare-Reference-IRGSP-1.0 genome (see Table S1).
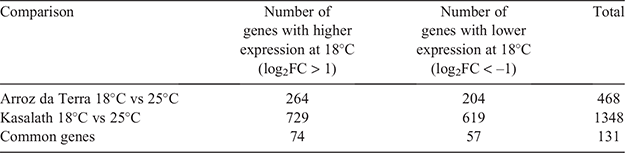
Differential expression analysis was performed on samples taken from both temperature treatments (Table 1). The total number of differentially expressed genes (DEGs) between the 25°C and 18°C treatments was 468 for Arroz da Terra, which was lower than the total number found for Kasalath (1348). Gene ontology (GO) enrichment analysis was conducted on genes with high or low expression in samples from the 18°C treatment. Significantly enriched GO categories for Arroz da Terra and Kasalath samples are listed (see Tables S3 and S4). In the analysis of genes with higher expressions in samples from the 18°C treatment than the 25°C treatment, it was found that 17 GO terms in Arroz da Terra and 40 GO terms in Kasalath were significantly enriched. In Arroz da Terra, the most significant GO terms for the higher expressed genes from the 18°C treatment for biological processes were ‘chitin metabolic process,’ ‘chitin catabolic process,’ ‘amyloglycan metabolic process’ and ‘amyloglycan catabolic process.’ Additionally, the most significant GO term for molecular function was ‘chitinase activity’ in Arroz da Terra. These GO terms related to chitin metabolism were also significantly enriched in genes with a higher expression in Kasalath samples from the 18°C treatment (see Table S4). Three chitinase genes (RAP-DB ID: Os04 g0493400, Os04 g0494100 and Os10 g0542900) belonging to these chitin metabolism-related GO terms were significantly higher in samples from the 18°C treatment than the 25°C treatment, in both Arroz da Terra and Kasalath (see Fig. S1).
|
However, the most significant GO terms representing biological processes were ‘response to stress’ and ‘response to stimulus’ in the highly expressed genes from Kasalath samples grown at 18°C compared with those grown at 25°C. These stress response-related GO terms were not significantly enriched in Arroz da Terra.
No GO terms were significantly enriched for genes with lower expressions at 18°C compared with 25°C in Arroz da Terra samples. However, in Kasalath samples, 43 GO terms were significantly enriched for the lower expressed genes at 18°C. The most significantly enriched GO term related to biological processes was ‘photosynthesis’ for the lower expressed genes in Kasalath samples grown at 18°C.
Differentially expressed genes between low temperature tolerant or sensitive cultivars
By comparing Arroz da Terra and Kasalath (Table 2), we found that the total number of DEGs was 2976 in the 18°C treatment samples, whereas it was 2104 in the 25°C treatment samples. Significantly enriched GO categories for DEGs between Arroz da Terra and Kasalath are listed in (see Table S5). At 25°C, two GO terms related to cellular components, ‘cytoplasmic part’ and ‘cytoplasm’ were significantly enriched in higher expressed genes found in Arroz da Terra. No significantly enriched GO terms were detected in the lower expressed genes found in Arroz da Terra samples from the 25°C treatment.

|
When grown at 18°C, no GO term was significantly enriched in genes that were more highly expressed in Arroz da Terra than Kasalath. However, 22 GO terms were significantly enriched in downregulated genes in Arroz da Terra. The most significantly enriched GO term related to biological processes was ‘response to stress’ for genes that were lowly expressed in Arroz da Terra compared with Kasalath.
Selection of transcripts related to chlorophyll content using F2 individuals grown at 18°C
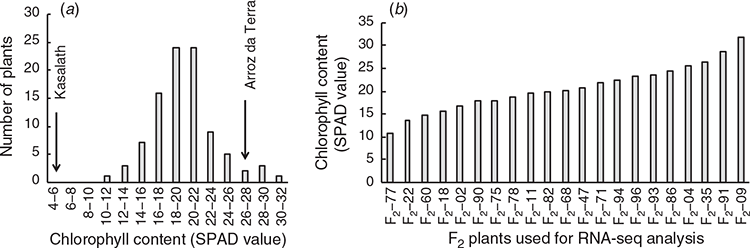
The chlorophyll content of 95 F2 individuals was measured after 21 days of growth at 18°C. The SPAD values of the second leaves of F2 plants ranged from 10.9 to 31.8 (Fig. 2a). To identify transcripts associated with chlorophyll content in plants grown at 18°C, 21 F2 individuals with varying SPAD values were analysed using RNA-Seq (Fig. 2b). As a result, 2.2–8.9 million reads per sample were obtained, and 84.8–87.3% of the clean reads were successfully mapped to the reference genome (see Table S1).
Transcripts whose expression levels were related to chlorophyll content were statistically selected using repeated regression analysis of random subsets of the transcriptome (Fukuda et al. 2018). The selection frequencies of genes for the regression model were determined, and 154 high frequency selected genes for each explanatory variable (frequencies >0.1) are listed (see Table S6). The expression levels of these 154 genes were significantly correlated with chlorophyll content in the 21 F2 plants (P < 0.05). Among them, the expression levels of 67 genes were significantly positively correlated with chlorophyll content. The expression levels of the other 87 genes were significantly negatively correlated with chlorophyll content. Among the 67 high frequency selected genes where expression levels were positively correlated with chlorophyll content, seven of these genes were classed as being more highly expressed in Arroz da Terra than in Kasalath under 18°C conditions. Of the 87 high frequency selected genes where expression levels negatively correlated with chlorophyll content, six were found to have lower expression levels in Arroz da Terra than in Kasalath under 18°C conditions.
Photosynthesis genes included in high frequency selected genes
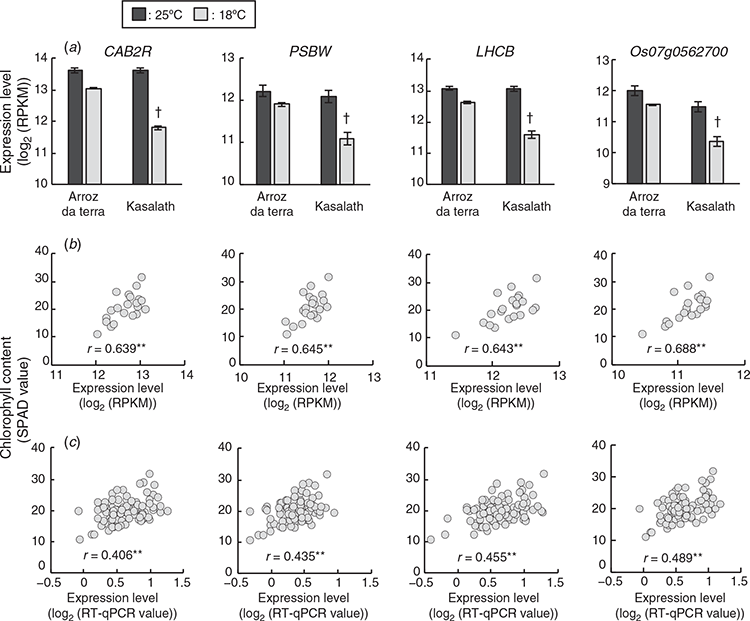
Among the 154 high frequency selected genes (see Table S6), four genes, CAB2R (RAP-DB ID: Os01 g0600900), PSBW (RAP-DB ID: Os01 g0773700), LHCB (RAP-DB ID: Os03 g0592500), and Os07 g0562700 were related to photosynthesis. These genes encoded chlorophyll a/b binding protein 2R, photosystem II reaction centre W protein, light-harvesting chlorophyll a/b-binding protein b2.1, and type III chlorophyll a/b-binding protein, respectively. The expression levels of four photosynthetic genes, CAB2R, PSBW, LHCB and Os07 g0562700 in the parental cultivars and the F2 plants are in Fig. 3. The expression levels of the above-listed photosynthetic genes were significantly positively correlated with the chlorophyll contents of leaves from the 21 F2 plants (Fig. 3b). Additionally, three genes, CAB2R, LHCB and Os07 g0562700 were more highly expressed in Arroz da Terra than in Kasalath under 18°C conditions (log2FC = 1.40 and FDR < 0.001, log2FC = 1.18 and FDR < 0.001, and log2FC = 1.35 and FDR < 0.001, respectively). PSBW was also expressed slightly higher in Arroz da Terra than in Kasalath at 18°C, although the log2 FC of expression values were lower than 1 (log2 FC = 0.97 and FDR < 0.001). CAB2R, PSBW, LHCB and Os07 g0562700 photosynthetic genes were not differentially expressed between Arroz da Terra and Kasalath at 25°C (log2FC = 0.03 and FDR = 1.00, log2FC = 0.18 and FDR = 0.24, log2FC = 0.07 and FDR = 0.88, and log2FC = 0.58 and FDR < 0.001, respectively). In Kasalath, these four genes showed significantly lower expression levels at 18°C than at 25°C (log2 FC = –2.10 and FDR < 0.001, log2 FC = –1.27 and FDR < 0.001, log2 FC = –1.72 and FDR < 0.001, and log2 FC = –1.37 and FDR < 0.001, respectively). These four photosynthetic genes were slightly lowly expressed at 18°C than at 25°C in Arroz da Terra, although the differences in expression levels were small (log2FC = –0.74 and FDR < 0.001, log2FC = –0.48 and FDR < 0.001, log2FC = –0.61 and FDR < 0.001, and log2FC = –0.61 and FDR < 0.001, respectively). To further evaluate the correlation between photosynthetic gene expression levels and chlorophyll content, all 95 F2 plants, including those that were not used for RNA-Seq, were analysed using quantitative real-time PCR. Results showed that the expression levels of the four above-listed photosynthetic genes were significantly positively correlated with chlorophyll content, even in the 95 F2 plants (Fig. 3c).
Stress response-related genes included in high frequency selected genes
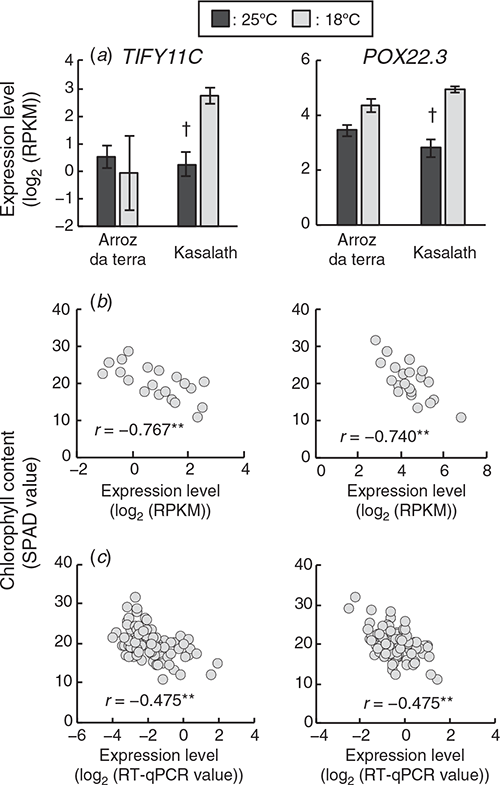
Two genes related to the stress response, TIFY11C (RAP-DB ID: Os03g0180900) (Ye et al. 2009; Hakata et al. 2017) and POX22.3 (RAP-DB ID: Os07g0677200) (Chittoor et al. 1997; Ning et al. 2010), were included in the 154 high frequency selected genes. The expression levels of TIFY11C and POX22.3 in the parental cultivars and the F2 plants are in Fig. 4. The expression levels of TIFY11 and POX22.3 were significantly negatively correlated with chlorophyll content in the 21 F2 plants (Fig. 4b). TIFY11C was one of the genes found to have lower expression levels in Arroz da Terra compared with Kasalath at 18°C (log2FC = –2.2 and FDR < 0.001). Expression levels of POX22.3 were not significantly different between Arroz da Terra and Kasalath at 18°C (log2FC = –0.39 and FDR = 0.059). TIFY11C and POX22.3 were not differentially expressed between the Arroz da Terra and Kasalath cultivars at 25°C (log2FC = 0.24 and FDR = 1.0 and log2FC = 0.66 and FDR = 0.097, respectively). The expression levels of TIFY11C in Arroz da Terra did not differ between the 18°C and 25°C treatments (log2FC = –0.22 and FDR = 1.0). The expression of POX22.3 was slightly higher at 18°C than at 25°C in Arroz da Terra (log2FC = 0.79 and FDR < 0.001). However, the expression levels of TIFY11C and POX22.3 were significantly higher in Kasalath plants at 18°C than at 25°C (log2FC = 2.3 and FDR < 0.001 and log2FC = 1.9 and FDR < 0.001, respectively). Furthermore, a correlation between the expression levels of stress response-related genes and chlorophyll content in 95 F2 plants was analysed using quantitative real-time PCR. The expression levels of TIFY11C and POX22.3 were significantly negatively correlated with chlorophyll content in these 95 F2 plants (this number includes plants that were not used for RNA-Seq) (Fig. 4c).
|
Discussion
The present study has clarified which transcripts are expressed in rice seedlings undergoing low-temperature stress (at 18°C). Our statistical approach, which used RNA-Seq data from 21 F2 plants indicated that four photosynthetic genes and two stress response genes were present in the high frequency selected genes related to chlorophyll content found in plants grown in 18°C conditions. Furthermore, the expression values of these six genes were significantly correlated with chlorophyll content, not only in plants used for RNA-Seq but in all 95 F2 plants. These results indicate that the statistical selection using 21 RNA-Seq samples was useful for predicting the effective genes related to chlorophyll contents within larger groups. The statistical approaches to select the expression biomarkers commonly require a large sample size, comprising more than hundreds of samples, to decrease ineffectiveness. In this study, we suggested that a small sample size, consisting of 21 F2 individuals, might be useful to select transcripts related to rice phenotypes.
Common genes enriched at 18°C in both Arroz da Terra and Kasalath
The GO term for genes that were more highly expressed at 18°C than at 25°C was related to chitinase activity, and it was significantly enriched in both Arroz da Terra and Kasalath. Several chitinase genes have been reported to be induced by infection with fungal pathogens or various abiotic stresses, suggesting that they are involved in the stress response (Takenaka et al. 2009). Additionally, several chitinases have been reported to display antifreeze activity in overwintering plants (Yeh et al. 2000). Chitinase may play a role in rice metabolism under low-temperature conditions in both tolerant and sensitive cultivars. However, even though three chitinase genes had significantly higher expressions in samples from the 18°C treatment than the 25°C treatment in both Arroz da Terra and Kasalath, their expression levels were not significantly correlated with chlorophyll content in 21 F2 plants at 18°C (see Fig. S1). Though chitinase might contribute to the metabolism at 18°C, it is considered that the expression levels of chitinase genes have small effect on the chlorophyll content at 18°C.
Photosynthetic genes related to chlorophyll content
Four photosynthetic genes, included in the high frequency selected genes, were significantly positively correlated with the chlorophyll contents of F2 plants grown at 18°C. Additionally, three out of the four photosynthetic genes were more highly expressed in the tolerant cultivar, Arroz da Terra, than in the sensitive cultivar, Kasalath, at 18°C. Furthermore, GO terms related to photosynthesis were significantly enriched for the lower expressed genes in Kasalath at 18°C than at 25°C. These results are consistent with previous studies that reported close correlations between SPAD and photosynthesis values in rice (Kumagai et al. 2009; Takai et al. 2010). This study also revealed that the genes related to photosynthesis are suppressed at the transcriptional level in plants that develop symptoms of chlorosis in low temperatures.
Stress response genes and chlorophyll content
Differential expression analysis between growth temperatures showed that GO terms related to the stress response were significantly enriched in genes with higher expression levels at 18°C than at 25°C, but this was only evident in Kasalath. The comparison between Arroz da Terra and Kasalath also revealed that GO terms related to the stress response were significantly enriched in genes with higher expression levels in Kasalath than in Arroz da Terra at 18°C. Statistical selection using F2 plants also revealed that the expression levels of the stress response genes, TIFY11C and POX22.3, were significantly negatively correlated with chlorophyll content at 18°C. TIFY11C proteins, as transcriptional regulators, are induced by jasmonic acid, which is related to the abiotic stress response (Ye et al. 2009; Hakata et al. 2017), while POX22.3 encodes peroxidase (Chittoor et al. 1997; Ning et al. 2010). These results suggest that low-temperature sensitive plants have higher expression levels of stress response genes than tolerant plants under long-term low temperature (18°C) stress. During a short-term (33–57 h) response to severe cold stress at 2°C, the opposite has been reported where more stress response pathways were enriched in a low-temperature tolerant rice cultivar than in a sensitive cultivar (Zhang et al. 2016). These studies indicate that the short-term response to severe cold stress and the long-term response to mild temperature stress are metabolically quite different.
During stress conditions, cellular homeostasis is disrupted and reactive oxygen species production is enhanced (Suzuki and Mittler 2006; Huang et al. 2012; You and Chan 2015). Several stress response genes are thought to scavenge reactive oxygen species, protecting cells from oxidative damage. It was reported that the expression levels of POX22.3 are downregulated in drought-sensitive mutant lines, suggesting that the regulation of reactive oxygen species is important for stress tolerance (Ning et al. 2010). However, reactive oxygen species are known to play a role in signal transduction in a plant’s stress response. During a short-term response to severe cold stress, accumulation of reactive oxygen species and upregulation of transcriptional factors mediating oxidative signals were observed in cold-tolerant rice cultivars (Zhang et al. 2016). During short-term responses to stress, a rapid increase in reactive oxygen species and transcription factors are important for controlling cell homeostasis (Suzuki and Mittler 2006; Huang et al. 2012; You and Chan 2015). In this study, a metabolic imbalance may have occurred in the cold-sensitive plants following long-term (mild) low-temperature stress, causing the accumulation of reactive oxygen species and induction of stress response genes. However, cold-tolerant plants are thought to maintain normal cell conditions following long-term (mild) low-temperature stress.
Furthermore, cold response transcription factors also function, in part, in the suppression of cold tolerance. One MYB transcriptional factor, OsMYB30, and a member of the TIFY family, OsJAZ9 (OsTIFY11A), can be induced by cold stress and enhance cold sensitivity (Lv et al. 2017). In our study, for the first time, the expression levels of TIFY11C were found to be negatively correlated with stress tolerance levels at 18°C. If TIFY11C has the effect of weakening the low-temperature tolerance of rice, it might be possible to increase the low-temperature tolerance of rice by suppressing its expression. Elucidating transcripts related to cold tolerance might lead to the development of strategies for enhancing cold tolerance. Further analysis of the functions of genes whose expression levels are related to tolerance is necessary to elucidate the mechanisms underlying the complex networks involved in the stress response.
Conclusions
We utilised differential expression analyses between the parental cultivars Arroz da Terra and Kasalath, and growth temperatures and incorporated these into a statistical approach using the RNA-Seq data of 21 F2 plants to select transcripts related to chlorophyll content under low-temperature conditions. Our analysis revealed that several photosynthetic genes were repressed at the transcriptional level in low-temperature sensitive plants at 18°C. Furthermore, low-temperature sensitive plants had higher expression levels of stress response genes, suggesting the presence of a metabolic imbalance at 18°C. Our findings also suggest that our statistical approach, which utilised small groups of F2 plants, was effective in isolating genes related to chlorophyll content at 18°C in larger groups. It suggested that the relatively small number of RNA-Seq samples might help select the transcripts related to rice phenotypes. These results indicate that RNA-Seq is a useful tool for clarifying metabolic mechanisms at low temperatures. In this study, the expression levels of two stress-responsive genes, TIFY11C and POX22.3, were found to be negatively correlated with stress-tolerance levels at 18°C for the first time. It indicates that plants with high expression levels of stress-responsive genes are not necessarily stress-tolerant. Due to low-temperature tolerance being an important trait for rice breeding, this study provides important information for strategies to improve stress tolerance.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This work was supported by the JSPS KAKENHI (grant number 16K07581) to Akari Fukuda, and the JSPS KAKENHI (grant number 19H02944) to Tatsuro Hirose, Akari Fukuda and Naohiro Aoki.
Acknowledgements
We are grateful to Ms. Setsuko Hayashi and Ms. Kyoko Mogami for their excellent technical assistance.
References
Ando T, Yamamoto T, Shimizu T, Ma XF, Shomura A, Takeuchi Y, Lin SY, Yano M (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theoretical and Applied Genetics 116, 881–890.| Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice.Crossref | GoogleScholarGoogle Scholar | 18274726PubMed |
Bai LWD, Liu J, Dai LF, Deng QW, Chen YL, Xie JK, Luo XD (2021) Identification and characterisation of cold stress-related proteins in Oryza rufipogon at the seedling stage using label-free quantitative proteomic analysis. Functional Plant Biology 48, 542–555.
| Identification and characterisation of cold stress-related proteins in Oryza rufipogon at the seedling stage using label-free quantitative proteomic analysis.Crossref | GoogleScholarGoogle Scholar |
Buti M, Pasquariello M, Ronga D, Milc JA, Pecchioni N, Ho VT, Pucciariello C, Perata P, Francia E (2018) Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp Japonica seedlings. Functional & Integrative Genomics 18, 627–644.
| Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp Japonica seedlings.Crossref | GoogleScholarGoogle Scholar |
Chittoor JM, Leach JE, White FF (1997) Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Molecular Plant-Microbe Interactions 10, 861–871.
| Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae.Crossref | GoogleScholarGoogle Scholar | 9304860PubMed |
da Maia LC, Cadore PRB, Benitez LC, Danielowski R, Braga EJB, Fagundes PRR, Magalhaes AM, de Oliveira AC (2017) Transcriptome profiling of rice seedlings under cold stress. Functional Plant Biology 44, 419–429.
| Transcriptome profiling of rice seedlings under cold stress.Crossref | GoogleScholarGoogle Scholar | 32480575PubMed |
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70.
| agriGO: a GO analysis toolkit for the agricultural community.Crossref | GoogleScholarGoogle Scholar | 20435677PubMed |
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software 33, 1–22.
| Regularization paths for generalized linear models via coordinate descent.Crossref | GoogleScholarGoogle Scholar | 20808728PubMed |
Fukuda A, Terao T (2015) QTLs for shoot length and chlorophyll content of rice seedlings grown under low-temperature conditions, using a cross between Indica and Japonica cultivars. Plant Production Science 18, 128–136.
| QTLs for shoot length and chlorophyll content of rice seedlings grown under low-temperature conditions, using a cross between Indica and Japonica cultivars.Crossref | GoogleScholarGoogle Scholar |
Fukuda A, Hirose T, Aoki N, Kondo S, Yonekura M, Kataoka T, Ohto C, Nagano AJ (2018) Selection of transcripts affecting initial growth rate of rice backcrossed inbred lines using RNA sequencing data. Frontiers in Plant Science 9, 1880
| Selection of transcripts affecting initial growth rate of rice backcrossed inbred lines using RNA sequencing data.Crossref | GoogleScholarGoogle Scholar | 30631334PubMed |
Hakata M, Muramatsu M, Nakamura H, Hara N, Kishimoto M, Iida-Okada K, Kajikawa M, Imai-Toki N, Toki S, Nagamura Y, Yamakawa H, Ichikawa H (2017) Overexpression of TIFY genes promotes plant growth in rice through jasmonate signaling. Bioscience, Biotechnology, and Biochemistry 81, 906–913.
| Overexpression of TIFY genes promotes plant growth in rice through jasmonate signaling.Crossref | GoogleScholarGoogle Scholar | 28079456PubMed |
Han B, Ma XD, Cui D, Wang YJ, Geng LY, Cao GL, Zhang H, Han LZ (2020) Comprehensive evaluation and analysis of the mechanism of cold tolerance based on the transcriptome of weedy rice seedlings. Rice (New York, N.Y.) 13, 12
| Comprehensive evaluation and analysis of the mechanism of cold tolerance based on the transcriptome of weedy rice seedlings.Crossref | GoogleScholarGoogle Scholar |
Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012) Signal transduction during cold, salt, and drought stresses in plants. Molecular Biology Reports 39, 969–987.
| Signal transduction during cold, salt, and drought stresses in plants.Crossref | GoogleScholarGoogle Scholar | 21573796PubMed |
Huner NPA, Oquist G, Hurry VM, Krol M, Falk S, Griffith M (1993) Photosynthesis, photoinhibition and low-temperature acclimation in cold tolerant plants. Photosynthesis Research 37, 19–39.
| Photosynthesis, photoinhibition and low-temperature acclimation in cold tolerant plants.Crossref | GoogleScholarGoogle Scholar |
Kamitani M, Kashima M, Tezuka A, Nagano AJ (2019) Lasy-Seq: a high-throughput library preparation method for RNA-Seq and its application in the analysis of plant responses to fluctuating temperatures. Scientific Reports 9, 7091
| Lasy-Seq: a high-throughput library preparation method for RNA-Seq and its application in the analysis of plant responses to fluctuating temperatures.Crossref | GoogleScholarGoogle Scholar | 31068632PubMed |
Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM (2016) Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Research 44, D574–D580.
| Ensembl Genomes 2016: more genomes, more complexity.Crossref | GoogleScholarGoogle Scholar | 26578574PubMed |
Kodama Y, Shumway M, Leinonen R (2012) International Nucleotide Sequence Database Collaboration. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Research 40, D54–D56.
| International Nucleotide Sequence Database Collaboration. The sequence read archive: explosive growth of sequencing data.Crossref | GoogleScholarGoogle Scholar | 22009675PubMed |
Kudapa H, Ramalingam A, Nayakoti S, Chen XP, Zhuang WJ, Liang XQ, Kahl G, Edwards D, Varshney RK (2013) Functional genomics to study stress responses in crop legumes: progress and prospects. Functional Plant Biology 40, 1221–1233.
| Functional genomics to study stress responses in crop legumes: progress and prospects.Crossref | GoogleScholarGoogle Scholar | 32481190PubMed |
Kumagai E, Araki T, Kubota F (2009) Correlation of chlorophyll meter readings with gas exchange and chlorophyll fluorescence in flag leaves of rice (Oryza sativa L.) Plants. Plant Production Science 12, 50–53.
| Correlation of chlorophyll meter readings with gas exchange and chlorophyll fluorescence in flag leaves of rice (Oryza sativa L.) Plants.Crossref | GoogleScholarGoogle Scholar |
Li M, Sui N, Lin L, Yang Z, Zhang YH (2019) Transcriptomic profiling revealed genes involved in response to cold stress in maize. Functional Plant Biology 46, 830–844.
| Transcriptomic profiling revealed genes involved in response to cold stress in maize.Crossref | GoogleScholarGoogle Scholar | 31217070PubMed |
Lv Y, Yang M, Hu D, Yang ZY, Ma SQ, Li XH, Xiong LZ (2017) The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiology 173, 1475–1491.
| The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression.Crossref | GoogleScholarGoogle Scholar | 28062835PubMed |
Mackill DJ, Lei XM (1997) Genetic variation for traits related to temperate adaptation of rice cultivars. Crop Science 37, 1340–1346.
| Genetic variation for traits related to temperate adaptation of rice cultivars.Crossref | GoogleScholarGoogle Scholar |
Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, Nagamura Y, Izawa T (2012) Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151, 1358–1369.
| Deciphering and prediction of transcriptome dynamics under fluctuating field conditions.Crossref | GoogleScholarGoogle Scholar | 23217716PubMed |
Nagata K, Fukuta Y, Shimizu H, Yagi T, Terao T (2002) Quantitative trait loci for sink size and ripening traits in rice (Oryza sativa L.). Breeding Science 52, 259–273.
| Quantitative trait loci for sink size and ripening traits in rice (Oryza sativa L.).Crossref | GoogleScholarGoogle Scholar |
Ning J, Li XH, Hicks LM, Xiong LZ (2010) A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology 152, 876–890.
| A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice.Crossref | GoogleScholarGoogle Scholar | 20007444PubMed |
Ohsumi A, Furuhata M, Matsumura O (2012) Varietal differences in biomass production of rice early after transplanting at low temperatures. Plant Production Science 15, 32–39.
| Varietal differences in biomass production of rice early after transplanting at low temperatures.Crossref | GoogleScholarGoogle Scholar |
Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, Ikeo K, Itoh T, Gojobori T, Sasaki T (2006) The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Research 34, D741–D744.
| The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information.Crossref | GoogleScholarGoogle Scholar | 16381971PubMed |
Onaga G, Wydra KD, Koopmann B, Sere Y, von Tiedemann A (2017) Elevated temperature increases in planta expression levels of virulence related genes in Magnaporthe oryzae and compromises resistance in Oryza sativa cv. Nipponbare. Functional Plant Biology 44, 358–371.
| Elevated temperature increases in planta expression levels of virulence related genes in Magnaporthe oryzae and compromises resistance in Oryza sativa cv. Nipponbare.Crossref | GoogleScholarGoogle Scholar | 32480570PubMed |
Pradhan SK, Pandit E, Nayak DK, Behera L, Mohapatra T (2019) Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biology 19, 352
| Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis.Crossref | GoogleScholarGoogle Scholar | 31412781PubMed |
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; Available from: https://www.R-project.org/.
Shen CX, Li D, He RH, Fang Z, Xia YM, Gao J, Shen H, Cao ML (2014) Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. Journal of Plant Biology 57, 337–348.
| Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress.Crossref | GoogleScholarGoogle Scholar |
Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiologia Plantarum 126, 45–51.
| Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction.Crossref | GoogleScholarGoogle Scholar |
Takai T, Kondo M, Yano M, Yamamoto T (2010) A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice (New York, N.Y.) 3, 172–180.
| A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice.Crossref | GoogleScholarGoogle Scholar |
Takenaka Y, Nakano S, Tamoi M, Sakuda S, Fukamizo T (2009) Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: chitinase inhibitor allosamidin enhances stress tolerance. Bioscience, Biotechnology, and Biochemistry 73, 1066–1071.
| Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: chitinase inhibitor allosamidin enhances stress tolerance.Crossref | GoogleScholarGoogle Scholar | 19420714PubMed |
Terao T, Nagata K, Morino K, Hirose T (2010) A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theoretical and Applied Genetics 120, 875–893.
| A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice.Crossref | GoogleScholarGoogle Scholar | 20151298PubMed |
Tibshirani R (1996) Regression shrinkage and selection via the Lasso. Journal of the Royal Statistical Society. Series B. Methodological 58, 267–288.
| Regression shrinkage and selection via the Lasso.Crossref | GoogleScholarGoogle Scholar |
Wang J, Jiang J, Oard JH (2000) Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.). Plant Science 156, 201–211.
| Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.).Crossref | GoogleScholarGoogle Scholar | 10936527PubMed |
Yamori W, Sakata N, Suzuki Y, Shikanai T, Makino A (2011) Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. The Plant Journal 68, 966–976.
| Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice.Crossref | GoogleScholarGoogle Scholar | 21848656PubMed |
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Molecular Biology 71, 291–305.
| Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice.Crossref | GoogleScholarGoogle Scholar | 19618278PubMed |
Yeh S, Moffatt BA, Griffith M, Xiong F, Yang DSC, Wiseman SB, Sarhan F, Danyluk J, Xue YQ, Hew CL, Doherty-Kirby A, Lajoie G (2000) Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiology 124, 1251–1264.
| Chitinase genes responsive to cold encode antifreeze proteins in winter cereals.Crossref | GoogleScholarGoogle Scholar | 11080301PubMed |
Yogendra KN, Kushalappa AC (2016) Integrated transcriptomics and metabolomics reveal induction of hierarchies of resistance genes in potato against late blight. Functional Plant Biology 43, 766–782.
| Integrated transcriptomics and metabolomics reveal induction of hierarchies of resistance genes in potato against late blight.Crossref | GoogleScholarGoogle Scholar | 32480502PubMed |
Yoshida R, Kanno A, Sato T, Kameya T (1996) Cool-temperature-induced chlorosis in rice plants. 1. Relationship between the induction and a disturbance of etioplast development. Plant Physiology 110, 997–1005.
| Cool-temperature-induced chlorosis in rice plants. 1. Relationship between the induction and a disturbance of etioplast development.Crossref | GoogleScholarGoogle Scholar | 8819872PubMed |
You J, Chan ZL (2015) ROS regulation during abiotic stress responses in crop plants. Frontiers in Plant Science 6, 1092
| ROS regulation during abiotic stress responses in crop plants.Crossref | GoogleScholarGoogle Scholar | 26697045PubMed |
Zhang SH, Zheng JS, Liu B, Peng SB, Leung H, Zhao JL, Wang XF, Yang TF, Huang ZH (2014) Identification of QTLs for cold tolerance at seedling stage in rice (Oryza sativa L.) using two distinct methods of cold treatment. Euphytica 195, 95–104.
| Identification of QTLs for cold tolerance at seedling stage in rice (Oryza sativa L.) using two distinct methods of cold treatment.Crossref | GoogleScholarGoogle Scholar |
Zhang J, Luo W, Zhao Y, Xu Y, Song S, Chong K (2016) Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytologist 211, 1295–1310.
| Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice.Crossref | GoogleScholarGoogle Scholar |