Post-translational regulation of the membrane transporters contributing to salt tolerance in plants
Amber Gupta A B * , Birendra Prasad Shaw
A B * , Birendra Prasad Shaw  A B * and Binod Bihari Sahu C
A B * and Binod Bihari Sahu C
A Abiotic Stress and Agro-Biotechnology Laboratory, Institute of Life Sciences, Nalco Square, Bhubaneswar, Odisha, 751023, India.
B Regional Centre for Biotechnology, Faridabad, Haryana, 121001, India.
C Department of Life Science, NIT Rourkela, Rourkela, Odisha, 769008, India.
Functional Plant Biology 48(12) 1199-1212 https://doi.org/10.1071/FP21153
Submitted: 13 May 2021 Accepted: 7 August 2021 Published: 20 October 2021
© 2021 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
This review article summarises the role of membrane transporters and their regulatory kinases in minimising the toxicity of Na+ in the plant under salt stress. The salt-tolerant plants keep their cytosolic level of Na+ up to 10–50 mM. The first line of action in this context is the generation of proton motive force by the plasma membrane H+-ATPase. The generated proton motive force repolarises the membrane that gets depolarised due to passive uptake of Na+ under salt stress. The proton motive force generated also drives the plasma membrane Na+/H+ antiporter, SOS1 that effluxes the cytosolic Na+ back into the environment. At the intracellular level, Na+ is sequestered by the vacuole. Vacuolar Na+ uptake is mediated by Na+/H+ antiporter, NHX, driven by the electrochemical gradient for H+, generated by tonoplast H+ pumps, both H+ATPase and PPase. However, it is the expression of the regulatory kinases that make these transporters active through post-translational modification enabling them to effectively manage the cytosolic level of Na+, which is essential for tolerance to salinity in plants. Yet our knowledge of the expression and functioning of the regulatory kinases in plant species differing in tolerance to salinity is scant. Bioinformatics-based identification of the kinases like OsCIPK24 in crop plants, which are mostly salt-sensitive, may enable biotechnological intervention in making the crop cultivar more salt-tolerant, and effectively increasing its annual yield.
Keywords: Na+/H+ antiporter, NHX-Na+/H+ ion exchanger, OsCIPK24, phosphorylation, plasma membrane H+-ATPase, salinity, salt overly sensitive 1, vacuolar H+-ATPase.
Introduction
Salinity is a major abiotic stresses that greatly affects plant growth resulting in loss in crop production. The concern for agriculture is not just the natural occurrence of saline soils, but also the increasing salinisation of the agricultural lands threatening the crop production in future. The agricultural lands in the coastal zone in addition are also affected by intrusion of sea water during cyclones and carrying of the ocean aerosols far inland. According to one estimate, more than 80 million ha of agricultural land, representing 40% of total irrigated land over the world, are affected by salinisation (Xiong and Zhu 2001). Further, more than 50% of all arable lands are likely to be affected soil salinisation by 2050 due to excessive ground water irrigation and climate change resulting in sea water intrusion in the coastal zones because of the rise in sea level (Wang et al. 2003; Smajgl et al. 2015).
The subject has been reviewed extensively (Munns and Tester 2008; Shahid et al. 2018; van Zelm et al. 2020; Chen et al. 2021) and it is now quite clear that salt tolerance is a quantitative trait with a complex phenomenon working at several levels of the cellular metabolic events. Nonetheless, salt tolerance can be broadly categorised as the ability of plants to reduce the effects of the osmotic and ionic stresses of a saline soil (Munns and Tester 2008). Exposure of the plants to high external salinity leads to change in the electrochemical gradient across the plasma membrane favouring passive transport of Na+ from the external environment into the root cytoplasm through ion transporters and channels, including the non-selective cation channels (NSCCs), high-affinity potassium transporters (HKTs) and H+/K+ symporters (HAKs). Any build-up of Na+ in the cytoplasm influences cellular metabolism by inhibiting enzyme activities, although the exact mechanism of the toxicity remains uncertain (Møller and Tester 2007; Munns and Tester 2008). Salt tolerance is thus avoidance of dehydration stress in the initial response to an increase in soil salinity, and avoidance of ion toxicity over time. However, it is difficult to draw a clear boundary between the two phases of the stress, particularly when the plant is exposed to high salinity (Munns and Tester 2008). Rather, a significant overlap between the response to the two stresses, including the early and downstream signalling has been suggested (van Zelm et al. 2020).
Even after decades of research on understanding the mechanism of salt tolerance, the scientific community has rarely been able to apply these efforts to bring salt affected agricultural lands under effective cultivation. The mechanism of salt tolerance has been reviewed many times covering various aspects. These include reviews mainly providing information on Na+ and K+ transport, ion relation and tissue tolerance (Møller and Tester 2007; Munns and Tester 2008; Roy et al. 2014; Munns et al. 2016; Almeida et al. 2017; van Zelm et al. 2020; Zhao et al. 2020; Chen et al. 2021) and osmolyte accumulation and osmotic adjustment (Roy et al. 2014; Munns et al. 2020; Zhao et al. 2020; Chen et al. 2021) along with molecular understanding of the process. Several reviews also provide information on the genes involved in salt stress sensing, response and signalling and their transcription regulation (Osakabe et al. 2013; Hanin et al. 2016; Almeida et al. 2017; Ismail and Horie 2017; van Zelm et al. 2020; Zhao et al. 2020; Chen et al. 2021; Ponce et al. 2021). Studies of functional analysis of genes involved in salt tolerance and molecular breeding and genetic manipulation of plants for salt tolerance have also been reviewed (Møller and Tester 2007; Roy et al. 2014; Hanin et al. 2016; Ismail and Horie 2017; van Zelm et al. 2020; Chen et al. 2021).
However, the importance of the work on post-translational modification of proteins post-signal perception and their role in salt tolerance has relatively remained unclear. Hence, the aim of this review is to bring to the notice of the readers the diversity and the importance of protein phosphorylation in salt tolerance in plants focusing only on the proteins involved in Na+ uptake, transportation and sequestration.
Na+ uptake and toxicity
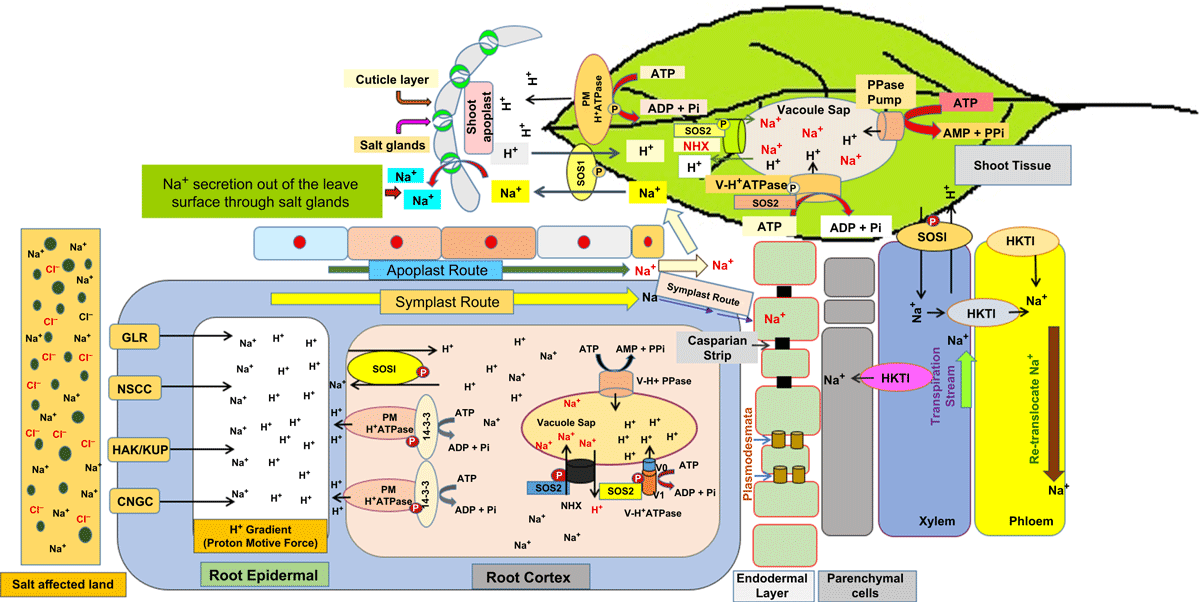
NSCCs, HKTs and HAKs are the primary routes of entry of Na+ inside the plant cells. The Na+ taken up passively finds its way to the xylem following both symplastic pathway using the plasmodesmata route and apoplastic pathway using the route through the cell wall and the intracellular spaces (Plett and Moller 2010; Adams and Shin 2014) (Fig. 1). The symplastic pathway is primarily used to cross the hurdle of the Casparian strips and suberin layers on the endodermis, but in certain plants, such as rice (Oryza sativa L.), the apoplastic flow may contribute to as much as 50% of the total Na+ uptake (Yeo et al. 1987; Ochiai and Matoh 2002). Endodermal tissue adjacent to stele is an important regulatory site for apoplast and transcellular transport because of the deposition of suberin and lignin.
Once inside the xylem, Na+ moves to different parts of shoots along with the water moving to the shoots as a result of the transpirational pull. While plants systematically work towards minimising the entry of Na+ into the xylem through the symplast, they have also developed mechanisms to retrieve back the Na+ ions out of the xylem restricting their movement into the sensitive aerial tissues and simultaneously facilitating their sequestration and accumulation primarily in the root tissues (Tester and Davenport 2003; Munns and Tester 2008; Almeida et al. 2017). A low level of accumulation of Na+ in shoots and its sequestration in roots are facilitated in Arabidopsis thaliana L. by AtHKT1 that unloads the ion from the xylem in root for preventing the movement of the ion to the shoot tissues (Almeida et al. 2017). The preferential expression of the plasma membrane-bound OsHKT1;5/OsHKT8 (locus SKC1) in the parenchyma cells surrounding the xylem vessels in rice (Ren et al. 2005; Kobayashi et al. 2017) eloquently speaks of its role in Na+ partitioning between roots and shoot (Almedia et al. 2017), although a recent study (Alnayef et al. 2020) has questioned a direct role of the transporter in removal of the ion, and suggested its expression to be linked to feedback regulation of other ion transporters for removal of Na+ from the xylem. Alnayef et al. (2020) also observed that the changes in the expression of OsHKT1;5 leads to alter the activity of the transporters that are involved in acquisition and homeostasis of K+ and Ca2+. Besides prevention of the salt toxicity through cellular sequestration of Na+ and its efflux from the root cells, examples of plants do exist where the protection against the ion occurs either by the exclusion of salt through the leaves, as in the recretohalphytes, Limonium bicolor(Bunge) Kuntze (Deng et al. 2015; Yuan et al. 2015), or its temporary storage in the bladder before getting excreted out upon encountering strong wing, as in Chenopodium quinoa Willd. (Kiani-Pouya et al. 2017; Bohm et al. 2018) (Fig. 1). In C. quinoa genotypes contrasting for salt tolerance, it has also been specifically demonstrated that under saline conditions, the salt-tolerant Q16 is able to keep the slow (SV) and fast (FV) tonoplast channel activity much lower compared with the salt sensitive Q5206 in order to prevent the leakage of Na+ accumulated in the vacuole (Bonales-Alatorre et al. 2013b). However, by and large, irrespective of the mechanisms involved, the ultimate objective in the prevention of salt toxicity in plants is the prevention of tissue accumulation of Na+ in the aerial part.
The key factors in salt tolerance
Research over the years have revealed that principal component of salinity stress is protection against Na+ toxicity (Pardo et al. 2006). A minimal accumulation of Na+ in the cytoplasm is the primary requirement by the plant for tolerance to salinity, as Na+ entering into the cells interferes with essential cytosolic metabolic enzymes by replacing K+ that is required for their functional activity as a cofactor (Duggleby and Dennis 1973). Thus the disturbances in K+ homeostasis due to its replacement by Na+ creates essential metabolic process impairment in the root and shoot tissue (Marschner 1995; PPI – Potash and Phosphate Institute 1998). Firstly, a low level of cellular accumulation of Na+ in plants challenged by salt is achieved by minimisation of entry of the ion into the cytoplasm via the ion channels and transporters and exclusion or efflux of the cytoplasmic Na+ into the external milieu. Secondly, excessive cytoplasmic Na+ is sequestered into the vacuole through an active ion transport mechanism operating at the tonoplast, a mechanism also referred to as tissue tolerance (Munns and Tester 2008). In root cells, this would minimise its transport to the younger tissue. Nevertheless, tissue tolerance to Na+ is also observed in leaves where they accumulate the ion to a high level and excrete it into the environment (Bonales-Alatorre et al. 2013a; Deng et al. 2015; Yuan et al. 2015). Another important factor in salt tolerance is the requirement of maintenance of optimum negative cytosolic osmotic potential to facilitate water uptake and prevent dehydration stress, and this is achieved by the biosynthesis and accumulation of compatible osmolytes in the cytoplasm (Peleg et al. 2011). The requirements under these categories for tolerance to salinity may vary from species to species, but these are operated simultaneously in the plant for survival under saline environment (Munns and Tester 2008; Roy et al. 2014; Pires et al. 2015). The cellular sequestration mechanism of the ions works in the aerial tissue as well (Fig. 1).
Essential membrane transporters operating in salt tolerance
Plants have developed a variety of mechanisms to keep the level of Na+ to minimal in the cytoplasm. The first ever proof of the genetic control of salt tolerance came from the forward genetic approach in which mutants of Arabidopsis generated by ethyle methane sulfonate and were tested for their salt hypersensitivity (Wu et al. 1996). Several mutants allelic to each other, named salt overly sensitive 1 (sos1), were identified and the SOS1 gene was mapped to chromosome 2 (Wu et al. 1996). Later on, the map-based cloning revealed that SOS1 encodes a plasma membrane-bound antiporter protein, Na+/H+ (Shi et al. 2000, 2002). Thus, the first level of protection against salt stress in the plant is at the level of the root by SOS1, which effluxes the Na+ entering into the root, particularly at the root tip where its expression has been observed in the epidermal cells (Shi et al. 2002; Shabala et al. 2005; Pardo et al. 2006). The role of SOS1 in the net exclusion of Na+ and its importance in salt tolerance is reflected from enhanced efflux of the ion in the salt-tolerant poplar species compared with the salt-sensitive ones when plants were exposed to NaCl (Sun et al. 2009). The importance of SOS1 in salt tolerance is realised from the fact that Arabidopsis overexpressing SOS1 shows increased salt tolerance (Shi et al. 2003). In addition, support for the role of SOS1 salt tolerance also comes from the down regulation of its expression in Thelluuingiella halophila (TNGHA – https://gd.eppo.int/taxon/TNGHA) that converted the halophyte to a salt-sensitive plant (Oh et al. 2007). Furthermore, grapevine (Vitis vinifera L.) overexpressing SOS1 was also found to maintain a lower cellular Na+ content and showed higher tolerance to salinity compared with the wild-type (Upadhyay et al. 2012), clearly indicating the cellular exclusion of the ion to be an important mechanism of salt tolerance in plants. In addition to expression at the root tips, SOS1 also expresses in the inner tissues roots, primarily in parenchyma and pericycle cells surrounding the vasculature, suggesting its role in the regulation of long-distance transport of Na+ in plants from root to shoot as well via xylem (Shi et al. 2002; Pardo et al. 2006). In stem and petiole, the expression of SOS1 is primarily confined to the parenchyma cells neighbouring the xylem vessel. It has been suggested that SOS1 plays a dual role in Na+ transport where it serves to load the ion into the xylem at low or moderate salt stress and it retrieves the ion from the xylem in root at severe salt stress (Shi et al. 2002). The purpose of such dual role of SOS1 is to facilitate the accumulation of Na+ in the leave tissues at low salt stress and to restrict movement of Na+ to shoot through xylem vessel to prevent its rapid accumulation in the leaves and other aerial tissues at high salt stress (Shi et al. 2002; Pardo et al. 2006). The role of SOS1 in the long-distance transport of Na+ also comes from the transgenic experiments in which the tobacco (Nicotiana tabacum L.) plant overexpressing SbSOS1 showed enhanced loading of Na+ into the xylem besides showing an increase in tolerance to salinity (Yadav et al. 2012). In addition to the role of SOS1 in the transport of Na+ across the cell membrane, its role has also been indicated as a possible sensor of the cytoplasmic level of the ion through it’s the long cytoplasmic tail, although the mechanism is not yet clear (Shi et al. 2000).
Vacuolar Na+/H+ antiporter
The second level of protection against salt stress is through sequestration of Na+ in the cellular vacuole mediated by another Na+/H+ antiporter, NHX, located in the tonoplast, and is a critical feature of salt tolerance in plants. The concept of the existence of Na+/H+ antiporter in the tonoplast was developed more than three decades ago by the observation of Na+-dependent change in pH of the tonoplast vesicles isolated from the storage tissue of Beta vulgaris L. (Blumwald and Poole 1985). However, the identity of the vascular Na+/H+ could only be established when the data bank of ESTs and the data on the whole-genome sequencing of Arabidopsis were available. AtNHX1 was the first cloned tonoplast Na+/H+ exchanger that was reported (Gaxiola et al. 1999), and currently at least six such exchangers have been annotated from the Arabidopsis genome sequence. Sequestration of Na+ in the vacuole ensures that its concentration in the cytoplasm lies in the range of 10–30 mM (Munns and Tester 2008), as enzymes are sensitive to Na+ and the activity of most of the enzymes starts getting inhibited at 100 mM NaCl, or even lesser concentration (Greenway and Osmond 1972; Flowers and Dalmond 1992). It has been observed that the expression of NHX1 and the capacity of Na+ sequestration in the vacuoles is greater in the salt-tolerant cultivars of wheat (Triticum aestivum L.) compared with the salt-sensitive ones (Cuin et al. 2011), eloquently speaking for the important role of the transporter in salt tolerance in plants mediated by sequestration of Na+ in the vacuole. In addition, overexpression of AtNHX1 in Arabidopsis thaliana and other plants has also been found to increase salt tolerance in the plant with increased Na+ compartmentation in the vacuole (Apse et al. 1999; Zhang and Shi 2013). Several exhaustive reviews on research on NHX supporting its role in salt tolerance in plants can be referred (Zhang and Shi 2013; Almeida et al. 2017; Assaha et al. 2017) that establish NHX as one of the most important effectors imparting salt resistance features to plants.
High affinity K+ transporter (HKTs)
The movement of Na+ inside the plant after the entry of the ion through the root is also controlled and regulated by a high-affinity potassium transporter (HKT) group of transporters, which primarily have two functions. In contrast to the current understanding of the role of HKT in the maintenance of cellular ion homeostasis, the first function of HKT in plant was considered earlier to be linked to transport of K+, as the name suggests (Schachtman and Schroeder 1994). It was also observed that the uptake of K+ by the wheat root, HKT1 was activated by micromolar concentration of Na+ and that at a high external concentration of Na+ the uptake of K+ was blocked and low-affinity Na+ uptake started, indicating an important role of HKT in Na+ uptake and toxicity in plants (Rubio et al. 1995). HKTs are active at the plasma membrane and are permeable only to Na+ or both Na+ and K+ (Schachtman and Schroeder 1994; Rodríguez-Navarro and Rubio 2006). The number of members in HKTs family varies with plant species. While Arabidopsis has a single member, AtHKT1;1, permeable only to Na+ (Uozumi et al. 2000), rice comprises seven to nine members depending on the cultivar (Garciadeblás et al. 2003) of which OsHKT4 (OsHKT1;1), OsHKT6 (OsHKT1;3) and OsHKT8 (OsHKT1;5) are permeable only to Na+ and OsHKT1 (OsHKT2;1) functions as Na+–K+ symport (Garciadeblás et al. 2003; Jabnoune et al. 2009). The functional role of a few of the OsHKTs has been elucidated, like OsHKT2;1 mediates influx of Na+ into roots under the condition of K+ starvation acting as Na+–K+ symporter (Horie et al. 2007), OsHKT1 (OsHKT2;1) and OsHKT4 (OsHKT1;1) are high affinity and low-affinity Na+ transporters, respectively, that facilitates uptake of the ion by root (Garciadeblas et al. 2003), OsHKT4 (OsHKT1;1) may also function as xylem transporter removing Na+ from xylem sap in roots and leaf sheaths (Garciadeblás et al. 2003; Suzuki et al. 2016; Kobayashi et al. 2017), OsHKT8/SKC1 (OsHKT1;5) functions in retrieval of Na+ from xylem apoplastic space by the xylem parenchyma in root and leaf sheath (Kobayashi et al. 2017) and Na+ exclusion in phloem parenchyma cells at the basal nodes (Kobayashi et al. 2017), and OsHKT2;2 (OsHKT2) drives Na+-dependent K+ uptake in root (Horie et al. 2007; Brini and Masmoudi 2012). The functional significance of other HKTs has not been elucidated so far.
Unlike HKTs, Na+/H+ antiporter, whether SOS1 or NHX, is functionally driven by the development of trans-membrane H+ gradient that is built up by the cells in the direction opposite to the movement of Na+; i.e. the direction of movement of Na+ is dependent on the difference in the concentrations of free Na+ and H+ across the membrane and the transport of Na+ across the membrane occurs towards lower the acidic environment (Martinoia et al. 2007; Munns and Tester 2008). The H+ gradient across the plasma membrane is, however, achieved by two different classes of membrane-bound enzymes; across the plasma membrane the pH gradient is achieved by the plasma membrane-bound H+ATPase, PM-H+ATPase, EC 3.6.1.35, and that across the tonoplast the function is fulfilled by the tonoplast-bound H+ATPase, V-H+ATPase, EC 3.6.1.34 and H+pyrophosphatase, V-H+PPase, EC 3.6.1.1 (Martinoia et al. 2007). The proton gradient generated across the tonoplast by the V-H+ATPase and V-H+PPase enables the vacuoles to serve as reservoirs for metabolites and ions necessary for the maintenance of general cell homeostasis and detoxification of cytoplasm (Taiz 1992; Martinoia et al. 2007). Both V-H+ATPase and V-H+PPase although have a similar function, structurally they are much different. While the former is comprised of multiple subunits with complex organisational structure (Sze et al. 2002), the latter is a single polypeptide (Martinoia et al. 2007). The importance of both the enzymes has been indicated in salt tolerance by overexpression of ‘c’ and ‘E1’ subunits of V-H+ATPase and V-H+PPase as a whole in Arabidopsis (Gaxiola et al. 2001; Zhou et al. 2016; Dabbous et al. 2017) with the plant overexpressing them showing higher salt tolerance than the wild-type.
CCC chloride-cation co-transporter
Although the efforts on understanding the mechanism of salt tolerance has remain largely centred on the cation transporters, the importance of the transporter facilitating movement of both cation and anion together, like cation chloride co-transporters (CCC) for co-transport of Cl−, Na+ and K+ has been recognised (Colmenero-Flores et al. 2007; Ishikawa et al. 2018). Based upon the mechanism and preference of symport for the combination of ions, CCC can be broadly divided into three categories: (1) K+:Cl− transporter, known as KCC group; (2) Na+:Cl− transporter known as NCC group; and (3) Na+:K+:Cl− co-transport, known as NKCC group. All these three members share the absolute requirement of Cl− and at least one cation (Na+ or K+) for its symport. The symport mechanism of Na+:K+:2Cl− is completely electroneutral or electrically silent (Geck et al. 1980; Russel 2000). It has been shown that CCC is preferentially expressed at symplast/xylem boundary in Arabidopsis (Colmenero-Flores et al. 2007), which suggests its importance in the long-distance ion transport. The functional significance of CCC in Na+:Cl− symport is recognised from the fact that efflux of Na+ from barley (Hordeum vulgare L.) stelar root gets significantly reduced due to the presence of the inhibitor bumetanide known to inhibit mammalian CCC (Zhu et al. 2017). In addition, it has also been reported that RNAi lines for CCC in rice accumulate less K+ compare to wild-type plant, indicating its significant role in K+ homeostasis as well (Kong et al. 2011).
The regulatory mechanism regarding CCC through the phosphorylation process (post-translational regulation) is still scant for the plant kingdom. There are few reports are available for NKCC in the animal kingdom where the activity of NKCC is regulated by phosphorylation mechanism. Although the effector regulatory kinase is still not characterised (Lytle and Forbush 1992; Lytle 1997).
NaCl signalling
The regulatory role of biomolecules, primarily proteins, starts at sensing the presence of excess Na+ in the external milieu, although excess is only a relative term when referred to in the context of glycophytes or halophytes. It is widely presumed that the perception of salinity stress in the environment happens as ionic stress, as the salt components are ionic, but mechanisms for sensing Na+ that are present mainly in animal, like Na+ selective ion channels and Na+ transporters (Maathuis 2014). However, the evidence of the presence of Na+ sensing mechanism does come from the halotropism observed in the root growth direction that is away from and specific to Na+ (Galvan-Ampudia et al. 2013). Besides, some other details on Na+ sensing mechanisms at the plasma membrane level are also available (Maathuis 2014; van Zelm et al. 2020). One among them is the Ca2+ wave-based where it has been shown that the moca1 (monocation-induced [Ca2+] increases 1) mutant of Arabidopsis does not show early calcium wave response to the monovalent ions like Na+, K+ and Li+ (Jiang et al. 2019). MOCA1 is a glucuronosyltransferase that leads to the production of glycosyl inositol phosphorylceramide (GIPC) sphingolipids at the plasma membrane that upon binding with the monovalent ions supposedly binds Ca2+ channel and open it to produce a wave of Ca2+ as the response (Jiang et al. 2019; van Zelm et al. 2020). Furthermore, the generation and propagation of long-range waves of calcium are exclusively induced by high salt, not by osmotic stress, and the response is almost instantaneous; the wave is initiated as early as 10 s after salt application and propagate throughout the plant system, including the leaves, within 30 s (Choi et al. 2014). Hence, although it is difficult to draw a clear boundary between osmotic and ionic phases of the salt stress, it is can be said with surety that the stress is a due to the excess of monovalent ions in the environment. The other well-documented mechanisms of Na+ sensing studied in some detail is that mediated by the receptor-like kinases (RLKs), which function by phosphorylating the intracellular serine/threonine kinase for the transduction of the extracellular signals to the downstream cellular effector molecules (Osakabe et al. 2013). Genome sequencing has revealed the presence of more than 600 RLK members in Arabidopsis and 1100 members in rice with the leucine-rich repeat constituting the largest group (Shiu and Bleecker 2001a, 2001b; Gish and Clark 2011). Not all these are involved in environmental signalling, they function in developmental processes as well. The possible role of RLKs in salt stress signalling, or abiotic stress signal transduction in general, is indicated from their responsiveness to the applied environmental stress in terms of expression. Any physical interaction of the abiotic factors with RLKs leading to sensing and transduction of the signal has not been reported so far. However, their important role in salt tolerance is evident from the biotechnological approaches, such as the alleviation of root growth inhibition in Medicago roots subjected to salt stress in transgenic roots with RNAi-mediated down-regulation of a salt-inducible LRR-RKL gene (SRLK), indicating a negative regulatory role (de Lorenzo et al. 2009). In contrast, however, Ouyang et al. (2010) reported a positive regulatory role of RLK in which it was observed that a salt- or drought-inducible gene, O. sativa stress-induced kinase gene 1 (OsSIK1) when overexpressed increased salt tolerance in the plant (rice), and the reason was attributed to a reduction in the accumulation of ROS with a concomitant increase in the ROS scavenging enzymes like peroxidases, superoxide dismutases and catalases. Thus, although RLKs have been reported to play important role in abiotic stress tolerance in plants, including salt stress, the functional details are little understood, making it a subject of further investigation.
The two-component signalling system, Histidine (His)-Aspartate (Asp) phosphorelay, has also been reported to sense and transduce the abiotic stress signal through phosphorylation (Tran et al. 2007; Jeon et al. 2010; Pham et al. 2012). In Arabidopsis, HKs have been categorised into two groups: one group, including ETR1, ERS1, AHK2, AHK3 and AHK4 functions as the receptors of the plant hormone ethylene and/or cytokinins (Schaller et al. 2008), while the other group forms the non- hormonal receptors, including AHK1, AHK5 and AKH4/CKI1 (Tran et al. 2007; Osakabe et al. 2013). The signal from the receptors passed downstream as phosphorelay involving five AHPs, the histidine-containing phosphotransmitter and numerous response regulators (ARRs), which are comprised of type-A and type-B ARRs that execute the perceived signal in the form of changes in gene expression or protein activity. Among the HKs the role of AHK1, AHK2, AHK3, AHK5 and CRE1 has been studied in some detail concerning abiotic stress response and tolerance. The possible role of AHK1 has been related to osmotic sensing since it was discovered to complement deletion of SLN1, an HK (osmosensor) in yeast (Urao et al. 1999; Tran et al. 2007). The ahk1 mutant developed further revealed that many stress-inducible genes, like DREB, ANAC and AREB1, were down-regulated in the plant along with the down-regulation of the downstream genes, indicating that AHK1 was a positive regulator of the abiotic stress-responsive genes (Tran et al. 2007). Furthermore, Arabidopsis plant overexpressing AHK1 showed greater tolerance to drought than the wild-type, and the ahk1 knockdown mutants exhibited severe sensitivity to drought stress, but the mutants were more sensitive to salinity stress than the wild-type, indicating that salt and drought tolerance follow different root somewhere in the signal transduction pathway (Tran et al. 2007). In contrast to AHK1, the AHK2 and AHK3 knockdown mutants were highly tolerant to both drought and salinity, and the ahkt2 ahk3 double mutants were even more tolerant, indicating their negative regulatory role in salt tolerance in oppose to AHK1 (Tran et al. 2007). The negative regulatory role in salt tolerance has also been demonstrated for AHK5 by generating its loss of function mutant ahk5 that conferred tolerance to high salinity to Arabidopsis, opposite to the non-hormonal HK receptor AHK1 (Pham et al. 2012). Thus, among the HKs, only AHK1 appears to be a sensor to drought stress, and others appear to perceive hormonal signal from other environmental factors, including abiotic and biotic, and could be involved in integrating multiple stress responses. The nature of signal perception for abiotic factors, however, remains unknown, including that of AHK1, which is reported to perceive osmotic stress. Nevertheless, AHK1 could be considered to be involved in salt stress signalling as the initial phase of salt stress is osmotic stress and the ahk1 mutant Arabidopsis becomes highly susceptible to salt stress compared with the wild-type (Tran et al. 2007). Furthermore, the knockout ahkt2 and ahkt3 showed down-regulation of expression of the type-A ARR4 and ARR7, suggesting their suppression of expression might overcome the negative regulatory role of AHKT2 and AHKT3 leading to enhanced salinity tolerance of the genetically modified plants (Tran et al. 2007). As a piece of circumstantial evidence to the supposition, the loss of function of ARR1 and ARR12 has been reported to enhance salt tolerance (Mason et al. 2010), similar to that of ARR4 and ARR7 (Tran et al. 2007). Overall, the collective reports support the premise that cross-talk between hormone and stress signalling networks plays an important role in plant stress response in which MOCA1 could be playing an important role. In all probability, it appears that the Na+ sensing mechanism works intracellular when Na+ enters through the HKTs and NASCs into the cytoplasm, which of course has not been elaborated so far (Ponce et al. 2021). The suggestion of the intracellular sensing of Na+ level is primarily based on the increase in the cytosolic level of Ca2+ in plants under salt stress (Gupta and Shaw 2021a) needs further experimental support. Further, the mechanism of binding of the GIPCs to the monovalent ions followed by their interaction with the Ca2+ channel is not yet sufficiently clear to demonstrate external sensing of Na+. However, it has been found that the propagation of the Ca2+ wave is facilitated by two-pore channel 1 (TPC1), which is tonoplast bound facilitating release of Ca2+ from the vacuole (Choi et al. 2014; Evans et al. 2016). Evans et al. (2016) further reported that ROS production by AtBROHD (Arabidopsis respiratory burst oxidase homologue D) mutually assists to amplify the Ca2+ wave transmission originated from TPC1-mediated Ca2+ releases.
Phosphorylation as the determinants of salt tolerance
The role of the effectors stated above, including SOS1, NHX1, PM-H+ATPase, V-H+ATPase, and HKTs in uptake, transport and sequestration of Na+ is well demonstrated, as is evident from the references cited. However, the research over the years has also indicated that the process of salt tolerance is quantitative; i.e. salt tolerance in plants is achieved by several intracellular biochemical events, besides the acts of the effectors, and phosphorylation of the effector proteins is one such event. The entire process is not so simple. Barring HKTs, the functioning of all other effectors are regulated through various biochemical processes. Many articles are documenting the regulatory mechanism of transporters like PM H+-ATPase, V-H+-ATPase, SOS1 and NHX1 through phosphorylation through kinases, which is a key factor in their functioning under saline stress conditions (Qiu et al. 2004; Klychnikov et al. 2007; Fuglsang et al. 2010; Brini and Masmoudi 2012; Ji et al. 2013; Fuglsang et al. 2014) (Fig. 1).
Phosphorylation of plasma membrane H+ ATPase
As the name suggests, PM-H+ATPase is a plasma membrane-bound enzyme, and it catalyses the breakdown of ATP that occurs on the cytoplasmic side. It is a single polypeptide unit comprised of 948 amino acid residues, but it is integrated into the membrane with the help of 10 trans-membrane α-helical hydrophobic segments with most of the remaining mass, including both N-terminal and C-terminals remaining exposed to the cytoplasm (Falhof et al. 2016). During the breakdown of ATP, H+ is exported outside the cell into the apoplast and environment, the reverse of ATP synthesis in which the flow of H+ occurs from the matrix to the outer compartment of the mitochondria while throwing H+ outside the cell. The PM-H+ATPase not only generates a trans-membrane gradient of protons (acidic outside), it also leads to the establishment of electrical gradient; i.e. membrane potential (inside negative and outside positive). The proton motive force generated by the PM-H+-ATPase, a P-type ATPase, contributes towards the transport of various ions and also organic molecules like sugar and amino acids across the plasma membrane that are required for various biochemical and physiological events including opening and closing of stomata, cell growth, nutrient uptake through the root and movement of ions from root to shoot and nutrients from shoot to root through xylem and phloem (Blumwald et al. 2000; Gaxiola et al. 2007; Mansour 2014). Apart from these physiological roles, PM-H+ATPase helps plants adapt to salinity stress as its proton pumping activity, which is induced in response to exposure to salinity, particularly in the salt-tolerant plants, leads to repolarisation of the plasma membrane depolarised in the presence of an excess of NaCl (Mansour 2014). The proton motive force generated across the plasma membrane also drives the Na+/H+ antiporter for the efflux of the Na+ back to the apoplast and to the environment (Gaxiola et al. 2007). This also implies that that the functioning of the PM-H+ATPase must be salt-inducible, which can be through the regulation of its expression. However, the functioning of many proteins, particularly the membrane-bound, are regulated by phosphorylation of their amino acid residue. Phosphorylation of PM H+-ATPase is a well-recognised way of post-translational regulation of its activity, and it happens by phosphorylation of the threonine (Thr) residue at the C-terminal end (Gaxiola et al. 2007; Mansour 2014). The regulatory site of the protein in Arabidopsis has been reported at its C-terminal end that consists of 100 amino acid residues, which forms the auto-inhibitory domain (Palmgren et al. 1991; Falhof et al. 2016).
Phosphosite mapping of the Arabidopsis PM-H+ATPase (AHA1, AHA2, AHA3, AHA4/11) has revealed a total of 11 phosphorylation site in the enzyme, including the several identified earlier (Niittylä et al. 2007; Rudashevskaya et al. 2012). The phosphorylation sites were mostly in the N and C terminal regulatory domains and seven out of nine phosphosites were found identical for AHA2 in plants and fungi (Rudashevskaya et al. 2012). EMBL-Clustal omega bioinformatics sequence alignment analysis revealed conservation of phosphorylation sites in the PM-H+ATPase (AHA2 of Arabidospis) among several plants, including wheat, rice, maize, and tomato (Fig. 2).
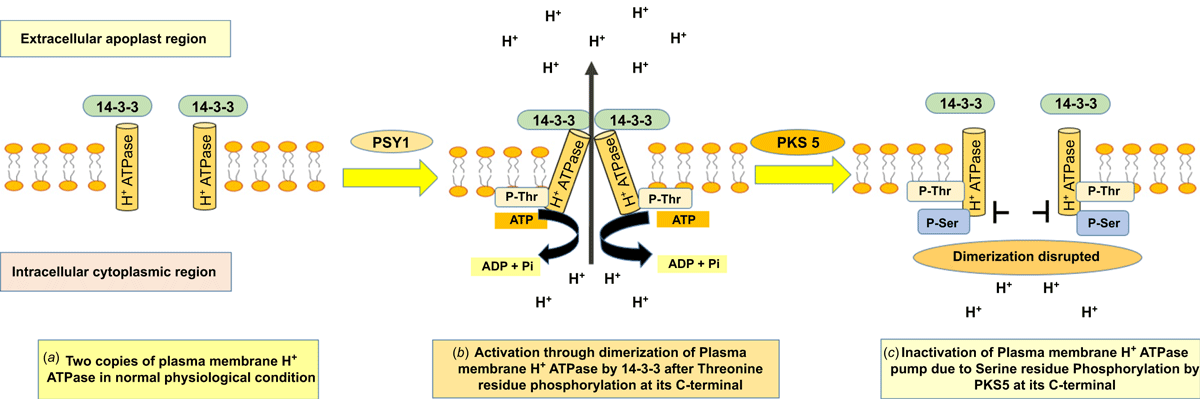
In Arabidopsis, the positive regulatory kinase enzyme (PSY1R) phosphorylates the protein at Thr-948 in AHA1 and Thr-947 in AHA2 at the C-terminal end (Fuglsang et al. 2010, 2014). However, the activation of the PM-H+ATPase function is not achieved merely by phosphorylation. The phosphorylation at the C-terminal end merely creates a binding site for the 14-3-3 proteins that releases autoinhibition of the enzyme. The binding of the 14-3-3 proteins further pairs with another phosphorylated and 14-3-3 protein-bound PM-H+ATPase leading to the formation of PM-H+ATPase-14-3-3/PM-H+ATPase-14-3-3 dimer, which is the active form of the enzyme (Fuglsang et al. 2007; Duby et al. 2009). Thus, phosphorylation of the Thr-948 is the beginning of initiation of making the dormant PM-H+ATPase active enabling proton pumping to create a proton gradient across the plasma membrane (Fig. 3) (Fuglsang et al. 2010, 2014).
|
Apart from activation through Thr phosphorylation, a study in Arabidopsis has further revealed that the activity of PM-H+ATPase is also regulated by phosphorylation of the Ser-931 residue by PKS5 kinase enzyme; the phosphorylation at the Ser-931 residue in AHA2 creates a steric hindrance for 14-3-3 interactions with PM H+-ATPase resulting in inactivation of the enzyme (Fig. 3) (Fuglsang et al. 2007; Duby et al. 2009).
Altogether it is well proven that phosphorylation events regulate the activity of the plasma membrane proton pump, at least in Arabidopsis. The evidence of the role of phosphorylation in the regulation of the activity of PM-H+ATPase is limited in other plant species including the crop plants because of a lack of information on the regulatory kinase in them. Nevertheless, recently Gupta and Shaw (2021b) working with four rice cultivars, including Nona Bokra and Pokkali and salt-sensitive IR-64 and IR-29, have shown that application of salt stress dose not lead to an increase in the expression of PM H+-ATPase either at the level of protein or mRNA, but the activity level of the enzyme increases several folds in both roots and shoot tissues of the salt-tolerant cultivars compared with the sensitive ones concomitant with a significant increase in Thr-phosphorylation of the enzyme in the former compared with the latter, clearly emphasize an important role of phosphorylation in regulation of the activity of the enzyme. In fact, it can very well be realised that response time to environmental stress in terms of enhancing the function of the protein directly linked to alleviation of the stress, such as that of NaCl, should be as less as possible, and it may not be possible for the plant to fulfil this requirement if it relies on the synthesis of new proteins. Hence, keeping the level of an effector protein to a connotationally high level and regulating its function based on the requirement upon getting the environmental signal would be the best option. Therefore, a high level of expression of PM H+-ATPase in a plant may not be the criteria for its salt tolerance. The criteria of salt tolerance rather would largely be the ability of the plant to activate the PM-H+ATPase by phosphorylating its Thr-residue at the C-terminal end (Kanczewska et al. 2005), which in the salt-tolerant cultivars of rice but not in the salt-sensitive cultivars (Gupta and Shaw 2021b). Thus, future research work on salt tolerance in terms of the PM-H+ATPase as a effector or the membrane-bound protein effectors should focus on the identification of the kinases that could be phosphorylating them for activation.
Regulation of vacuolar-H+-ATPase by phosphorylation
Vacuolar H+-ATPase is the proton gradient generating pump that is well reported at the endomembrane system (mainly vacuolar membrane, trans-Golgi network, and endoplasmic reticulum) (Ratajczak 2000). Vacuolar H+-ATPase is structurally composed of two sub complexes, V1 and V0, for which this enzyme has been categorised under multi-subunit structures. The peripheral V1 complex is an actual site for ATP hydrolysis while V0 is an integral membrane complex that carries out the proton translocation (Gaxiola et al. 2007).
V-H+-ATPase maintains the cytosolic pH homeostasis that plays an important role in cell elongation and growth (Hanitzsch et al. 2007). An increase in the activity of V-H+ATPase has been reported in response to salt stress in many plants where this pump facilitates the Na+ sequestration in vacuole with the Na+/H+ antiporter NHX driven by H+ gradient generated across the tonoplast by the V-H+ATPase (Jiang et al. 2010; Bassil and Blumwald 2014). Being a multi-subunit enzyme, data on the expression of V-H+ATPase in response to salt stress is not expected, but an increase in expression through the transgenic approach of a few of its subunits, like V-H+ATPase A, a, C, c, D, d, F, G, and H subunit in response to salt stress has been reported (He et al. 2014). However, the regulation of the activity of V-H+ATPase at the level of the enzyme becomes more significant than that of the PM-H+ATPase, as the former is being a multi-subunit structure, and hence regulation of its activity by regulation of expression may not be feasible. Although not much data on the regulation of activity of V-H+ATPase by phosphorylation is available, the report of Batelli et al. (2007) demonstrating the interaction of Ser/Thr kinase (SOS2) with the B1 and B2 subunit of V-H+ATPase through yeast two-hybrid experiment leading to stimulation of its proton pumping activity in Arabidopsis does support the regulation of activity of the enzyme by phosphorylation. However, the details of the amino acid residues phosphorylated are lacking. In addition, Klychnikov et al. (2007) reported that 14-3-3 also regulates the activity of V-H+-ATPase, the details are of course lacking.
Regulation of SOS1-Na+/H+ activity through phosphorylation
Salt overly sensitive 1 protein (SOS1) is the major Na+/H+ antiporter that facilitates Na+ efflux in plants. SOS1 mediates the Na+ efflux outside of the cell into the environment from the epidermal tissue of the root together with regulating the Na+ transportation from root to shoot (Shi et al. 2002; Olías et al. 2009). Approximately 70–90% of the Na+ entering into the xylem is retrieved from the transpiration stream by SOS1 back into the xylem parenchyma for the sequestration in the vacuoles of the root cells via the V-H+ATPase/NHX sequestration mechanism (Tester and Davenport 2003; Munns 2005). The ion pumping activity of SOS1 is largely dependent on the proton gradient generated by PM H+-ATPase. SOS1 functional activity is regulated by the SOS2 (CIPK24: CBL-interacting serine/threonine-protein kinase) and SOS3 (CBL4: Calcineurin B-like protein). In the presence of Ca2+, the N-terminal of SOS3 gets modified through myristoylation due to the binding of Ca2+ in the EF-hand of SOS3. The activated SOS3 protein interacts with CIPK 24 (SOS2) that is classified under SnRK (sucrose non-fermenting-related serine/threonine kinase) protein family. The SOS2-SOS3 protein complex phosphorylates the amino acid residue at the C-terminal site of SOS1 that leads to its activation (Pardo 2010; Brini and Masmoudi 2012; Hasegawa 2013; Ji et al. 2013). It has been shown that a mutation in the SOS2 (CIPK) kinase gene in the yeast system leads to disruption of SOS1 activation, illustrating the importance of phosphorylation for its ion exchange capability (Quintero et al. 2011). In a monocot species like rice, OsCIPK24 (LOC_Os06g40370) plays the major role of SOS2 (Gupta and Shaw 2021a), and its CDS sequence is conserved in most of the monocots species, including Oryza coarctata L., Oryza brachyantha L., H. vulgare subsp. Spontaneum, Sorghum bicolor L., Panicum virgatum L., Setaria viridis (L.) P. Beauv., Triticum dicoccoides L., T. aestivum and Zea mays L. The similarity of OsCIPK24 with the dicots like Glycine max L., Solanum lycopersicum L., and Vigna radiata L. is, however, less than 80% and thus, it is very likely that the role of OsCIPK24 might be being played by a different member of CIPKs, which is yet to be identified.
Regulation of NHX pump through phosphorylation
NHX-Na+/H+ antiporter helps in mitigating the Na+ toxicity through sequestration of the Na+ in the vacuole ensuring the concentration of the ion in the cytosol remain between 10 and 30 mM. NHX mediates the sequestration of Na+ into the vacuole by utilising the electrochemical proton gradient in the vacuole sap that is created by V-H+ATPase and V-H+PPase (Jiang et al. 2010; Bassil and Blumwald 2014). In Arabidopsis, the activity of NHX1 is regulated through its C-terminal tail by the interaction of calmodulin-like protein 15 (AtCaM15) in the presence of Ca2+ and optimum pH (Yamaguchi et al. 2003). Under the control physiological acidic pH at the vacuolar lumen and high Ca2+ concentration, AtCaM15 interacts preferentially with AtNHX1 for K+/H+ exchange activity over Na+/H+ (Yamaguchi et al. 2003), but under saline stress that causes alkalisation of vacuole, AtNHX1 starts favouring Na+/H+ activity when compared to K+/H+ transport because of reduced binding of AtCaM15 to the antiporter (Yamaguchi et al. 2003; Rodríguez-Rosales et al. 2009).
The activity of NHX is regulated by the phosphorylation of its amino acid residues (Qiu et al. 2004). In OsNHX3 the phosphorylation has been reported to occur at the residue S471 located at the C-terminus of the enzyme, and this residue shows conservation in several NHXs of rice, including OsNHX1, 2 and 4 (Bassil et al. 2012). The conservation of S471 residue at the C-terminus of the enzyme is also found in Arabidopsis NHX like NHX3, 5 and 6 (Bassil et al. 2012). The regulation of activity of OsNHX1 has also been proven by mutation study in which non-synonymous mutation of serine to asparagine (S477N) leads to loss of the phosphorylation event in OsNHX1 that impairs the SOS2 interaction leading to a lower antiporter functional activity of the antiporter (Negrão et al. 2013). However, the biological role of the phosphorylation event at S477 is questionable as both salt-susceptible (IR29 and IR64) and salt-tolerant (FL478) rice varieties show mutation of S477 to S477N (serine to aspargine) (Negrao et al. 2013).
Conclusion and perspectives
Developing crop cultivars resistant to salinity is required to bring the salt-affected lands into productive cultivation. Through ion relation studies, it has been realised that the maintenance of a low cytosolic Na+ level is the primary requirement for the resistance of a plant to salinity.
The primary factors in preventing the build-up of cytosolic Na+ level in plants exposed to an excess of external Na+ are the control of its entry into the cell, followed by its efflux and sequestration into the vacuoles. While the entry of the ion through the ion channel is reduced by repolarisation of the depolarised plasma membrane by efficient pumping out of H+ by the PM-H+ATPase, its built-in cytoplasm is further reduced by the action of SOS1 driven by the proton motive force generated by the action of the PM-H+ATPase, the second level of protection against prevention of accumulation of the ion in the cytoplasm. The third level of protection against the accumulation of Na+ in the cytosol is by the action of NHX that is driven by V-H+ATPase. The important role of CCC in the co-transport of Na+:Cl− and K+:Cl− has also been recognised. Because of the occurrence of these multiple levels of protection, it has not been possible to successfully develop or make a salt-sensitive crop cultivar into a salt-tolerant one, although several investigations have worked on the overexpression of the H+ transporters (PM H+ATPase and Vacuolar H+ ATPase) and the antiporters (NHX and SOS1) for improving salt tolerance in several of the crop species by the way of improving Na+ efflux and sequestration (Chen et al. 2021), despite Na+ sequestration being considered as very important since the leaves of the salt-tolerant plants function normally even after high tissue built-up of the ion (Munns and Tester 2008).
The overexpression of these transporters has failed to produce the desired result probably because that their activity is regulated by the post-translational phosphorylation events. Hence, any amount of constitutional expression of these transporters may not provide the desired result. There is no need for overexpression of a protein that is available constitutionally at a high level in the tissue, such as that of PM-H+ATPase (Gupta and Shaw 2021b). However, investigations have shown that the level of Thr phosphorylation in PM-H+ATPase does show a significantly greater increase in the salt-tolerant rice cultivars compared with the salt-tolerant ones (Gupta and Shaw 2021b). Hence, it can be inferred that by regulating the expression of the kinases that phosphorylates the effectors, other than CCC, information on which is scant, it could be possible to improve the salt tolerance of a salt-sensitive plant. However, the task is not straightforward. Firstly, it is necessary to understand the long-distance salt stress signalling mechanism, information on which is limited (Munns and Tester 2008; van Zelm et al. 2020); precisely, we still lack a clear understanding of the perception of salt and the signalling pathway that directs an effector to act (van Zelm et al. 2020). Secondly, to achieve the goal of intervention at the level of the regulatory kinases, it would be necessary to identify the kinases that regulate the effectors. So far our information on the kinases phosphorylating the effectors involved in salt tolerance is limited. In silico approach in the direction of identification of the regulatory kinases of the effectors may be of great help. In this context, the CIPK24 and PSY1 already identified for their role in the phosphorylation of PM-H+ATPase and SOS1 in rice and Arabidopsis, respectively, may be good as a starting point. Biological intervention in the line of raising a transgenic plant engineered with the identified and characterised interactive kinases expressed under salt responsive promoter element could prove a promising and successful approach for converting a high yielding salt-sensitive crop cultivar into a salt-resistance transgenic (Roy et al. 2014), which could lead to increase in total yield of the crop by bringing into the salt-affected land into cultivation.
Data availability statement
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
Author has no conflict of interest.
Declaration of funding
AG duly acknowledges DST, New Delhi for providing financial assistance.
Acknowledgements
AG and BPS jointly conceptualised the content of this review article. BPS wrote the manuscript. AG, BPS and BBS jointly revised the manuscript and finalised the figures. AG, BPS and BBS have edited and reviewed this manuscript carefully. AG duly acknowledges Director, ILS, Bhubaneswar for providing research facilities. This invited review article is awarded Gold Open Access free of charge.
References
Adams E, Shin R (2014) Transport, signaling, and homeostasis of potassium and sodium in plants. Journal of Integrative Plant Biology 56, 231–249.| Transport, signaling, and homeostasis of potassium and sodium in plants.Crossref | GoogleScholarGoogle Scholar | 24393374PubMed |
Almeida DM, Margarida Oliveira M, Saibo NJM (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genetics and Molecular Biology 40, 326–345.
| Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants.Crossref | GoogleScholarGoogle Scholar | 28350038PubMed |
Alnayef M, Solis C, Shabala L, Ogura T, Chen Z, Bose J, Maathuis FJM, Venkataraman G, Tanoi K, Yu M, Zhou M, Horie T, Shabala S (2020) Changes in expression level of OsHKT1;5 alters activity of membrane transporters involved in K+ and Ca2+ acquisition and homeostasis in salinized rice roots. International Journal of Molecular Sciences 21, 4882
| Changes in expression level of OsHKT1;5 alters activity of membrane transporters involved in K+ and Ca2+ acquisition and homeostasis in salinized rice roots.Crossref | GoogleScholarGoogle Scholar |
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258.
| Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 10455050PubMed |
Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Frontiers in Physiology 8, 509
| The role of Na+ and K+ transporters in salt stress adaptation in glycophytes.Crossref | GoogleScholarGoogle Scholar | 28769821PubMed |
Bassil E, Blumwald E (2014) The ins and outs of intracellular ion homeostasis: NHX-type cation/H(+) transporters. Current Opinion in Plant Biology 22, 1–6.
| The ins and outs of intracellular ion homeostasis: NHX-type cation/H(+) transporters.Crossref | GoogleScholarGoogle Scholar | 25173972PubMed |
Bassil E, Coku A, Blumwald E (2012) Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. Journal of Experimental Botany 63, 5727–5740.
| Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development.Crossref | GoogleScholarGoogle Scholar | 22991159PubMed |
Batelli G, Verslues PE, Agius F, Qiu Q, Fujii H, Pan S, Schumaker KS, Grillo S, Zhu JK (2007) SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Molecular and Cellular Biology 27, 7781–7790.
| SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity.Crossref | GoogleScholarGoogle Scholar | 17875927PubMed |
Blumwald E, Poole RJ (1985) Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiology 78, 163–167.
| Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris.Crossref | GoogleScholarGoogle Scholar | 16664191PubMed |
Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochimica et Biophysica Acta 1465, 140–151.
| Sodium transport in plant cells.Crossref | GoogleScholarGoogle Scholar | 10748251PubMed |
Böhm J, Messerer M, Müller HM, Scholz-Starke J, Gradogna A, Scherzer S, Maierhofer T, Bazihizina N, Zhang H, Stigloher C, Ache P, Al-Rasheid KAS, Mayer KFX, Shabala S, Carpaneto A, Haberer G, Zhu JK, Hedrich R (2018) Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Current Biology 28, 3075–3085.e7.
| Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa.Crossref | GoogleScholarGoogle Scholar | 30245105PubMed |
Bonales-Alatorre E, Pottosin I, Shabala L, Chen ZH, Zeng F, Jacobsen SE, Shabala S (2013a) Differential activity of plasma and vacuolar membrane transporters contributes to genotypic differences in salinity tolerance in a halophyte species, Chenopodium quinoa. International Journal of Molecular Sciences 14, 9267–9285.
| Differential activity of plasma and vacuolar membrane transporters contributes to genotypic differences in salinity tolerance in a halophyte species, Chenopodium quinoa.Crossref | GoogleScholarGoogle Scholar | 23629664PubMed |
Bonales-Alatorre E, Shabala S, Chen ZH, Pottosin I (2013b) Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, quinoa. Plant Physiology 162, 940–952.
| Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, quinoa.Crossref | GoogleScholarGoogle Scholar | 23624857PubMed |
Brini F, Masmoudi K (2012) Ion transporters and abiotic stress tolerance in plants. ISRN Molecular Biology 2012, 927436
| Ion transporters and abiotic stress tolerance in plants.Crossref | GoogleScholarGoogle Scholar | 27398240PubMed |
Chen T, Shabala S, Niu Y, Chen ZH, Shabala L, Meinke H, Venkataraman G, Pareek A, Xu J, Zhou M (2021) Molecular mechanisms of salinity tolerance in rice. The Crop Journal 9, 506–520.
| Molecular mechanisms of salinity tolerance in rice.Crossref | GoogleScholarGoogle Scholar |
Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proceedings of the National Academy of Sciences of the United States of America 111, 6497–6502.
| Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants.Crossref | GoogleScholarGoogle Scholar | 24706854PubMed |
Colmenero-Flores JM, Martínez G, Gamba G, Vázquez N, Iglesias DJ, Brumós J, Talón M (2007) Identification and functional characterization of cation-chloride cotransporters in plants. The Plant Journal 50, 278–292.
| Identification and functional characterization of cation-chloride cotransporters in plants.Crossref | GoogleScholarGoogle Scholar | 17355435PubMed |
Craig Plett D, Moller IS (2010) Na+ transport in glycophytic plants: what we known and would like to know. Plant, Cell & Environment 33, 612–626.
| Na+ transport in glycophytic plants: what we known and would like to know.Crossref | GoogleScholarGoogle Scholar |
Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S (2011) Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant, Cell & Environment 34, 947–961.
| Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods.Crossref | GoogleScholarGoogle Scholar |
Dabbous A, Saad RB, Brini F, Farhat-Khemekhem A, Zorrig W, Abdely C, Hamed KB (2017) Over-expression of a subunit E1 of a vacuolar H+-ATPase gene (Lm VHA-E1) cloned from the halophyte Lobularia maritima improves the tolerance of Arabidopsis thaliana to salt and osmotic stresses. Environmental and Experimental Botany 137, 128–141.
| Over-expression of a subunit E1 of a vacuolar H+-ATPase gene (Lm VHA-E1) cloned from the halophyte Lobularia maritima improves the tolerance of Arabidopsis thaliana to salt and osmotic stresses.Crossref | GoogleScholarGoogle Scholar |
de Lorenzo L, Merchan F, Laporte P, Thompson R, Clarke J, Sousa C, Crespi M (2009) A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. The Plant Cell 21, 668–680.
| A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress.Crossref | GoogleScholarGoogle Scholar | 19244136PubMed |
Deng Y, Feng Z, Yuan F, Guo J, Suo S, Wang B (2015) Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands. Plant Physiology and Biochemistry 97, 20–27.
| Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands.Crossref | GoogleScholarGoogle Scholar | 26397201PubMed |
Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M (2009) Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. Journal of Biological Chemistry 284, 4213–4221.
| Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region.Crossref | GoogleScholarGoogle Scholar |
Duggleby RG, Dennis DT (1973) Pyruvate kinase, a possible regulatory enzyme in higher plants. Plant Physiology 52, 312–317.
| Pyruvate kinase, a possible regulatory enzyme in higher plants.Crossref | GoogleScholarGoogle Scholar | 16658554PubMed |
Evans MJ, Choi WG, Gilroy S, Morris RJ (2016) A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC cation channel propagates the systemic response to salt stress. Plant Physiology 171, 1771–1784.
| A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC cation channel propagates the systemic response to salt stress.Crossref | GoogleScholarGoogle Scholar | 27261066PubMed |
Falhof J, Pedersen JT, Fuglsang AT, Palmgren M (2016) Plasma membrane H(+)-ATPase regulation in the center of plant physiology. Molecular Plant 9, 323–337.
| Plasma membrane H(+)-ATPase regulation in the center of plant physiology.Crossref | GoogleScholarGoogle Scholar | 26584714PubMed |
Flowers TJ, Dalmond D (1992) Protein synthesis in halophytes: the influence of potassium, sodium and magnesium in vitro. Plant and Soil 146, 153–161.
| Protein synthesis in halophytes: the influence of potassium, sodium and magnesium in vitro.Crossref | GoogleScholarGoogle Scholar |
Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, Palmgren MG, Zhu JK (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634.
| Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein.Crossref | GoogleScholarGoogle Scholar | 17483306PubMed |
Fuglsang AT, Paez-Valencia J, Gaxiola RA (2010) Plant proton pumps: regulatory circuits involving H+-ATPase and H+-PPase. In ‘Transporters and pumps in plant signaling’. (Eds M Geisler, K Venema) pp. 39–64. (Springer: Berlin, Germany)
Fuglsang AT, Kristensen A, Cuin TA, Schulze WX, Persson J, Thuesen KH, Ytting CK, Oehlenschlæger CB, Mahmood K, Sondergaard TE, Shabala S, Palmgren MG (2014) Receptor kinase mediated control of primary active proton pumping at the plasma membrane. The Plant Journal 80, 951–964.
| Receptor kinase mediated control of primary active proton pumping at the plasma membrane.Crossref | GoogleScholarGoogle Scholar | 25267325PubMed |
Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, Haring MA, Munnik T, Vernoux T, Testerink C (2013) Halotropism is a response of plant roots to avoid a saline environment. Current Biology 23, 2044–2050.
| Halotropism is a response of plant roots to avoid a saline environment.Crossref | GoogleScholarGoogle Scholar | 24094855PubMed |
Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. The Plant Journal 34, 788–801.
| Sodium transport and HKT transporters: the rice model.Crossref | GoogleScholarGoogle Scholar | 12795699PubMed |
Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proceedings of the National Academy of Sciences of the United States of America 96, 1480–1485.
| The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast.Crossref | GoogleScholarGoogle Scholar | 9990049PubMed |
Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proceedings of the National Academy of Sciences of the United States of America 98, 11444–11449.
| Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump.Crossref | GoogleScholarGoogle Scholar | 11572991PubMed |
Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Letters 581, 2204–2214.
| Plant proton pumps.Crossref | GoogleScholarGoogle Scholar | 17412324PubMed |
Geck P, Pietrzyk C, Burckhardt BC, Pfeiffer B, Heinz E (1980) Electrically silent cotransport on Na+, K+ and Cl− in Ehrlich cells. Biochimica et Biophysica Acta 600, 432–447.
| Electrically silent cotransport on Na+, K+ and Cl− in Ehrlich cells.Crossref | GoogleScholarGoogle Scholar | 7407122PubMed |
Gish LA, Clark SE (2011) The RLK/Pelle family of kinases. The Plant Journal 66, 117–127.
| The RLK/Pelle family of kinases.Crossref | GoogleScholarGoogle Scholar | 21443627PubMed |
Greenway H, Osmond CB (1972) Salt responses of enzymes from species differing in salt tolerance. Plant Physiology 49, 256–259.
| Salt responses of enzymes from species differing in salt tolerance.Crossref | GoogleScholarGoogle Scholar | 16657936PubMed |
Gupta A, Shaw BP (2021a) Augmenting salt tolerance in rice by regulating uptake and tissue specific accumulation of Na+ through Ca2+ induced alteration of biochemical events. Plant Biology 23, 122–130.
| Augmenting salt tolerance in rice by regulating uptake and tissue specific accumulation of Na+ through Ca2+ induced alteration of biochemical events.Crossref | GoogleScholarGoogle Scholar | 33768704PubMed |
Gupta A, Shaw BP (2021b) Biochemical and molecular characterisations of salt tolerance components in rice varieties tolerant and sensitive to NaCl: the relevance of Na+ exclusion in salt tolerance in the species. Functional Plant Biology 48, 72–87.
| Biochemical and molecular characterisations of salt tolerance components in rice varieties tolerant and sensitive to NaCl: the relevance of Na+ exclusion in salt tolerance in the species.Crossref | GoogleScholarGoogle Scholar |
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Frontiers in Plant Science 7, 1787
| New insights on plant salt tolerance mechanisms and their potential use for breeding.Crossref | GoogleScholarGoogle Scholar | 27965692PubMed |
Hanitzsch M, Schnitzer D, Seidel T, Golldack D, Dietz KJ (2007) Transcript level regulation of the vacuolar H+-ATPase subunit isoforms VHA-a, VHA-E and VHA-G in Arabidopsis thaliana. Molecular Membrane Biology 24, 507–518.
| Transcript level regulation of the vacuolar H+-ATPase subunit isoforms VHA-a, VHA-E and VHA-G in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 17710654PubMed |
Hasegawa PM (2013) Sodium Na+ homeostasis and salt tolerance of plants. Environmental and Experimental Botany 92, 19–31.
| Sodium Na+ homeostasis and salt tolerance of plants.Crossref | GoogleScholarGoogle Scholar |
He X, Huang X, Shen Y, Huang Z (2014) Wheat V H+-ATPase subunit genes significantly affect salt tolerance in Arabidopsis thaliana. PLoS One 9, e86982
| Wheat V H+-ATPase subunit genes significantly affect salt tolerance in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 24498005PubMed |
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. The EMBO Journal 26, 3003–3014.
| Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth.Crossref | GoogleScholarGoogle Scholar | 17541409PubMed |
Ishikawa T, Cuin TA, Bazihizina N, Shabala S (2018) Xylem ion loading and its implications for plant abiotic stress tolerance. Advances in Botanical Research 87, 267–301.
| Xylem ion loading and its implications for plant abiotic stress tolerance.Crossref | GoogleScholarGoogle Scholar |
Ismail AM, Horie T (2017) Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annual Review of Plant Biology 68, 405–434.
| Genomics, physiology, and molecular breeding approaches for improving salt tolerance.Crossref | GoogleScholarGoogle Scholar | 28226230PubMed |
Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, Véry AA (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiology 150, 1955–1971.
| Diversity in expression patterns and functional properties in the rice HKT transporter family.Crossref | GoogleScholarGoogle Scholar | 19482918PubMed |
Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, Kim J (2010) A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. The Journal of Biological Chemistry 285, 23371–23386.
| A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 20463025PubMed |
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Molecular Plant 6, 275–286.
| The salt overly sensitive (SOS) pathway: established and emerging roles.Crossref | GoogleScholarGoogle Scholar | 23355543PubMed |
Jiang X, Leidi EO, Pardo JM (2010) How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal & Behavior 5, 792–795.
| How do vacuolar NHX exchangers function in plant salt tolerance?Crossref | GoogleScholarGoogle Scholar |
Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, Xiang Y, Niu Y, Liu F, Li C, Ye R, Byeon B, Xue Y, Zhao H, Wang HN, Crawford BM, Johnson DM, Hu C, Pei C, Zhou W, Swift GB, Zhang H, Vo-Dinh T, Hu Z, Siedow JN, Pei ZM (2019) Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346.
| Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx.Crossref | GoogleScholarGoogle Scholar | 31367039PubMed |
Kanczewska J, Marco S, Vandermeeren C, Maudoux O, Rigaud JL, Boutry M (2005) Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proceedings of the National Academy of Sciences of the United States of America 102, 11675–11680.
| Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer.Crossref | GoogleScholarGoogle Scholar | 16081536PubMed |
Kiani-Pouya A, Roessner U, Jayasinghe NS, Lutz A, Rupasinghe T, Bazihizina N, Bohm J, Alharbi S, Hedrich R, Shabala S (2017) Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant, Cell & Environment 40, 1900–1915.
| Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species.Crossref | GoogleScholarGoogle Scholar |
Klychnikov OI, Li KW, Lill H, de Boer AH (2007) The V-ATPase from etiolated barley (Hordeum vulgare L.) shoots is activated by blue light and interacts with 14-3-3 proteins. Journal of Experimental Botany 58, 1013–1023.
| The V-ATPase from etiolated barley (Hordeum vulgare L.) shoots is activated by blue light and interacts with 14-3-3 proteins.Crossref | GoogleScholarGoogle Scholar | 17185742PubMed |
Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef MA, Shabala S, An G, Ma JF, Horie T (2017) OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. The Plant Journal 91, 657–670.
| OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice.Crossref | GoogleScholarGoogle Scholar | 28488420PubMed |
Kong XQ, Gao XH, Sun W, An J, Zhao YX, Zhang H (2011) Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Molecular Biology 75, 567–578.
| Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1.Crossref | GoogleScholarGoogle Scholar | 21369877PubMed |
Lytle C (1997) Activation of the avian erythrocyte Na–K–Cl cotransport protein by cell shrinkage, cAMP, fluoride, and calyculin-A involves phosphorylation at common sites. Journal of Biological Chemistry 272, 15069–15077.
| Activation of the avian erythrocyte Na–K–Cl cotransport protein by cell shrinkage, cAMP, fluoride, and calyculin-A involves phosphorylation at common sites.Crossref | GoogleScholarGoogle Scholar |
Lytle C, Forbush B (1992) The Na–K–Cl cotransport protein of shark rectal gland. II. Regulation by direct phosphorylation. Journal of Biological Chemistry 267, 25438–25443.
| The Na–K–Cl cotransport protein of shark rectal gland. II. Regulation by direct phosphorylation.Crossref | GoogleScholarGoogle Scholar |
Maathuis FJ (2014) Sodium in plants: perception, signalling, and regulation of sodium fluxes. Journal of Experimental Botany 65, 849–858.
| Sodium in plants: perception, signalling, and regulation of sodium fluxes.Crossref | GoogleScholarGoogle Scholar | 24151301PubMed |
Mansour MM (2014) The plasma membrane transport systems and adaptation to salinity. Journal of Plant Physiology 171, 1787–1800.
| The plasma membrane transport systems and adaptation to salinity.Crossref | GoogleScholarGoogle Scholar | 25262536PubMed |
Marschner H (1995) Mineral nutrition of higher plants. Annals of Botany 78, 527–528.
Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. Journal of Experimental Botany 58, 83–102.
| Vacuolar transporters and their essential role in plant metabolism.Crossref | GoogleScholarGoogle Scholar | 17110589PubMed |
Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Eric Schaller G (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. The Plant Journal 64, 753–763.
| Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots.Crossref | GoogleScholarGoogle Scholar | 21105923PubMed |
Møller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals? Trends in Plant Science 12, 534–540.
| Salinity tolerance of Arabidopsis: a good model for cereals?Crossref | GoogleScholarGoogle Scholar | 18023242PubMed |
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytologist 167, 645–663.
| Genes and salt tolerance: bringing them together.Crossref | GoogleScholarGoogle Scholar |
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681.
| Mechanisms of salinity tolerance.Crossref | GoogleScholarGoogle Scholar | 18444910PubMed |
Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD (2016) Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Functional Plant Biology 43, 1103–1113.
| Tissue tolerance: an essential but elusive trait for salt-tolerant crops.Crossref | GoogleScholarGoogle Scholar | 32480530PubMed |
Munns R, Passioura JB, Colmer TD, Byrt CS (2020) Osmotic adjustment and energy limitations to plant growth in saline soil. The New Phytologist 225, 1091–1096.
| Osmotic adjustment and energy limitations to plant growth in saline soil.Crossref | GoogleScholarGoogle Scholar | 31006123PubMed |
Negrão S, Almadanim MC, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McNally KL, Oliveira MM (2013) New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnol Journal 11, 87–100.
| New allelic variants found in key rice salt-tolerance genes: an association study.Crossref | GoogleScholarGoogle Scholar |
Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Molecular & Cellular Proteomics 6, 1711–1726.
| Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Ochiai K, Matoh T (2002) Characterization of the Na+ delivery from roots to shoots in rice under saline stress: excessive salt enhances apoplastic transport in rice plants. Soil Science and Plant Nutrition 48, 371–378.
| Characterization of the Na+ delivery from roots to shoots in rice under saline stress: excessive salt enhances apoplastic transport in rice plants.Crossref | GoogleScholarGoogle Scholar |
Oh DH, Gong Q, Ulanov A, Zhang Q, Li Y, Ma W, Yun DJ, Bressan RA, Bohnert HJ (2007) Sodium stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNA interference line. Journal of Integrative Plant Biology 49, 1484–1496.
| Sodium stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNA interference line.Crossref | GoogleScholarGoogle Scholar |
Olías R, Eljakaoui Z, Li J, De Morales PA, Marín-Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for one copy per V1 complex. Plant, Cell & Environment 32, 904–916.
| The plasma membrane Na+/H+ antiporter SOS1 is essential for one copy per V1 complex.Crossref | GoogleScholarGoogle Scholar |
Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP (2013) Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. Journal of experimental botany 64, 445–458.
| Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress.Crossref | GoogleScholarGoogle Scholar | 23307915PubMed |
Ouyang SQ, Liu YF, Liu P, Lei G, He SJ, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. The Plant Journal 62, 316–329.
| Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants.Crossref | GoogleScholarGoogle Scholar | 20128882PubMed |
Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H (+)-ATPase. Journal of Biological Chemistry 266, 20470–20475.
| Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H (+)-ATPase.Crossref | GoogleScholarGoogle Scholar |
Pardo JM (2010) Biotechnology of water and salinity stress tolerance. Current Opinion in Biotechnology 21, 185–196.
| Biotechnology of water and salinity stress tolerance.Crossref | GoogleScholarGoogle Scholar | 20189794PubMed |
Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. Journal of Experimental Botany 57, 1181–1199.
| Alkali cation exchangers: roles in cellular homeostasis and stress tolerance.Crossref | GoogleScholarGoogle Scholar | 16513813PubMed |
Peleg Z, Apse MP, Blumwald E (2011) Chapter 12 - Engineering salinity and water-stress tolerance in crop plants: getting closer to the field. Advances in Botanical Research 57, 405–443.
| Chapter 12 - Engineering salinity and water-stress tolerance in crop plants: getting closer to the field.Crossref | GoogleScholarGoogle Scholar |
Pham J, Liu J, Bennett MH, Mansfield JW, Desikan R (2012) Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytologist 194, 168–180.
| Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection.Crossref | GoogleScholarGoogle Scholar |
Pires IS, Negrão S, Oliveira MM, Purugganan MD (2015) Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiologia Plantarum 155, 43–54.
| Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress.Crossref | GoogleScholarGoogle Scholar | 26082319PubMed |
Ponce KS, Meng L, Guo L, Leng Y, Ye G (2021) Advances in sensing, response and regulation mechanism of salt tolerance in rice. International Journal of Molecular Sciences 22, 2254
| Advances in sensing, response and regulation mechanism of salt tolerance in rice.Crossref | GoogleScholarGoogle Scholar | 33668247PubMed |
PPI – Potash and Phosphate Institute (1998) Potassium for agriculture. In ‘Better crops with plant food. Vol. 32(3)’. (Potash and Phosphate Institute: Atlanta, GA, USA)
Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. Journal of Biological Chemistry 279, 207–215.
| Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway.Crossref | GoogleScholarGoogle Scholar |
Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, Pardo JM (2011) Activation of the plasma membrane Na+/H+ antiporter salt-overlysensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proceedings of the National Academy of Sciences of the United States of America 108, 2611–2616.
| Activation of the plasma membrane Na+/H+ antiporter salt-overlysensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain.Crossref | GoogleScholarGoogle Scholar | 21262798PubMed |
Ratajczak R (2000) Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochimica et Biophysica Acta (BBA) - Biomembranes 1465, 17–36.
| Structure, function and regulation of the plant vacuolar H+-translocating ATPase.Crossref | GoogleScholarGoogle Scholar |
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37, 1141–1146.
| A rice quantitative trait locus for salt tolerance encodes a sodium transporter.Crossref | GoogleScholarGoogle Scholar | 16155566PubMed |
Rodríguez-Navarro A, Rubio F (2006) High-affinity potassium and sodium transport systems in plants. Journal of Experimental Botany 57, 1149–1160.
| High-affinity potassium and sodium transport systems in plants.Crossref | GoogleScholarGoogle Scholar | 16449373PubMed |
Rodríguez-Rosales MP, Galvez FJ, Huertas R, Aranda MN, Baghour M, Cagnac O, Venema K (2009) Plant NHX cation/proton antiporters. Plant Signaling & Behavior 4, 265–276.
| Plant NHX cation/proton antiporters.Crossref | GoogleScholarGoogle Scholar |
Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Current Opinion in Biotechnology 26, 115–124.
| Salt resistant crop plants.Crossref | GoogleScholarGoogle Scholar | 24679267PubMed |
Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663.
| Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance.Crossref | GoogleScholarGoogle Scholar | 7502075PubMed |
Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG (2012) Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. Journal of Biological Chemistry 287, 4904–4913.
| Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments.Crossref | GoogleScholarGoogle Scholar |
Russell JM (2000) Sodium-potassium-chloride cotransport. Physiological Reviews 80, 211–276.
| Sodium-potassium-chloride cotransport.Crossref | GoogleScholarGoogle Scholar | 10617769PubMed |
Schachtman DP, Schroeder J I (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658.
| Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants.Crossref | GoogleScholarGoogle Scholar | 8065452PubMed |
Schaller GE, Kieber JJ, Shiu SH (2008) Two-component signaling elements and histidyl-aspartyl phosphorelays. The Arabidopsis Book 6, e0112
| Two-component signaling elements and histidyl-aspartyl phosphorelays.Crossref | GoogleScholarGoogle Scholar | 22303237PubMed |
Shabala L, Cuin TA, Newman IA, Shabala S (2005) Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222, 1041–1050.
| Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants.Crossref | GoogleScholarGoogle Scholar | 16079998PubMed |
Shahid SA, Zaman M, Heng L (2018) Introduction to soil salinity, sodicity and diagnostics techniques. In: ‘Guideline for salinity assessment, mitigation and adaptation using nuclear and related techniques’. (Springer: Cham, Switzerland)
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences of the United States of America 97, 6896–6901.
| The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter.Crossref | GoogleScholarGoogle Scholar | 10823923PubMed |
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477.
| The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants.Crossref | GoogleScholarGoogle Scholar | 11884687PubMed |
Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology 21, 81–85.
| Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 12469134PubMed |
Shiu SH, Bleecker AB (2001a) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences of the United States of America 98, 10763–10768.
| Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases.Crossref | GoogleScholarGoogle Scholar | 11526204PubMed |
Shiu SH, Bleecker AB (2001b) Plant receptor-like kinase gene family: diversity, function, and signaling. Science’s STKE 2001, re22
| Plant receptor-like kinase gene family: diversity, function, and signaling.Crossref | GoogleScholarGoogle Scholar | 11752632PubMed |
Smajgl A, Toan T Q, Nhan DK, Ward J, Trung NH, Tri LQ, Tri VPD, Vu PT (2015) Responding to rising sea levels in the Mekong Delta. Nature Climate Change 5, 167–174.
| Responding to rising sea levels in the Mekong Delta.Crossref | GoogleScholarGoogle Scholar |
Sun J, Chen S, Dai S, Wang R, Li N, Shen X, Zhou X, Lu C, Zheng X, Hu Z, Zhang Z, Song J, Xu Y (2009) NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiology 149, 1141–1153.
| NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species.Crossref | GoogleScholarGoogle Scholar | 19028881PubMed |
Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, Schroeder JI, Ma JF, Horie T (2016) OsHKT1;4-mediated Na(+) transport in stems contributes to Na(+) exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biology 16, 22
| OsHKT1;4-mediated Na(+) transport in stems contributes to Na(+) exclusion from leaf blades of rice at the reproductive growth stage upon salt stress.Crossref | GoogleScholarGoogle Scholar | 26786707PubMed |
Sze H, Schumacher K, Muller ML, Padmanaban S, Taiz L (2002) A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends in Plant Science 7, 157–161.
| A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase.Crossref | GoogleScholarGoogle Scholar | 11950611PubMed |
Taiz L (1992) The plant vacuole. Journal of Experimental Biology 172, 113–122.
| The plant vacuole.Crossref | GoogleScholarGoogle Scholar |
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Annals of Botany 91, 503–527.
| Na+ tolerance and Na+ transport in higher plants.Crossref | GoogleScholarGoogle Scholar | 12646496PubMed |
Tran LSP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104, 20623–20628.
| Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in Xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae. Plant Physiology 122, 1249–1259.
| The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in Xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae.Crossref | GoogleScholarGoogle Scholar | 10759522PubMed |
Upadhyay A, Upadhyay AK, Bhirangi RA (2012) Expression of Na+/H+ antiporter gene in response to water and salinity stress in grapevine rootstocks. Biologia Plantarum 56, 762–766.
| Expression of Na+/H+ antiporter gene in response to water and salinity stress in grapevine rootstocks.Crossref | GoogleScholarGoogle Scholar |
Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. The Plant Cell 11, 1743–1754.
| A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor.Crossref | GoogleScholarGoogle Scholar | 10488240PubMed |
van Zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annual Review of Plant Biology 71, 403–433.
| Salt tolerance mechanisms of plants.Crossref | GoogleScholarGoogle Scholar | 32167791PubMed |
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14.
| Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance.Crossref | GoogleScholarGoogle Scholar | 14513379PubMed |
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. The Plant Cell 8, 617–627.
| SOS1, a genetic locus essential for salt tolerance and potassium acquisition.Crossref | GoogleScholarGoogle Scholar | 12239394PubMed |
Xiong L, Zhu JK (2001) Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiologia Plantarum 112, 152–166.
| Abiotic stress signal transduction in plants: molecular and genetic perspectives.Crossref | GoogleScholarGoogle Scholar | 11454221PubMed |
Yadav NS, Shukla PS, Jha A, Agarwal PK, Jha B (2012) The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biology 12, 188
| The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco.Crossref | GoogleScholarGoogle Scholar | 23057782PubMed |
Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C-terminus that regulates antiporter cation selectivity. Proceedings of the National Academy of Sciences of the United States of America 100, 12510–12515.
| Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C-terminus that regulates antiporter cation selectivity.Crossref | GoogleScholarGoogle Scholar | 14530406PubMed |
Yeo AR, Yeo ME, Flowers TJ (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. Journal of Experimental Botany 38, 1141–1153.
| The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions.Crossref | GoogleScholarGoogle Scholar |
Yuan F, Lyu MJ, Leng BY, Zheng GY, Feng ZT, Li PH, Zhu XG, Wang BS (2015) Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant, Cell & Environment 38, 1637–1657.
| Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation.Crossref | GoogleScholarGoogle Scholar |
Zhang JL, Shi H (2013) Physiological and molecular mechanisms of plant salt tolerance. Photosynthesis Research 115, 1–22.
| Physiological and molecular mechanisms of plant salt tolerance.Crossref | GoogleScholarGoogle Scholar | 23539361PubMed |
Zhao C, Zhang H, Song C, Zhu JK, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. The Innovation 1, 100017
| Mechanisms of plant responses and adaptation to soil salinity.Crossref | GoogleScholarGoogle Scholar | 34557705PubMed |
Zhou A, Bu Y, Takano T, Zhang X, Liu S (2016) Conserved V-ATPase c subunit plays a role in plant growth by influencing V-ATPase-dependent endosomal trafficking. Plant Biotechnology Journal 14, 271–283.
| Conserved V-ATPase c subunit plays a role in plant growth by influencing V-ATPase-dependent endosomal trafficking.Crossref | GoogleScholarGoogle Scholar | 25917395PubMed |
Zhu M, Zhou M, Shabala L, Shabala S (2017) Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant, Cell & Environment 40, 1009–1020.
| Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance.Crossref | GoogleScholarGoogle Scholar |