Long-read sequencing in fungal identification
Minh Thuy Vi Hoang A B , Laszlo Irinyi A B C and Wieland Meyer A B C D E *A Molecular Mycology Research Laboratory, Centre for Infectious Diseases and Microbiology, Faculty of Medicine and Health, Sydney Medical School, Westmead Clinical School, The University of Sydney, Sydney, NSW 2006, Australia.
B Molecular Mycology Research Laboratory, Centre for Infectious Diseases and Microbiology, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia.
C Sydney Institute for Infectious Diseases, The University of Sydney, Sydney, NSW 2006, Australia.
D Westmead Hospital (Research and Education Network), Westmead, NSW 2145, Australia.
E Curtin Medical School, Curtin University, Perth, WA 6102, Australia.

Minh Thuy Vi Hoang is a PhD student at the University of Sydney at the Sydney Medical School. Her doctoral work explores the application of long-read sequencers to the diagnosis of fungal diseases. Prior, she received a Bachelor of Medical Science (First Class Honours) in Microbiology at the University of Sydney. |

Dr Laszlo Irinyi is a Postdoctoral Fellow at Westmead Institute for Medical Research, Westmead, NSW, Australia and his research focuses on the identification and molecular taxonomy of human and animal pathogenic fungi. Currently his focus is on the adaptation of new generation sequencing technologies in routine diagnostics for fungal identification. |

Professor Wieland Meyer is a Molecular Medical Mycologist and academic at the Faculty of Medicine and Health, Sydney Medical School; The University of Sydney, Associate Dean of Curtin Medical School, Curtin University, and the Fundação Oswaldo Cruz (FIOCRUZ) in Rio de Janeiro, Brazil, heading the MMRL within the CIDM, Westmead Institute for Medical Research, with a PhD in fungal genetics from the Humboldt University of Berlin, Germany. His research focuses on phylogeny, molecular identification, population genetics, molecular epidemiology, and virulence mechanisms of human and animal pathogenic fungi. He is the Convener of the Mycology Interest Group of ASM, and the President of the International Mycological Association (IMA). |
Microbiology Australia 43(1) 14-18 https://doi.org/10.1071/MA22006
Submitted: 23 December 2021 Accepted: 11 February 2022 Published: 8 April 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the ASM. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Long-read sequencing is currently supported by sequencing platforms from Pacific Biosciences and Oxford Nanopore Technologies, both of which generate ultra-long reads. Metabarcoding and metagenomics are the two approaches used when implementing sequencing. Metabarcoding involves the amplification and sequencing of selected nucleic acid regions, while in a metagenomic approach extracted nucleic acids are sequenced directly without prior amplification. Both approaches have associated advantages and disadvantages, which, in combination with long-read sequencing, provide a promising new approach for fungal identification and diagnosis of mycoses, on which we will reflect in this short review.
Keywords: diagnostics, DNA barcoding, fungal identification, long-read sequencing, metabarcoding, metagenomics, mycoses, next generation sequencing.
Long-read sequencing
Long-read sequencing technologies are characterised by the potential to generate ultra-long reads over 10 kb in one run.1 Pacific Biosciences (PacBio) first released their long-read sequencing instrument in 2011 and most recently released the Sequel IIe (Fig. 1).2 PacBio sequencing is based on the conversion of fluorescent signals produced when nucleotides are bound to a template strand via a polymerase. The template double-stranded DNA has two hairpin adaptors bound to each end and so sequencing continues around the template for the duration of the polymerase lifetime. PacBio sequencing has achieved high accuracy, which is reported to be 99.8%, and produces a high throughput with parallel sequencing of millions of template strands.3 However, PacBio sequencing is a trade-off between read length and read quality: as longer template strands are sequenced, there is often a less accurate consensus sequence. Additionally, library preparation is estimated to be 1 day, which may not be suitable for time sensitive applications. Oxford Nanopore Technologies (ONT) began commercial release in 2015 with their MinION sequencer and has since released instruments and flow cells for a variety of throughput requirements (Fig. 2).4 The basis of ONT sequencers are biological nanopores fixed in membranes, of which nucleotide strands travel through to cause a current change, which is then translated into a sequence. ONT sequencing read length is only limited by sample DNA length, quantity, and purity and as they provide a wide array of library preparation kits and throughput options it can be scaled to any potential use intended. Additionally, the low initial investment and small portable size of some ONT sequencers allow sequencing to be performed in locations outside the laboratory.2 The major drawback of ONT is the lower read accuracy reported, although computation intensive bioinformatic tools are available to increase accuracy to 99%.5 Samples that result in low DNA quantity, quality, and short read lengths do not take full advantage of ONT sequencing and impede sequencing by blocking and inactivating pores. These sequencing technologies are further discussed in other studies.2

|

|
Studies using PacBio and ONT long-read sequencers to sequence and identify fungal species using metabarcoding and metagenomic approaches are described below to demonstrate the potential use for long-read sequencers in fungal identification.
Metabarcoding
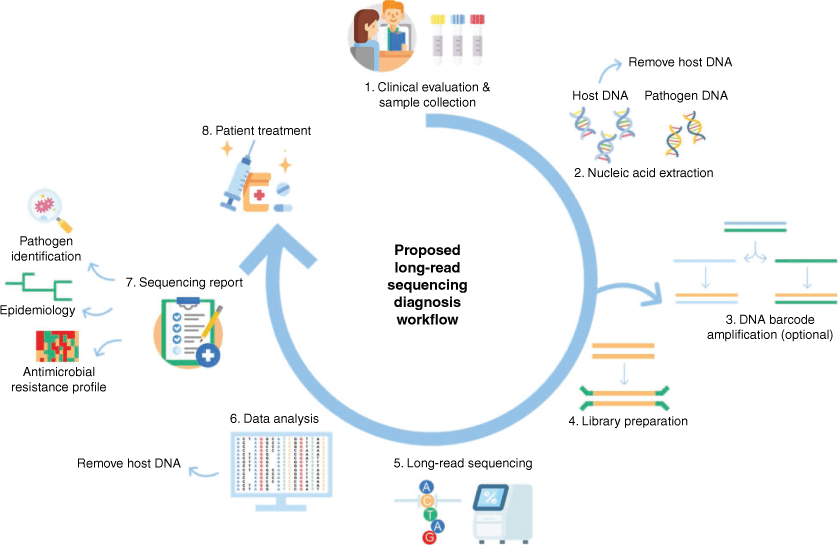
Metabarcoding involves the sequencing of targeted nucleic acid regions within environmental and clinical samples. This approach combines DNA barcoding with high throughput sequencing of specific taxonomic regions (barcodes). These barcodes, which have high taxonomic coverage and high resolution, are amplified and sequenced (Fig. 3).6 The generated barcode sequences are clustered into operational taxonomic units (OTUs) based on sequence similarity and compared against databases containing reference barcode sequences to provide an accurate representation of the microbial population. The advantages of this approach are the low level of genomic material required and faster and less complex computational analysis. However, as only specific regions are sequenced, information is limited to identification, although this may not be an issue for all studies. Prior amplification of a sample also potentially introduces bias, which may result in the inaccurate representation of the microbial community.6 Previous microbial metabarcoding studies with short-read sequencing technologies used micro-barcodes that spanned less than 600 bp, which were often shorter than the full-length barcoding regions.7 Long-read sequencers can span beyond the full-length barcode regions and resolve longer structural variations that are challenging for short-read sequencers, leading to higher discriminatory power.
|
Long-read fungal metabarcoding studies primarily use the full-length internal transcribed spacer (ITS) region of the rRNA gene cluster to identify fungal species, although shorter barcode regions are also used. To validate the ability of long-read sequencers to identify fungal species, ONT and PacBio have been used to identify members of mock fungal communities. The full-length ITS and the ITS1 regions were shown to accurately identify 16 and 26 fungal species within mock communities respectively using PacBio sequencing.8,9 The full-length ITS region in conjunction with ONT sequencing has also successfully identified fungal species in mock communities.10,11 The ITS region was found to be the superior locus for fungal identification in nanopore sequencing although, in a mock community with varying abundance of 16 fungal species, species level identification was only achieved for 1/3 of fungal species.7 Metabarcoding of clinical samples with nanopore sequencing of the ITS region, has been explored extensively. Pathogens were identified from nine positive blood culture bottles which were then verified by routine diagnosis, one of which was a Candida albicans infection.5 Type strains of five Candida species were also identified to sufficient (100–200×) coverage and nanopore sequencing errors did not affect correct species identification.5 Full-length ITS nanopore sequencing has also identified potential pathogens in patient samples previously negative by traditional diagnostic methods.11,12 PacBio has also been used in the clinical space to characterise the gut mycobiome of 14 healthy individuals with the ITS1 locus.9 PacBio sequencing has primarily been applied to metabarcoding of environmental samples and has been demonstrated to outperform nanopore sequencing in such applications due to nanopore sequencing errors.13 Metabarcoding of samples, such as tree roots,14 soil,13 mangrove sediments,15 and lake water,7 have revealed the potential of PacBio targeting the full-length ITS region to be used in broad ecological studies requiring accurate characterisation of the mycobiome of complex environmental samples. Metabarcoding of fungi using long-read sequencing has been established to be a promising avenue for the identification and characterisation of fungal species in clinical and environmental samples.
Metagenomics
Metagenomic sequencing, also known as shotgun sequencing, aims to sequence all genetic material within a sample. In this approach, all genetic material (DNA or RNA) is extracted from the primary samples, which are then fragmented and undergo library preparation to suit the sequencing technology. The sequencing library then undergoes in-depth sequencing and data analysis (Fig. 3).16 An advantage of metagenomics over metabarcoding is higher resolution, as more parts of the genomes of every organism in the sample are sequenced, potentially generating information beyond identification, such as antimicrobial resistance and virulence. The direct sequencing of genetic material eliminates the need for prior culturing and amplification, reducing the time from sample collection to identification, eliminating any bias that may occur due to these additional steps, resulting in a more accurate representation of the community composition.17 In metagenomic sequencing studies, the overwhelming amount of background DNA compared to microbial DNA remains a challenge and methods to enrich microbial DNA have been developed.18 Additionally, the cost and bioinformatic requirements of metagenomic sequencing is generally higher than those of metabarcoding studies. For full use of metagenomic sequencing reads, robust reference whole genome sequences are required. Long-reads generated in metagenomic studies are more correctly mapped to reference genomes and give high discriminatory power for accurate identification. However, the main current hurdles for long-read metagenomics based fungal identification are the fact that current genome databases lack adequate coverage for all fungal species.19
Metagenomic studies with long-read sequencers to identify fungal species are currently limited. However, preliminary studies have demonstrated their promising potential. PacBio sequencing of skin samples using a metagenomic approach identified a similar microbial community to short-read sequencing.20 Additionally, metagenomic PacBio sequencing has been used in conjunction with short-read sequencing for genome assembly of fungal species in complex lichen samples.21,22 Metagenomic nanopore sequencing identified pathogens from 87 patient samples in a single hospital study from a range of infections, including fungal infections, achieving sensitivity and specificity of 90.9 and 100% respectively, outperforming short-read sequencing.23 The same approach was applied to three patient samples positive for Pneumocystis jirovecii and three negative respiratory samples.19 All positive samples returned reads identified as P. jirovecii. However, P. jirovecii was also detected in negative samples. Furthermore, fungal species were identified that are geographically restricted to areas that did not align with the patient’s travel histories. These likely misidentifications were attributed to issues applying the bioinformatics tools to fungal identification. Additionally, all samples reported 77–95% Homo sapiens reads aside from one outlier (10%) demonstrating the high abundance of human background DNA.19 Studies involving methods to overcome the limitations of metagenomic sequencing have emerged. The combination of metagenomic sequencing and whole genome amplification has been utilised to increase DNA quantity whilst maintaining the community composition to characterise the microbiome on the surface of oil paintings.24 A method to deplete human DNA in clinical samples has been applied to respiratory samples and confirmed an Aspergillus infection previously diagnosed by culturing.25 Additionally, this method also identified fungemia caused by Candida glabrata within 24 h whilst an extended culturing time (48–72 h to actionable results) was required for traditional identification.25 Although there are currently drawbacks, with methods to overcome its limitations, long-read metagenomics is a favourable prospect for fungal identification.
Conclusion
Sequencing of genetic material has taken a significant step forward with the release of PacBio and ONT long-read sequencers. Their initial applications to fungal identification have indicated the promising potential for routine use in clinical diagnostics, microbiome characterisation, whole genome assembly and more, if the limiting factors of DNA extraction, low fungal-human sample DNA range, lack of reference sequences (DNA barcodes and whole genomes), and lack of bioinformatic tools, can be overcome. Implementation of long-read sequencing to diagnosis of fungal infections additionally requires a standardised workflow (Fig. 3). If successfully introduced into routine diagnosis of fungal infections it would drastically reduce turnaround time, from currently several days/weeks to less than 24 h (Fig. 3), enabling a timely and accurate induction of antifungal treatment, reducing mortalities, treatment and hospitalisation costs.
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This study was supported by a National Health and Medical Research Council of Australia (NHMRC; grant no. APP1121936) to Wieland Meyer.
References
[1] Pollard, MO et al.. (2018) Long reads: their purpose and place. Hum Mol Genet 27, R234–R241.| Long reads: their purpose and place.Crossref | GoogleScholarGoogle Scholar | 29767702PubMed |
[2] Tedersoo, L (2021) Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl Environ Microbiol 87, e0062621.
| Perspectives and benefits of high-throughput long-read sequencing in microbial ecology.Crossref | GoogleScholarGoogle Scholar | 34132589PubMed |
[3] Tedersoo, L et al.. (2018) PacBio metabarcoding of fungi and other eukaryotes: errors, biases and perspectives. New Phytol 217, 1370–1385.
| PacBio metabarcoding of fungi and other eukaryotes: errors, biases and perspectives.Crossref | GoogleScholarGoogle Scholar | 28906012PubMed |
[4] Deamer, D et al.. (2016) Three decades of nanopore sequencing. Nat Biotechnol 34, 518–524.
| Three decades of nanopore sequencing.Crossref | GoogleScholarGoogle Scholar | 27153285PubMed |
[5] Ashikawa, S et al.. (2018) Rapid identification of pathogens from positive blood culture bottles with the MinION nanopore sequencer. J Med Microbiol 67, 1589–1595.
| Rapid identification of pathogens from positive blood culture bottles with the MinION nanopore sequencer.Crossref | GoogleScholarGoogle Scholar | 30311873PubMed |
[6] Taberlet, P et al.. (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21, 2045–2050.
| Towards next-generation biodiversity assessment using DNA metabarcoding.Crossref | GoogleScholarGoogle Scholar | 22486824PubMed |
[7] Heeger, F et al.. (2018) Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments. Mol Ecol Resour 18, 1500–1514.
| Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments.Crossref | GoogleScholarGoogle Scholar | 30106226PubMed |
[8] Mafune, KK et al.. (2020) A rapid approach to profiling diverse fungal communities using the MinION nanopore sequencer. Biotechniques 68, 72–78.
| A rapid approach to profiling diverse fungal communities using the MinION nanopore sequencer.Crossref | GoogleScholarGoogle Scholar | 31849245PubMed |
[9] Motooka, D (2017) Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front Microbiol 8, 238.
| Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals.Crossref | GoogleScholarGoogle Scholar | 28261190PubMed |
[10] D’Andreano, S et al.. (2021) Rapid and real-time identification of fungi up to species level with long amplicon nanopore sequencing from clinical samples. Biol Methods Protoc 6, bpaa026.
| Rapid and real-time identification of fungi up to species level with long amplicon nanopore sequencing from clinical samples.Crossref | GoogleScholarGoogle Scholar | 33506108PubMed |
[11] Wang, M et al.. (2020) Same-day simultaneous diagnosis of bacterial and fungal infections in clinical practice by nanopore targeted sequencing. medRxiv , 2020.04.08.20057604.
| Same-day simultaneous diagnosis of bacterial and fungal infections in clinical practice by nanopore targeted sequencing.Crossref | GoogleScholarGoogle Scholar |
[12] Chan, WS et al.. (2020) Potential utility of targeted Nanopore sequencing for improving etiologic diagnosis of bacterial and fungal respiratory infection. Diagn Pathol 15, 41.
| Potential utility of targeted Nanopore sequencing for improving etiologic diagnosis of bacterial and fungal respiratory infection.Crossref | GoogleScholarGoogle Scholar | 32340617PubMed |
[13] Loit, K et al.. (2019) Relative performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) third-generation sequencing instruments in identification of agricultural and forest fungal pathogens. J Appl Environ Microbiol 85, e01368–19.
| Relative performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) third-generation sequencing instruments in identification of agricultural and forest fungal pathogens.Crossref | GoogleScholarGoogle Scholar |
[14] Marčiulyniene, D et al.. (2021) DNA-metabarcoding of belowground fungal communities in bare-root forest nurseries: focus on different tree species. Microorganisms 9, 150.
| DNA-metabarcoding of belowground fungal communities in bare-root forest nurseries: focus on different tree species.Crossref | GoogleScholarGoogle Scholar | 33440909PubMed |
[15] Zhang, ZF et al.. (2021) High-level diversity of basal fungal lineages and the control of fungal community assembly by stochastic processes in mangrove sediments. Appl Environ Microbiol 87, e0092821.
| High-level diversity of basal fungal lineages and the control of fungal community assembly by stochastic processes in mangrove sediments.Crossref | GoogleScholarGoogle Scholar | 34190611PubMed |
[16] Thomas, T et al.. (2012) Metagenomics – a guide from sampling to data analysis. Microb Inform Exp 2, 3.
| Metagenomics – a guide from sampling to data analysis.Crossref | GoogleScholarGoogle Scholar | 22587947PubMed |
[17] Laudadio, I et al.. (2018) Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS 22, 248–254.
| Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome.Crossref | GoogleScholarGoogle Scholar | 29652573PubMed |
[18] Charalampous, T et al.. (2019) Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol 37, 783–792.
| Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection.Crossref | GoogleScholarGoogle Scholar | 31235920PubMed |
[19] Irinyi, L et al.. (2020) Long-read sequencing based clinical metagenomics for the detection and confirmation of Pneumocystis jirovecii directly from clinical specimens: a paradigm shift in mycological diagnostics. Med Mycol 58, 650–660.
| Long-read sequencing based clinical metagenomics for the detection and confirmation of Pneumocystis jirovecii directly from clinical specimens: a paradigm shift in mycological diagnostics.Crossref | GoogleScholarGoogle Scholar | 31758176PubMed |
[20] Tsai, YC et al.. (2016) Resolving the complexity of human skin metagenomes using single-molecule sequencing. mBio 7, e01948–15.
| Resolving the complexity of human skin metagenomes using single-molecule sequencing.Crossref | GoogleScholarGoogle Scholar | 26861018PubMed |
[21] Greshake Tzovaras, B et al.. (2020) What is in Umbilicaria pustulata? A metagenomic approach to reconstruct the holo-genome of a lichen. Genome Biol Evol 12, 309–324.
| What is in Umbilicaria pustulata? A metagenomic approach to reconstruct the holo-genome of a lichen.Crossref | GoogleScholarGoogle Scholar | 32163141PubMed |
[22] Wang, B et al.. (2021) Chromosome-scale genome assembly of Fusarium oxysporum strain Fo47, a fungal endophyte and biocontrol agent. Mol Plant Microbe Interact 33, 1108–1111.
| Chromosome-scale genome assembly of Fusarium oxysporum strain Fo47, a fungal endophyte and biocontrol agent.Crossref | GoogleScholarGoogle Scholar |
[23] Gu, W et al.. (2021) Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med 27, 115–124.
| Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids.Crossref | GoogleScholarGoogle Scholar | 33169017PubMed |
[24] Piñar, G et al.. (2020) Rapid diagnosis of biological colonization in cultural artefacts using the MinION nanopore sequencing technology. Int Biodeterior Biodegradation 148, 104908.
| Rapid diagnosis of biological colonization in cultural artefacts using the MinION nanopore sequencing technology.Crossref | GoogleScholarGoogle Scholar |
[25] Yang, L et al.. (2019) Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir Res 20, 265.
| Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study.Crossref | GoogleScholarGoogle Scholar | 31775777PubMed |


