Glycine max acyl–acyl carrier protein thioesterase B gene overexpression alters lipid content and fatty acid profile of Arabidopsis seeds
Shihui Zhao A # , Fan Yan A # , Yajing Liu A , Monan Sun A , Ying Wang A , Jingwen Li A , Xinsheng Zhang A , Xuguang Yang A * and Qingyu Wang
A * and Qingyu Wang  A *
A *
A
Abstract
The fatty acyl–acyl carrier protein thioesterase B (FATB) gene, involved in the synthesis of saturated fatty acids, plays an important role in the content of fatty acid and composition of seed storage lipids. However, the role of FATB in soybeans (Glycine max) has been poorly characterised. This paper presents a preliminary bioinformatics and molecular biological investigation of 10 hypothetical FATB members. The results revealed that GmFATB1B, GmFATB2A and GmFATB2B contain many response elements involved in defense and stress responses and meristem tissue expression. Moreover, the coding sequences of GmFATB1A and GmFATB1B were significantly longer than those of the other genes. Their expression varied in different organs of soybean plants during growth, with GmFATB2A and GmFATB2B showing higher relative expression. In addition, subcellular localisation analysis revealed that they were mainly present in chloroplasts. Overexpression of GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B in transgenic Arabidopsis thaliana plants increased the seed oil content by 10.3%, 12.5%, 7.5% and 8.4%, respectively, compared to that in the wild-type and led to significant increases in palmitic and stearic acid content. Thus, this research has increased our understanding of the FATB family in soybeans and provides a theoretical basis for subsequent improvements in soybean quality.
Keywords: acyl–acyl carrier protein thioesterase B (FATB), fatty acid, gene cloning, oil content, oleic acid, overexpression, soybean (Glycine max L.), subcellular localisation.
Introduction
Soybean (Glycine max) is one of the important oil crops, with soybean oil accounting for 31% of the total edible oil production worldwide (Kim and Krishnan 2004). Although most legumes are relatively low in fat, soybeans have an exceedingly high fat content (Fischer et al. 2020), which can be divided into saturated and unsaturated fatty acids. The hydrocarbon groups of saturated fatty acids (such as palmitic and stearic acids) are alkanes consisting of C–C bonds (Thelen and Ohlrogge 2002), while the hydrocarbon groups of unsaturated fatty acids (such as oleic, linoleic and linolenic acids) are alkenyl groups containing C=C bonds (Bonaventure et al. 2003). In soybeans, linoleic acid (18-carbon chain with two double bonds, C18:2) is the most prevalent, accounting for approximately 53% of the total fatty acid content (Messina 1997). Furthermore, linoleic acid, oleic acid (C18:1), palmitic acid (C16:0), linolenic acid (C18:3) and stearic acid (C18:0) are found in large quantities in soybean, together accounting for 98% of the total fatty acid content (Prabakaran et al. 2018). The proportion of different fatty acids determines the quality of the soybean oil (Thelen and Ohlrogge 2002). It has been shown that unsaturated fatty acids, which constitute a large proportion of soybean oil, reduce its shelf life (Indiarto and Qonit 2020).
In oilseed crops, thioesterases are key enzymes that determine the concentration and composition of fatty acids (Voelker 1996). In plants, plastidial acyltransferases can terminate de novo fatty acid synthesis, and the acyl group of acyl-acyl carrier protein (acyl-ACP) can be used to produce glycerolipids in plastids (prokaryotic pathway) or alternatively, acyl-ACP thioesterases (FATs) can release free fatty acids (Yuan et al. 1995). Thus, FATs play an important role in the activity of plastid-localised fatty acid synthases and are key enzymes in the de novo synthesis of free fatty acids in higher plant plastids, as they can determine the chain length (Ma et al. 2021; Liu et al. 2022). Based on amino acid sequence and substrate specificity, plant FATs are classified into the FATA and FATB subgroups (Jones et al. 1995). FATA encodes a C18:l-ACP thioesterase, whereas the FATB encodes a thioesterase that prefers acyl-ACP with a saturated acyl group (Voelker et al. 1997; Bonaventure et al. 2003). The type B fatty acyl-ACP thioesterase encoded by the FATB gene has high affinity for saturated fatty acids as substrates (Bonaventure et al. 2003; Zheng et al. 2014), particularly for C16:0-ACP (Jones et al. 1995). Thus, substrate preference determines the fatty acid composition of different crop plants. These two types of FATs play different roles in the accumulation of plant oil. Several FAT genes have been cloned from different plants (Facciotti and Yuan 1998; Bonaventure et al. 2003; Pathak et al. 2004; Cahoon and Schmid 2008; Aznar-Moreno et al. 2016) and some of these studies have shown a significant increase in oil content in Arabidopsis thaliana seeds overexpressing the FAT genes. In these studies, the content of saturated fatty acids was remarkably decreased, while the content of unsaturated fatty acids was significantly increased (Dong et al. 2014; Ozseyhan et al. 2018). However, in transgenic lines, overexpressing the GmFATB gene, saturated fatty acid content was significantly increased, whereas unsaturated fatty acid content was significantly decreased (Dörmann et al. 2000). This indicates that the FATs regulate both saturated and unsaturated fatty acid content.
Many studies have been conducted on the soybean FATB gene. Thapa et al. (2016), Bachleda et al. (2016) and Goettel et al. (2016) showed that the mutation or deletion of GmFATB1A leads to the reduction of palmitic acid content in soybean seeds. In Arabidopsis, a GmFATB knockout mutant showed not only low saturated fatty acid content, but also slow seedling growth and low seed vigour (Bonaventure et al. 2003). Recently, CRISPR/Cas9 technology has been used to improve soybean quality (Kar et al. 2022). Kim et al. (2021) used a CRISPR/Cas9 vector with stepwise or simultaneous Golden Gate cloning and found that knockdown of GmFATB resulted in inactivation of thioesterase and prevented the accumulation of saturated fatty acids in soybean seeds. Ma et al. (2021) used CRISPR/Cas9-mediated GmFATB knockdown to significantly reduce the saturated fatty acid content in soybean seeds, demonstrating that the GmFATB gene has great potential for improving soybean oil quality and promoting human health. However, the effect of overexpression of the FATB gene in soybeans has not been studied.
In this study, we constructed transgenic Arabidopsis plants constitutively overexpressing FATB. The expression of the genes in different parts of the plants and at different growth stages was analysed, and the location of the genes in the cells was determined using subcellular localisation analysis. In addition, both the fatty acid content and the composition of the four overexpressing lines were analysed using Soxhlet extraction and gas chromatography.
Materials and methods
Plant material
Soybean (Glycine max L.) variety Williams 82 (w82) used in this experiment was provided by the Institute of Plant Sciences of Jilin University and planted at its variety trial station (43°55′22.63″N, 125°16′3216′33.00″E). Roots, stems, leaves, flowers and seeds of soybean plants cultivated under conventional field management were collected 10, 20, 30 and 40 days after flowering for subsequent analysis. Tissue samples were frozen in liquid nitrogen and stored in refrigerators at −80°C.
Identification of target FATB genes in soybean
To identify all FATB genes in soybeans, data (G. max Wm82.a4.v1) were downloaded from the SoyBase protein database (https://soybase.org/soyseq/). A local BLASTP search was performed using an Arabidopsis thaliana L. FATB protein sequence (AT1G08510) as the reference sequence, with a cut-off of E ≤ e−50. Furthermore, the conserved motifs of the FATB protein sequences in soybean were analysed using the MEME software (ver. 4.11.4). The exon structure of the candidate FATB was analysed using the TBtools software (ver. 06734), and the conserved structural domain of the FATB sequence was analysed using Pfam (ver. 35.0). In addition, cis-acting elements were analysed using TBtools. Conserved domains of FATB proteins were aligned using ClustalX, and the phylogenetic tree was constructed by MEGAX 2.0, using the maximum-likelihood method (1000 bootstrap) based on the best fit model of JTT + I + G. A phylogenetic tree based on 41 legumes and five glycine FATB proteins was reconstructed using MEGAX 2.0.
RNA extraction, gene cloning and reverse transcription quantitative PCR (RT-qPCR) assays
Samples from each of the soybean tissues were subjected to total RNA extraction using Kit 1 (PrimeScript RT Master Mix, Takara, Shiga, Japan), followed by reverse transcription to complementary DNA (cDNA) using Kit 2 (PrimeScript RT Master Mix, Takara). The candidate FATB gene was amplified using PCR with specific primers (see Supplementary Table S1; Sangon Biotech, Shanghai, China) and using cDNA as a template. The PCR procedure was as follows: initial denaturation at 95°C for 30 s, followed by 32 cycles of denaturation at 95°C for 30 s, annealing at a primer specific temperature for 30 s, extension at 72°C for 2 min, with a final extension at 72°C for 10 min. PCR products were detected using 1% agarose gel electrophoresis. The PCR products were ligated to the pMD18-T vector and validated by bidirectional sequencing. Primers used for RT-qPCR analysis are in Table S2. The GmActin gene (geneid: glyma04g39380.1) was used as an internal reference gene. RT-qPCR was performed using Power SYBR Green PCR Master Mix (Takara) with the following thermal cycle conditions: pre-denaturation at 95°C for 30 s, followed by a total of 40 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 34 s. For each sample, three biological replicates and three technical replicates were taken. The obtained GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B were fused to the N-terminal of green fluorescent proteins (GFP) in the pCAMBIA3301 vector. Further, plasmids for the above four genes were constructed using the pCHF3300 vector that contains the CaMV35S promoter (Li et al. 2014) and transformed into wild-type (WT) A. thaliana (cv. Columbia) using the Agrobacterium-mediated floral dip method for overexpression.
Subcellular localisation of GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B
Protoplasts of A. thaliana mesophyll cells were isolated from leaves for transient transformation as previously described (Vráblová et al. 2020). The plasmids (pCAMBIA3301 35S:GmFATB1A-GFP, 35S:GmFATB1B-GFP, 35S:GmFATB2A-GFP and 35S:GmFATB2B-GFP) and an empty vector were transformed into A. thaliana protoplasts using a PEG-calcium-mediated method. After 18–24 h, observations and photographs were taken at 560–630 nm using a confocal microscope.
Fatty acid analysis of GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B
Positive A. thaliana plants containing overexpression vectors were identified using PCR. T3 seeds of three independent overexpression (OE) lines, WT plants and plants containing homozygous mutation (FATB-ko) were used to analyse the fatty acid content.
Total fatty acids were extracted using the Soxhlet method. Subsequently, the composition of the fatty acids was analysed using an Agilent gas chromatography system (7890 or 8860) equipped with a flame ionisation detector and HP-5 capillary column (30 m × 0.32 mm × 0.10 μm).
Standard mixtures used were C16:0, C18:0, C18:1, C18:2 and C18:3 FAME mix (Sigma, New York, NY, USA). Samples with three replications were prepared independently for fatty acid analysis.
Results
Cloning and characterisation of the FATB gene
Ten potential FATB genes with complete open reading frames, designated as GmFATB1A to GmFATB5B, were obtained from G. max. Nine of these genes (except GmFATB4A) were then successfully cloned from W82 using PCR (Fig. S1). The sequencing results of the nine cloned genes were compared with the results of transcriptome analysis. The NJ tree (Fig. 1a) showed that all four soybean cultivars have approximately 10 FATB proteins, which can be clustered into five clades. Furthermore, each clade contains two proteins except for the FATB2A clade, which contains five proteins. In this study, the individual clade is referred to as a subgroup, and each subgroup includes two branches (Fig. 1a); branch A and B. Moreover, the FATB genes of the four cultivars were distributed among the clades, indicating that the FATB genes in soybean are highly homologous and conserved, and that the FATB genes existed before the differentiation of soybean species.
(a) Phylogenetic analysis of FATB proteins from four soybean cultivars (Williams 82, Fiskeby, Lee and Glycine soja). (b) Motif distribution and exon–intron structures of FATBs. i. Phylogenetic tree based on the full-length sequences of soybean FATB proteins constructed using MEGAX software. ii. MEME motif distributions in FATB proteins. iii. The exon–intron structures of the FATB genes.
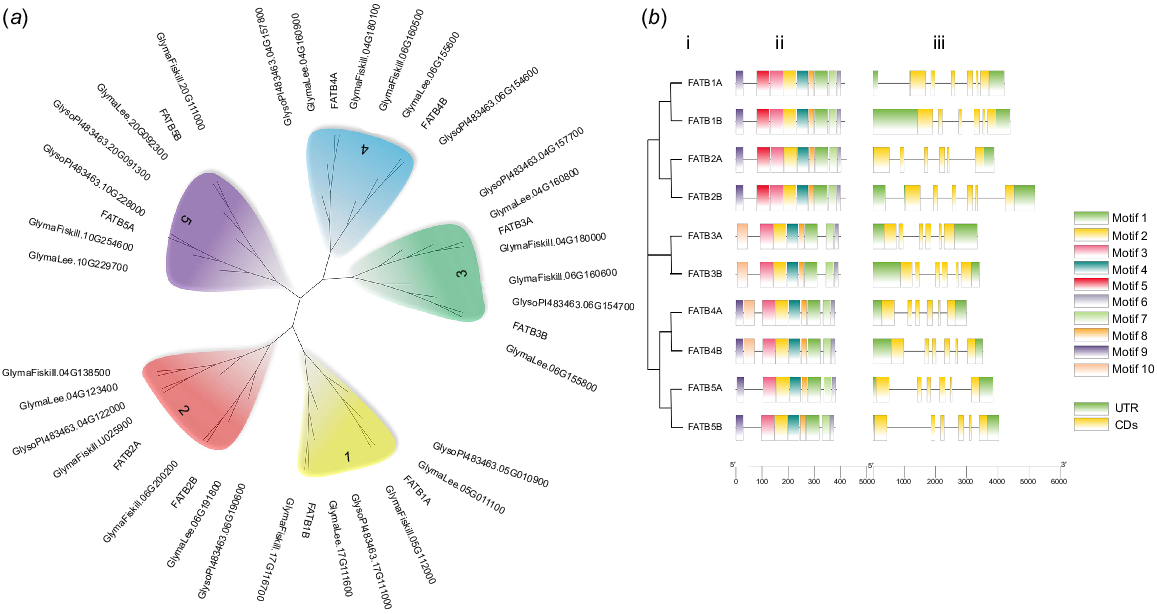
In total, 10 conserved patterns of 14–50 amino acids were identified (Fig. S4). Motifs 1, 2, 3, 4, 6, 7 and 8 were present in all GmFATB proteins (Fig. 1b), whereas motif 9 was not present in the GmFATB5 subgroup; motif 5 was only present in the GmFATB1 and GmFATB2 subgroups; and motif 10 was only present in the GmFATB3 and GmFATB5 subgroups. According to Facciotti’s research, the catalytic active region of FATB is in motif 1, Cys-49 and His-14, localise to the same region of the model as catalytic residues found in other enzymes with hot dog folds. Asn-12 mutation to Ala results in a reduction of enzyme activity by >200 times. Fig. 1b shows that the lengths of the FATB genes ranged from 3 to 5 kb, and all these genes contained six exons and five introns. Previous studies (Facciotti and Yuan 1998) have shown that most plant FATBs contain about 60 amino acid Signal peptide sequences. In addition, it shows that FATB1 and FATB2 subgroup members contain the acyl-thio_N domain, and all FATB members contain the acyl-ACP_TE domain (Fig. S3).
Analysis of cis-acting elements showed that the entire family of genes can be divided into three categories: hormone responses, biotic and abiotic responses, and development and metabolism. Most FATB genes contained cis-acting elements involved in responses to plant hormones, such as jasmonic acid, abscisic acid and gibberellin (Fig. S2).
Expression pattern of soybean FATB gene family members
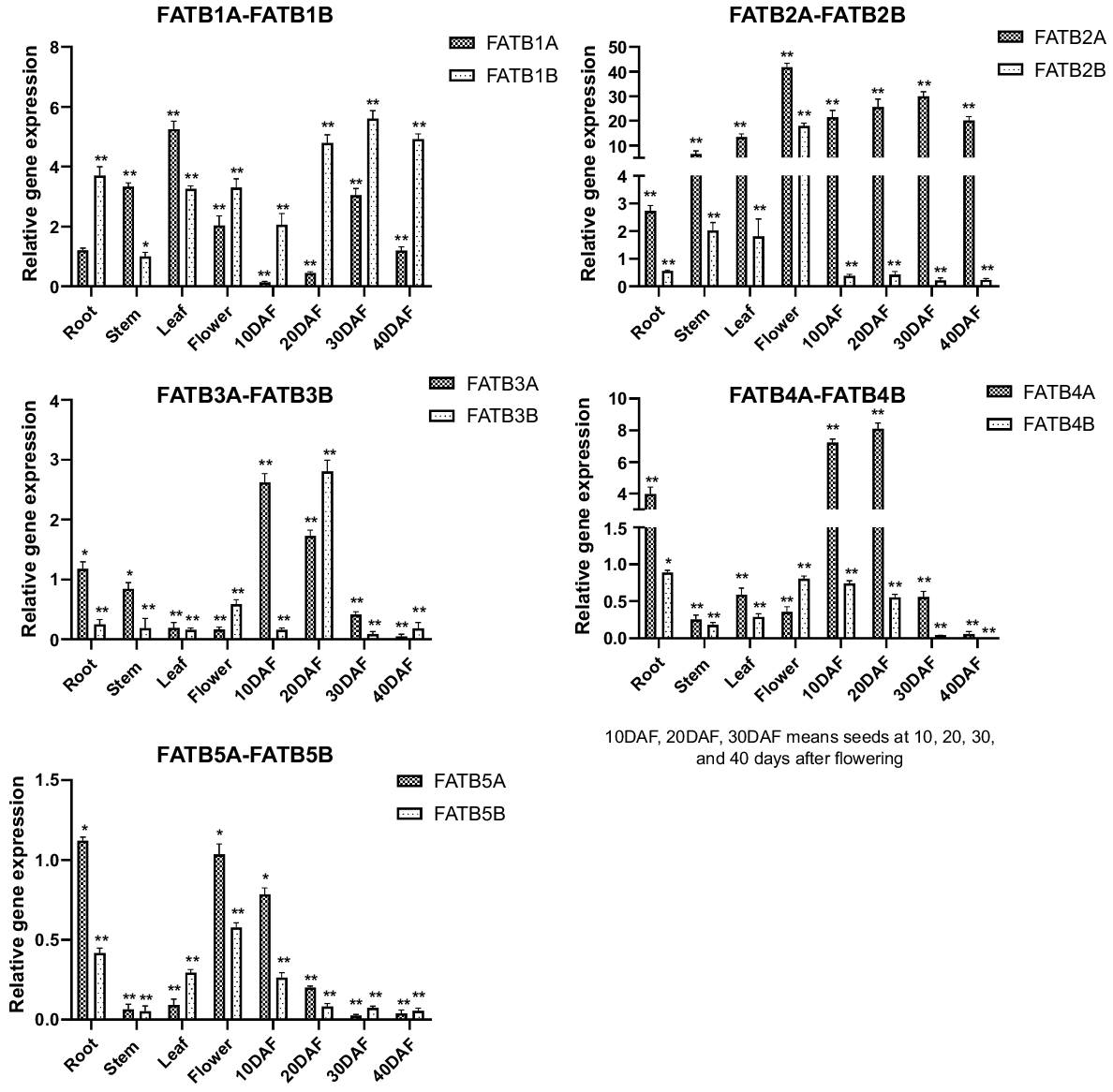
Relative gene expression of all GmFAT genes were compared to the expression level of GmFATB1A root as control samples. The patterns of expression of GmFATB genes differed in various tissues and organs, as well as in different developmental stages of the seeds. Significant differences occurred in the expression of various genes in the GmFATB gene family in different organs and tissues. As shown in Fig. 2, GmFATB2A and GmFATB2B are commonly expressed in many tissues, but their expression levels are very high, followed by GmFATB1A and GmFATB1B. The expression of GmFAT3B is low in all organs and tissues. GmFATB2A is highly expressed during flowering and seed maturation. During seed maturation, the expression level of GmFATB2B is lower than that of GmFAT2A. The expression of GmFATB4A in roots is higher than that in other tissues and organs. Meanwhile, other members of the GmFATB gene family were not expressed at high levels in the organs and tissues studied.
Subcellular localisation of FATB genes
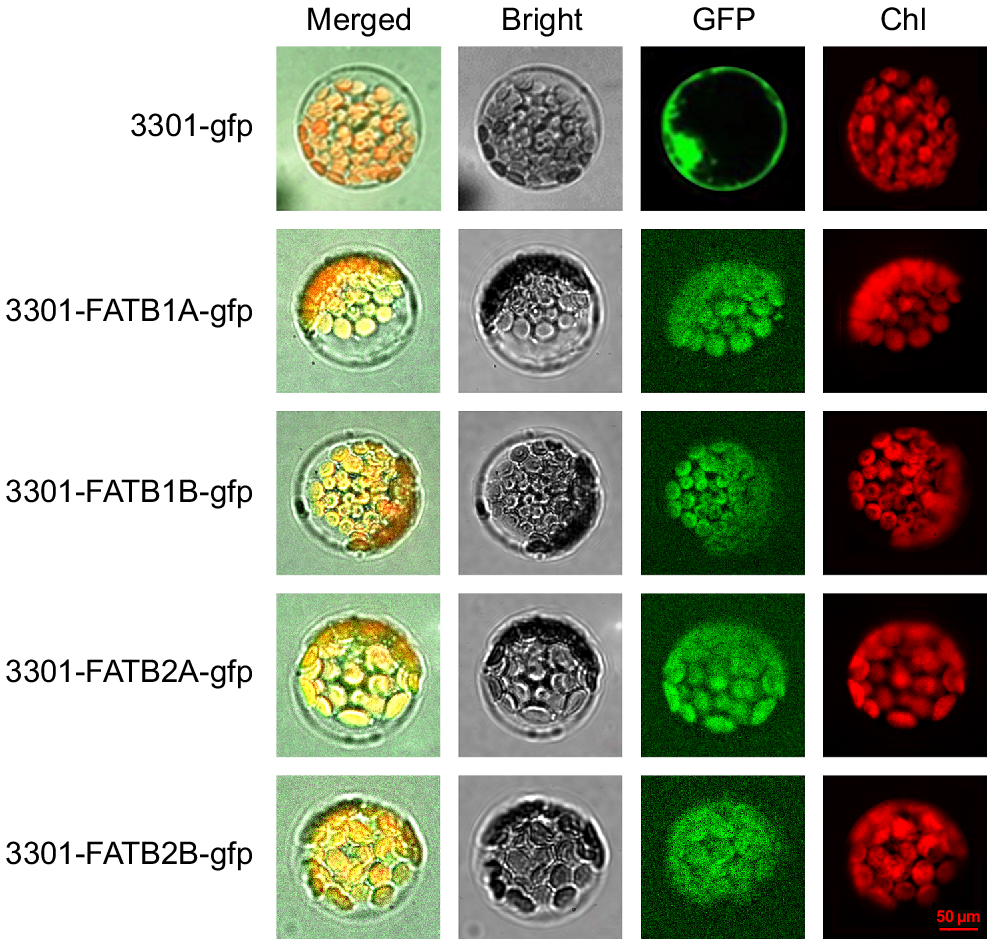
To further investigate the location of FATB genes in vivo, four highly expressed genes were selected for subcellular localisation studies, as follows. The successful construction of the plant recombinant vectors (pCAMBIA-3301-FATB1A-gfp, pCAMBIA-3301-FATB1B-gfp, pCAMBIA-3301-FATB2A-gfp and pCAMBIA-3301-FATB2B-gfp) was verified using bacterial fluid PCR. The pCAMBIA3301-GFP used as a positive protein control was detected in the the cell nuclear membrane and cell membrane. The subcellular localisation analysis of Arabidopsis protoplast species showed that the green fluorescence emitted by these four recombinant fusion proteins was consistent with the red autofluorescence of chloroplasts in red, indicating that all four of these genes are located within chloroplasts (Fig. 3).
Cellular localisation of GmFATB1A, GmFATB1B, GmFATB2A and GmFATB1B isoforms in Arabidopsis protoplasts. Bright means fields in transformed protoplasts. The green fluorescence of the GFP signal is shown in green, and the red fluorescence of plastid autofluorescence is shown in red. The scale bar is 50 μm, and all panels are at the same magnification for subcellular localisation.
Ectopic expression of GmFATB in A. thaliana
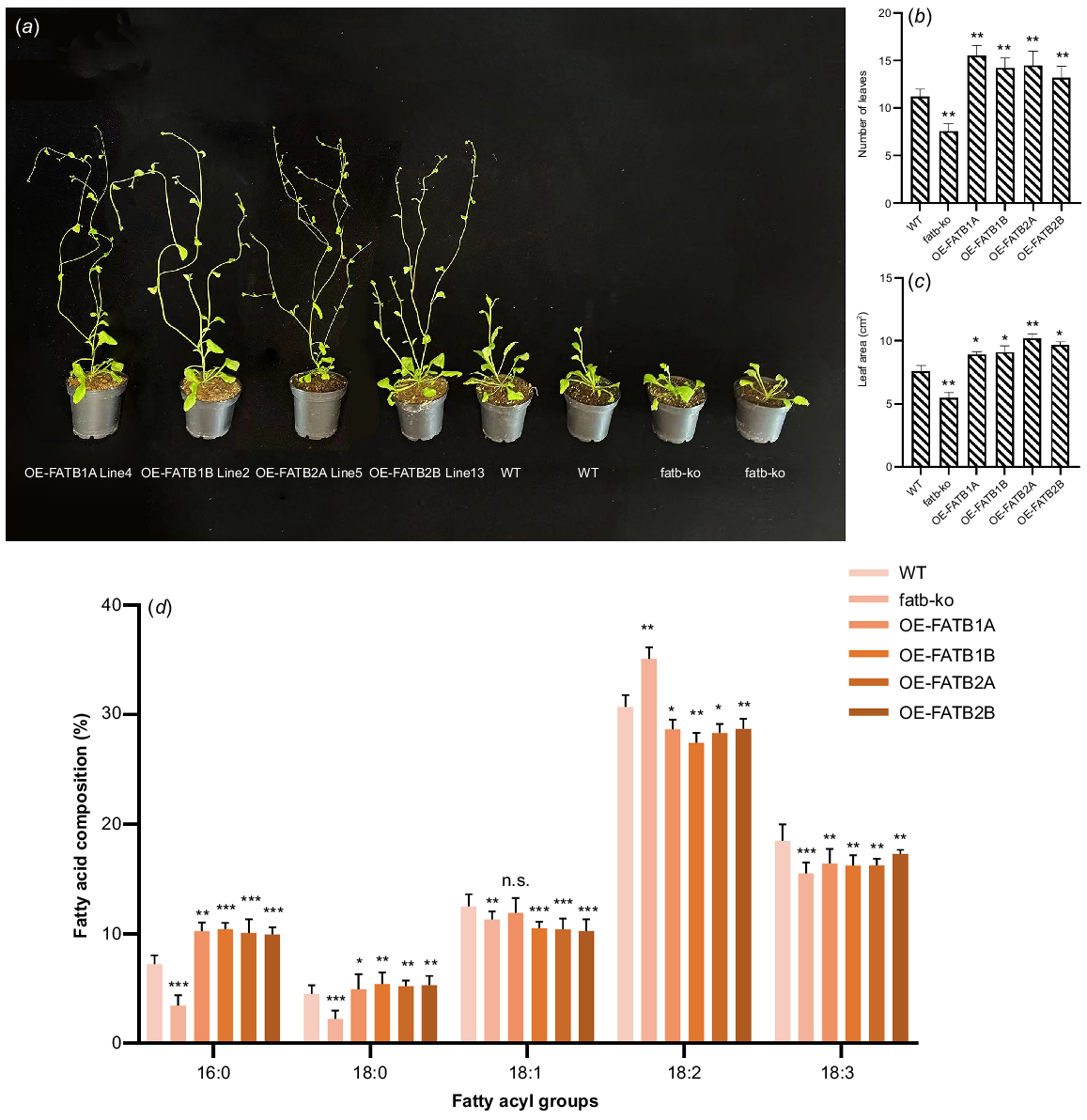
Plants that constitutively express GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B (35S:: GmFATB) were analysed. The plants carrying the GmFATB gene developed significantly earlier than WT Arabidopsis. As shown in Fig. 4a, under the same treatment, after 24 days of sowing, plants overexpressing GmFAT genes have entered the flowering stage, WT is still in the vegetative development stage, and mutant plants develop slowly. Early studies have shown that the number of rosette leaves in the flowering process of Arabidopsis can reflect the developmental time of its nutritional stage (Lièvre et al. 2016). At their first bloom time, we counted the rosette leaf number overexpressed plants, WT and mutant Arabidopsis (Fig. 4b). Additionally, the result showed the rosette leaf number of overexpressed plants lines was significantly higher than in WT Arabidopsis. (average 2.5) (Fig. 4b; t-test, P < 0.05 or P < 0.01). In addition, 35S::GmFATB-expressing plants had greater leaf area at the germination stage than was seen with the control plants (average, 1.85 cm2) (Fig. 4c; t-test, P < 0.05 or P < 0.01).
(a) Early flowering phenotype of 35S::GmFATB transgenic Arabidopsis plants. (b) Number of rosette leaves. (c) Pre-flowering leaf area. (d) Analysis of the differences in total fatty acid content in different transgenic plants in comparison to the WT. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001 (based on t-test).
Effects of GmFatB gene expression on fatty acid content
Compared with that of WT Arabidopsis, the total lipid content of transgenic plants was significantly (t-test, P < 0.05) increased. GmFATB1A-, GmFATB1B-, GmFATB2A- and GmFATB2B-overexpressing plants showed increased oil content in seeds by 10.3%, 12.5%, 7.5% and 8.4%, respectively, compared with that seen in the control.
The fatty acid contents in plants overexpressing GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B, as well as in WT and fatb-ko A. thaliana mutant (Fig. 4d). The contents of two saturated fatty acids (palmitic and stearic acids) were significantly higher in A. thaliana strains overexpressing GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B than in WT Arabidopsis. The proportion of saturated fatty acids, including palmitic and stearic acids, decreased significantly in the mutant compared to the WT Arabidopsis. In strains overexpressing GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B, the content of oleic acid decreased slightly, and that of linoleic acid also decreased, compared with that in the WT Arabidopsis. Overexpression of each of the GmFATB1A genes in Arabidopsis can change the composition of fatty acids in Arabidopsis seeds, which shows that the soybean GmFATB1A gene plays an important role in regulating the proportion of fatty acids in fatty acid synthesis.
Discussion
The soybean FATB gene family contains several GmFATB genes, but only one gene is present in Arabidopsis, which may be related to the whole-genome replication in soybeans and their role in the accumulation of oil in soybean seeds. We designed primers for 10 genes, and notably did not clone GmFATB4A. Since GmFATB4A was not expressed in flowers and seeds and only slightly expressed in leaves, we speculated that GmFATB4A may be non-functional. We identified similar numbers and compositions of motifs across the different subgroups. However, only members of the GmFATB1 and GmFATB2 groups had an acylthio-N structural domain. Current research has shown that the FATB1a gene can affect seed palmitic acid content, but no other studies have shown that GmFATB1A has other effects on plant growth and development, and the role of its homologue (FATB1B) is also unclear (Bonaventure et al. 2003). Therefore, more detailed studies are needed to gain insight into these genes.
The results of the promoter sequence analysis revealed that the following four types of responsive elements were present in the highest numbers: (1) abscisic acid-responsive; (2) element essential for anaerobic induction; (3) photoresponsive, and (4) light-responsive element. There is no significant difference in the distribution and abundance of response elements among different soybean fat B subgroups. RT-qPCR results of GmFATB gene family were consistent with the expression trend of Transcriptome of Phytozome soybean. The expression levels of soybean FATB vary among different subgroups, with similar expression levels in the same subgroup and similar expression trends in various tissues. Furthermore, the expression patterns showed that although GmFATB2A and GmFATB2B showed relatively high expression in flowers, GmFATB2B showed considerably low expression in seeds. This suggests that different genes in the GmFATB gene family may play regulatory roles in separate parts of the plant.
Overexpression of GmFATB1A, GmFATB1B, GmFATB2A and GmFATB2B changed the free fatty acid content in Arabidopsis seeds significantly. By contrast, previous knockout experiments using FATB1 have revealed that FATB1A and FATB1B single mutants have reduced palmitic (11%) and stearic acid (21%) content in leaves. The double mutant FATB1A:1B had reduced palmitic (42%) and stearic acid (35%) content, showed growth defects, and had sterile male organs. These findings demonstrated the important role of the FATB1 gene in the regulation of fatty acids in seeds. In this study, further analysis revealed that GmFATB1B yielded the greatest effect while GmFATB2B yielded the least effect.
Previous studies (Cardinal et al. 2007; Bachleda et al. 2016; Zhou et al. 2019) have mainly focused on the effects of FATB mutants on the fatty acid composition of soybean oil. However, this study investigated the effect of overexpressing members of the FATB gene family on fatty acid content and the function of FATB genes using different methods. To gain an insight into the mechanisms by which FATB genes regulate fatty acid content and composition, further studies are needed to investigate the transcription factors involved in the regulation of the FATB gene family, and to explore the relationships between the individual genes of the entire gene family.
Data availability
The data that support this study are available in the article and accompanying online supplementary material.
Declaration of funding
This research was funded by the Major Science and Technology Sponsored Program for Transgenic Biological Breeding (No. 2016ZX08004-003), the National Natural Science Foundation of China (No. 32001572), and the Science and Technology Department of Jilin Province (No. 20180201030).
References
Aznar-Moreno JA, Venegas-Calerón M, Martínez-Force E, Garcés R, Salas JJ (2016) Acyl carrier proteins from sunflower (Helianthus annuus L.) seeds and their influence on FatA and FatB acyl-ACP thioesterase activities. Planta 244, 479-490.
| Crossref | Google Scholar | PubMed |
Bachleda N, Pham A, Li Z (2016) Identifying FATB1a deletion that causes reduced palmitic acid content in soybean N87-2122-4 to develop a functional marker for marker-assisted selection. Molecular Breeding 36, 45.
| Crossref | Google Scholar |
Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB (2003) Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. The Plant Cell 15, 1020-1033.
| Crossref | Google Scholar | PubMed |
Cahoon EB, Schmid KM (2008) Metabolic engineering of the content and fatty acid composition of vegetable oils. Advances in Plant Biochemistry and Molecular Biology 1, 161-200.
| Crossref | Google Scholar |
Cardinal AJ, Burton JW, Camacho-Roger AM, Yang JH, Wilson RF, Dewey RE (2007) Molecular analysis of soybean lines with low palmitic acid content in the seed oil. Crop Science 47, 304-310.
| Crossref | Google Scholar |
Dong S, Huang J, Li Y, Zhang J, Lin S, Zhang Z (2014) Cloning, characterization, and expression analysis of acyl–acyl carrier protein (ACP)-thioesterase B from seeds of Chinese Spicehush (Lindera communis). Gene 542, 16-22.
| Crossref | Google Scholar | PubMed |
Dörmann P, Voelker TA, Ohlrogge JB (2000) Accumulation of palmitate in Arabidopsis mediated by the acyl-acyl carrier protein thioesterase FATB1. Plant Physiology 123, 637-644.
| Crossref | Google Scholar | PubMed |
Facciotti MT, Yuan L (1998) Molecular dissection of the plant acyl-acyl carrier protein thioesterases. European Journal of Lipid Science and Technology 100, 167-172.
| Crossref | Google Scholar |
Fischer E, Cachon R, Cayot N (2020) Pisum sativum vs Glycine max, a comparative review of nutritional, physicochemical, and sensory properties for food uses. Trends in Food Science & Technology 95, 196-204.
| Crossref | Google Scholar |
Goettel W, Ramirez M, Upchurch RG, An Y-QC (2016) Identification and characterization of large DNA deletions affecting oil quality traits in soybean seeds through transcriptome sequencing analysis. Theoretical and Applied Genetics 129, 1577-1593.
| Crossref | Google Scholar | PubMed |
Indiarto R, Qonit MAH (2020) A review of soybean oil lipid oxidation and its prevention techniques. International Journal of Advanced Science and Technology 29, 5030-5037.
| Google Scholar |
Jones A, Davies HM, Voelker TA (1995) Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. The Plant Cell 7, 359-371.
| Crossref | Google Scholar | PubMed |
Kar SR, Choudhury S, Chakraborty A (2022) CRISPR/Cas9 for soybean improvement: a review. Asia Pacific Journal of Molecular Biology and Biotechnology 30, 40-56.
| Crossref | Google Scholar |
Kim W-S, Krishnan HB (2004) Expression of an 11 kDa methionine-rich delta-zein in transgenic soybean results in the formation of two types of novel protein bodies in transitional cells situated between the vascular tissue and storage parenchyma cells. Plant Biotechnology Journal 2, 199-210.
| Crossref | Google Scholar | PubMed |
Kim W-N, Kim H-J, Chung Y-S, Kim H-U (2021) Construction of multiple guide RNAs in CRISPR/Cas9 vector using stepwise or simultaneous Golden Gate cloning: case study for targeting the FAD2 and FATB multigene in soybean. Plants 10, 2542.
| Crossref | Google Scholar | PubMed |
Li JT, Yu G, Sun XH, Jia CG, Du Q, Li QY, Pan HY (2014) Modification of vectors for functional genomic analysis in plants. Genetics and Molecular Research 13, 7815-7825.
| Crossref | Google Scholar | PubMed |
Lièvre M, Granier C, Guédon Y (2016) Identifying developmental phases in the Arabidopsis thaliana rosette using integrative segmentation models. New Phytologist 210, 1466-1478.
| Crossref | Google Scholar | PubMed |
Liu Y, Han J, Li Z, Jiang Z, Luo L, Zhang Y, Chen M, Yang Y, Liu Z (2022) Heterologous expression of Jatropha curcas fatty acyl-ACP thioesterase A (JcFATA) and B (JcFATB) affects fatty acid accumulation and promotes plant growth and development in Arabidopsis. International Journal of Molecular Sciences 23, 4209.
| Crossref | Google Scholar |
Ma J, Sun S, Whelan J, Shou H (2021) CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. International Journal of Molecular Sciences 22, 3877.
| Crossref | Google Scholar | PubMed |
Ozseyhan ME, Li P, Na G, Li Z, Wang C, Lu C (2018) Improved fatty acid profiles in seeds of Camelina sativa by artificial microRNA mediated FATB gene suppression. Biochemical and Biophysical Research Communications 503, 621-624.
| Crossref | Google Scholar | PubMed |
Pathak MK, Bhattacharjee A, Ghosh D, Ghosh S (2004) Acyl–acyl carrier protein (ACP)-thioesterase from developing seeds of Brassica campestris cv. B-54 (Agrani). Plant Science 166, 191-198.
| Crossref | Google Scholar |
Prabakaran M, Lee J-H, Ahmad A, Kim S-H, Woo K-S, Kim M-J, Chung I-M (2018) Effect of storage time and temperature on phenolic compounds of soybean (Glycine max L.) flour. Molecules 23, 2269.
| Crossref | Google Scholar |
Thapa R, Carrero-Colón M, Hudson KA (2016) New alleles of FATB1A to reduce palmitic acid levels in soybean. Crop Science 56, 1076-1080.
| Crossref | Google Scholar |
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metabolic Engineering 4, 12-21.
| Crossref | Google Scholar | PubMed |
Voelker T (1996) Plant acyl-ACP thioesterases: chain-length determining enzymes in plant fatty acid biosynthesis. Genetic Engineering 18, 111-133.
| Crossref | Google Scholar | PubMed |
Voelker TA, Jones A, Cranmer AM, Davies HM, Knutzon DS (1997) Broad-range and binary-range acyl-acyl-carrier protein thioesterases suggest an alternative mechanism for medium-chain production in seeds. Plant Physiology 114, 669-677.
| Crossref | Google Scholar | PubMed |
Vráblová M, Vrábl D, Sokolová B, Marková D, Hronková M (2020) A modified method for enzymatic isolation of and subsequent wax extraction from Arabidopsis thaliana leaf cuticle. Plant Methods 16, 129.
| Crossref | Google Scholar | PubMed |
Yuan L, Voelker TA, Hawkins DJ (1995) Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proceedings of the National Academy of Sciences of the United States of America 92, 10639-10643.
| Crossref | Google Scholar |
Zheng P, Babar MDA, Parthasarathy S, Gibson R, Parliament K, Flook J, Patterson T, Friedemann P, Kumpatla S, Thompson S (2014) A truncated FatB resulting from a single nucleotide insertion is responsible for reducing saturated fatty acids in maize seed oil. Theoretical and Applied Genetics 127, 1537-1547.
| Crossref | Google Scholar | PubMed |
Zhou Z, Lakhssassi N, Cullen MA, El Baz A, Vuong TD, Nguyen HT, Meksem K (2019) Assessment of phenotypic variations and correlation among seed composition traits in mutagenized soybean populations. Genes 10, 975.
| Crossref | Google Scholar | PubMed |