Distribution and conservation of known secondary metabolite biosynthesis gene clusters in the genomes of geographically diverse Microcystis aeruginosa strains
Leanne A. Pearson A B , Nicholas D. Crosbie A and Brett A. Neilan B CA Applied Research, Melbourne Water Corporation, 990 La Trobe Street, Docklands, Vic. 3008, Australia.
B School of Environmental and Life Sciences, SR233, Social Sciences Building, The University of Newcastle, University Drive, Callaghan, NSW 2308, Australia.
C Corresponding author. Email: brett.neilan@newcastle.edu.au
Marine and Freshwater Research 71(5) 701-716 https://doi.org/10.1071/MF18406
Submitted: 23 October 2018 Accepted: 12 July 2019 Published: 17 October 2019
Journal Compilation © CSIRO 2020 Open Access CC BY-NC-ND
Abstract
The cyanobacterium Microcystis aeruginosa has been linked to toxic blooms worldwide. In addition to producing hepatotoxic microcystins, many strains are capable of synthesising a variety of biologically active compounds, including protease and phosphatase inhibitors, which may affect aquatic ecosystems and pose a risk to their use. This study explored the distribution, composition and conservation of known secondary metabolite (SM) biosynthesis gene clusters in the genomes of 27 M. aeruginosa strains isolated from six different Köppen–Geiger climates. Our analysis identified gene clusters with significant homology to nine SM biosynthesis gene clusters spanning four different compound classes: non-ribosomal peptides, hybrid polyketide–non-ribosomal peptides, cyanobactins and microviridins. The aeruginosin, microviridin, cyanopeptolin and microcystin biosynthesis gene clusters were the most frequently observed, but hybrid polyketide–non-ribosomal peptide biosynthesis clusters were the most common class overall. Although some biogeographic relationships were observed, taxonomic markers and geography were not reliable indicators of SM biosynthesis cluster distribution, possibly due to previous genetic deletions or horizontal gene transfer events. The only cyanotoxin biosynthesis gene cluster identified in our screening study was the microcystin synthetase (mcy) gene cluster, suggesting that the production of non-microcystin cyanotoxins by this taxon, such as anatoxin-a or paralytic shellfish poison analogues, is either absent or rare.
Additional keywords: biogeography, cyanobacteria, cyanobactin, cyanotoxins, microviridin, non-ribosomal peptide, phylogeny, polyketide.
Introduction
Microcystis aeruginosa is a cosmopolitan bloom-forming species of cyanobacteria notorious for its production of hepatotoxic microcystins. These small cyclic peptides are toxic in very low doses and pose a threat to human health, agriculture and freshwater ecosystems worldwide. Although microcystin is the most toxic metabolite produced by M. aeruginosa, recent genome sequencing efforts have revealed that a variety of other secondary metabolite (SM) pathways are also encoded within the M. aeruginosa pan-genome (Humbert et al. 2013; Fig. 1). Whether the corresponding biologically active compounds pose a threat to water quality is uncertain. However, solely focusing on microcystin when assessing water quality could prove to be an oversight.
|
Interestingly, the ability to produce microcystins appears to be sporadically distributed across the species (Tillett et al. 2001), with toxic and non-toxic M. aeruginosa strains coexisting in many ecosystems. The apparent lack of phylogenetic linkage between microcystin production and traditional taxonomic markers (16S rDNA, phycocyanin, etc.; Tillett et al. 2001) initially hampered molecular differentiation of toxic and non-toxic M. aeruginosa strains. However, the discovery of the microcystin synthetase (mcy) gene cluster in PCC 7806 (Dittmann et al. 1997; Tillett et al. 2000) enabled the development of highly specific and sensitive polymerase chain reaction (PCR)-based methods for assessing the toxigenicity of bloom samples (e.g. Jungblut and Neilan 2006; Al-Tebrineh et al. 2010, 2011; Baker et al. 2013). These methods are now widely used by scientists and water quality authorities around the world.
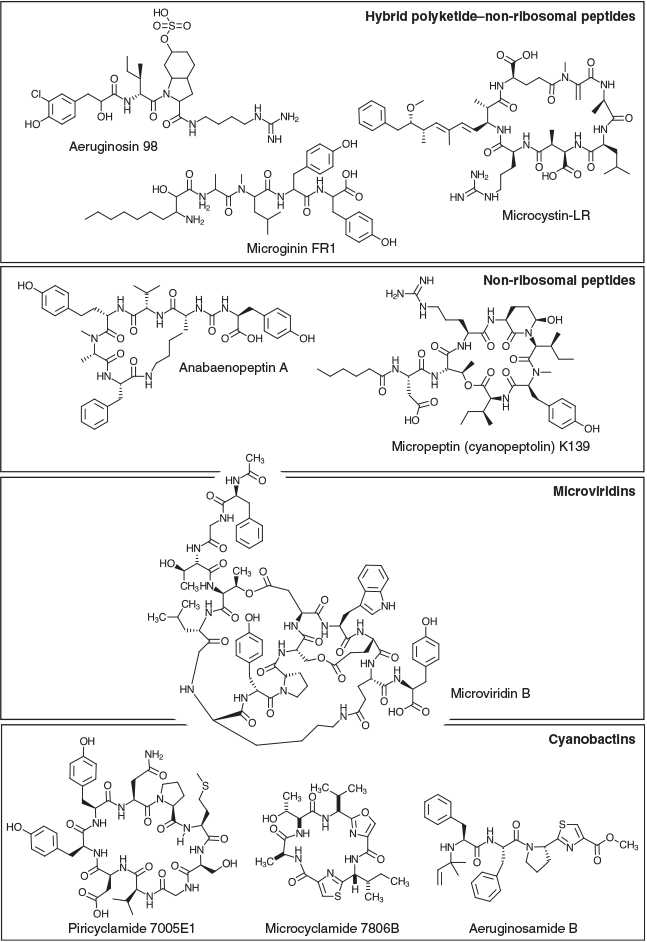
Although it has long been recognised that M. aeruginosa is capable of producing a wide range of other SMs, including non-ribosomal peptides, hybrid polyketide–non-ribosomal peptides (PK-NRPs), cyanobactins and microviridins (Fig. 2), these compounds have received much less attention than the microcystins in terms of their effects on water quality (Janssen 2019). However, many have demonstrable effects on growth and viability in laboratory test organisms, such as mice and zebrafish, as well as in cell cultures, highlighting their potential to disrupt aquatic ecosystems. Furthermore, the biogeography of SM biosynthesis pathways in M. aeruginosa has not been investigated in detail, and it is unclear whether their distribution is scattered, as for microcystin, or segregated according to the geoclimatic origin of isolates. Characterisation of the distribution, composition and conservation of the various SM biosynthesis gene clusters in M. aeruginosa strains will provide insight into their potential SM profiles and is an important first step towards holistic molecular monitoring of freshwater blooms.
|
Known SMs produced by M. aeruginosa
Hybrid PK-NRPs
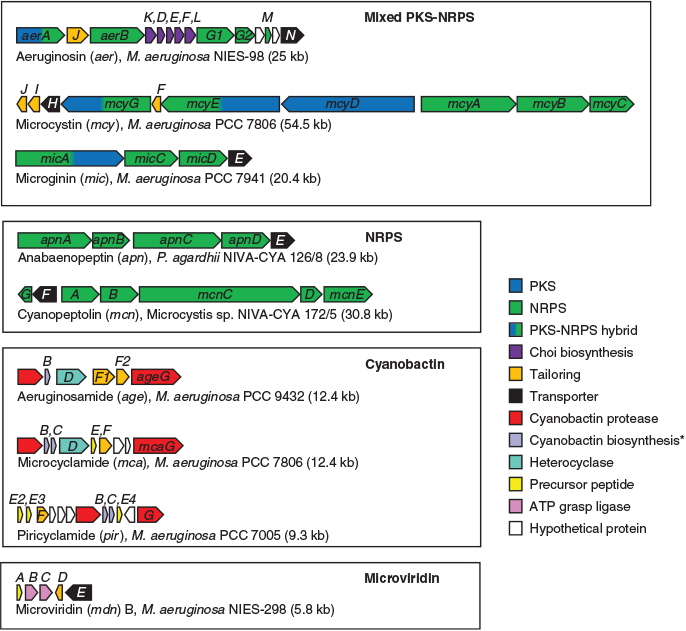
Hybrid PK-NRPs produced by M. aeruginosa include the microcystins, aeruginosins and microginins. The microcystins are a structurally diverse family of non-ribosomal heptapeptides sharing a common monocyclic structure and a unique β-amino acid (Adda) side-chain (Botes et al. 1985). Microcystins inhibit eukaryotic serine/threonine protein phosphatases 1 and 2A (MacKintosh et al. 1990) and are toxic to a variety of animals and plants. Bioaccumulation of the toxin may also occur in resistant organisms (Ferrão-Filho and Kozlowsky-Suzuki 2011). In mammals, the primary target of microcystin is the liver, where disruption of the cytoskeleton causes hepatic haemorrhage and organ failure (Dawson 1998). Chronic subacute exposure to the toxin may additionally initiate and promote tumour development (Fujiki and Suganuma 2011). The gene cluster responsible for microcystin biosynthesis in M. aeruginosa PCC 7806 spans 55 kb and encodes 10 genes (mcyA–J), including three non-ribosomal peptide synthetases (NRPSs), one polyketide synthase (PKS), two hybrid NRPS-PKSs, three auxiliary enzymes and an ABC transporter (Tillett et al. 2000; Fig. 1). The mcy gene cluster has since been identified in several other cyanobacterial genera (Christiansen et al. 2003; Rouhiainen et al. 2004) and is highly conserved in terms of composition and sequence identity compared with other SM biosynthesis clusters.
Aeruginosins are linear non-ribosomal tetrapeptides characterised by a central L-2-carboxy-6-hydroxyoctahydroindole (L-Choi) moiety, a C-terminal arginine derivative (e.g. argininal, argininol, agmatine, 1-amidino-2-ethoxy-3-aminopiperidine) and an N-terminal hydroxyphenyl lactic acid or phenyl lactic acid group (Ersmark et al. 2008; Scherer et al. 2016). Most aeruginosins are potent inhibitors of serine proteases, including the human blood coagulation cascade enzyme thrombin and the digestive enzyme trypsin (Ersmark et al. 2008). Certain variants may also be highly toxic to invertebrates, including crustaceans (Scherer et al. 2016). The gene cluster responsible for aeruginosin biosynthesis in M. aeruginosa PCC 7806 spans 25 kb and encodes 14 genes (aerA–N), including three NRPSs, one hybrid NRPS-PKS, six putative Choi biosynthesis genes, a halogenase, an ABC transporter, an oxidoreductase (OxRed) and several cryptic genes (Ishida et al. 2009; Fig. 1).
The microginins are a family of linear non-ribosomal lipopeptides that contain four to six amino acids, including one or more tyrosines at the C-terminus and the characteristic N-terminal side chain 3-amino-2-hydroxy decanoic acid (Ahda; Strangman and Wright 2016). Microginins inhibit angiotensin-converting enzyme (ACE; Okino et al. 1993), a central component of the renin–angiotensin system that controls blood pressure. The gene cluster responsible for microginin biosynthesis in Planktothrix NIVA-CYA98 spans 21 kb and encodes four genes (micA–E), including a PKS-NRPS, two NRPSs and an ABC transporter (Rounge et al. 2009; Fig. 1).
Non-ribosomal peptides
Non-ribosomal peptides produced by M. aeruginosa include the cyanopeptolins and anabaenopeptins. Cyanopeptolins are a family of cyclic depsipeptides characterised by the presence of 3-amino-6-hydroxy-2-piperidone (Ahp; Martin et al. 1993). Members of the cyanopeptolin family have been shown to inhibit several serine proteases. The gene cluster responsible for cyanopeptolin biosynthesis in Microcystis NIVA-CYA172/5 spans 31 kb and encodes seven genes (mcnA–G), including four NRPSs, a halogenase and an ABC transporter (Tooming-Klunderud et al. 2007). The micropeptin cluster in M. aeruginosa K-139 and PCC 7806 is highly similar in structure and composition to the cyanopeptolin cluster, although it lacks the halogenase (mcnD) and the cryptic enzyme (mcnG; Nishizawa et al. 2011; Fig. 1).
Anabaenopeptins are a family of diverse cyclic hexapeptides characterised by several conserved motifs, including a ureido bond, N-methylation in Position 5, and D-Lys in Position 2 (Christiansen et al. 2011). Anabaenopeptins are potent inhibitors of carboxypeptidase-A (CPA) and thrombin-activatable fibrinolysis inhibitor (TAFIa), enzymes involved in blood coagulation (Schreuder et al. 2016). The anabaenopeptin (apnA–E) gene cluster in Planktothrix agardhii CYA126/8 spans 24 kb and encodes four NRPSs and an ABC transporter (Christiansen et al. 2011; Fig. 1). In some cyanobacterial genomes, the apn gene cluster is colocated with the hph gene cluster, which is believed to provide additional substrates (e.g. L-homotyrosine and L-homophenylalanine) for the biosynthesis of anabaenopeptins (Lima et al. 2017).
Cyanobactins
M. aeruginosa produces a variety of modified ribosomal peptides known as cyanobactins, which are synthesised by the proteolytic cleavage and cyclisation of precursor peptides. Structural diversity within this family of compounds is achieved by heterocyclisation and oxidation of amino acids, or their post-translational prenylation (Sivonen et al. 2010). Cyanobactins exhibit a diverse range of bioactivities, including cytotoxic, antimalarial and allelopathic activities. Although quite diverse in their size (6–20 kb), composition and arrangement, cyanobactin biosynthesis gene clusters always encode two proteases (homologues of patellamide biosynthesis genes, PatA and PatG) that cleave and cyclise the precursor peptide as well as proteins participating in post-translational modifications (Sivonen et al. 2010). Some cyanobactin biosynthesis clusters (e.g. microcyclamide, mcaA–G; and aeruginosamide, ageA–G) also contain heterocyclase enzymes, whereas others (e.g. piricyclamides, pirA-G) do not (Leikoski et al. 2013).
Microviridins
Another class of post-translationally modified peptides produced by M. aeruginosa are the microviridins, which are tricyclic depsipeptides with an unusual cage-like architecture composed of two lactone rings and one lactam ring (Ishitsuka et al. 1990). Microviridins are potent inhibitors of serine proteases, including elastase, chymotrypsin and trypsin (Murakami et al. 1997). A particular variant, microviridin J, has been shown to be toxic to zooplankton (Rohrlack et al. 2004). The gene cluster responsible for microviridin B biosynthesis in M. aeruginosa NIES-843 contains five genes (mdnA–E), encoding the leader peptide (MdnA), two dedicated ATP-grasp ligases (MdnB and MdnC) a Gcn5-related N-acetyltransferase (GNAT)-type acetyltransferase (MdnD) and an ABC transporter (MdnE; Ziemert et al. 2008a; Philmus et al. 2008; Weiz et al. 2011; Fig. 1). MdnA may be distally encoded in other strains (Gatte-Picchi et al. 2014).
Although the aforementioned compounds have been isolated from several M. aeruginosa cultures and bloom samples, the corresponding biochemical pathways and biosynthesis gene clusters have only been examined in a few cyanobacterial strains, in some cases from distantly related genera. In order to better understand the corresponding genetic diversity and evolutionary history of these pathways in M. aeruginosa, we examined the distribution, composition and conservation of known SM biosynthesis gene clusters in 27 M. aeruginosa strains isolated from different geoclimatic regions. A comprehensive multilocus phylogenetic analysis was also undertaken to understand the biogeography of SM production in this taxon. The results are discussed with regard to predicting SM profiles in this species and designing future molecular diagnostic tests for potentially harmful strains.
Materials and methods
Climate zone allocation
Climate zones were allocated to M. aeruginosa strains based on an updated version of the Köppen–Geiger climate classification system (Kottek et al. 2006) accessed through the kgc package (C. Bryant, N. R. Wheeler, F. Rubel, and R. H. French, see https://CRAN.R-project.org/package=kgc, accessed September 2018), which classifies regions of the Earth by their relative heat and humidity through the year. Climate zones are divided into five main groups: A (tropical), B (dry), C (temperate), D (continental) and E (polar). The groups are then subdivided based on seasonal precipitation and temperature. Geographical coordinates used to determine Köppen–Geiger climate zones were based on reports in the literature for each isolate.
Genome assemblies
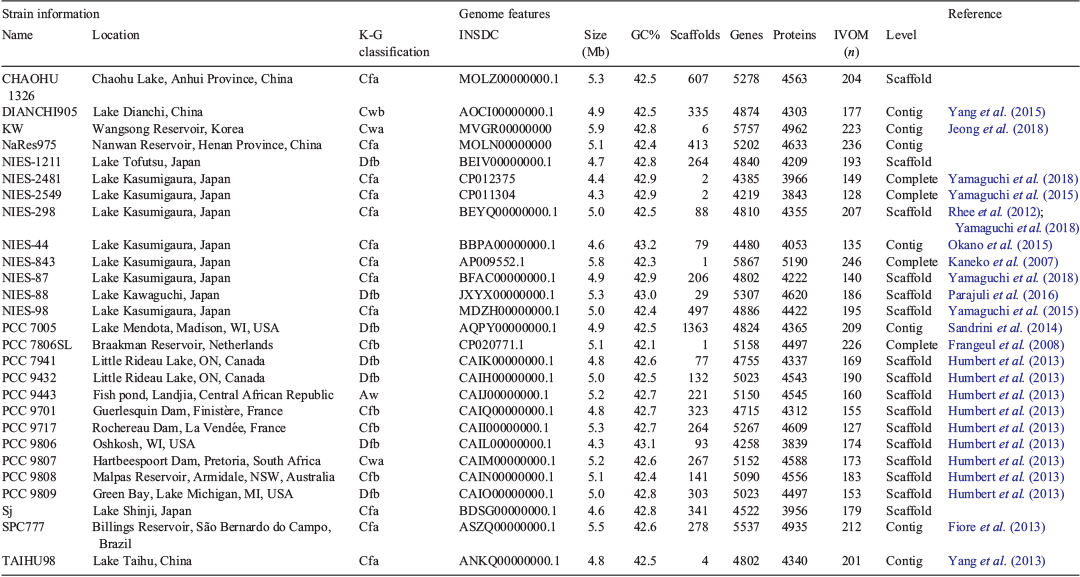
M. aeuriginosa genome assemblies with RefSeq (Haft et al. 2018) membership (n = 27) were downloaded from RefSeq and GenBank (Benson et al. 2018) between 2 February 2017 and 23 June 2018 in fna (used in average nucleotide identity, ANI, and phylogenetic analyses) and gbff (used in the prediction of secondary metabolite biosynthesis gene clusters) format respectively. General genome features were also obtained from GenBank. Of the two available NIES-298 whole-genome sequences, the assembly with the higher genome size and gene count (BEYQ01) was used.
Average nucleotide identity
ANI values were calculated according to the orthologous average nucleotide identity (OrthoANI) algorithm (Lee et al. 2016) implemented with USEARCH (OrthoANIu; Yoon et al. 2017).
Identification and analysis of SM biosynthesis gene clusters
Putative SM biosynthesis gene clusters encoded within the 27 M. aeruginosa genomes were identified and analysed using Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH, ver. 4.10, see https://antismash.secondarymetabolites.org, accessed Sept 2018; Blin et al. 2017), with ClusterFinder enabled (minimum cluster size 4; minimum probability 60%). Results were verified by comparing the inferred peptide sequence identity (ID; minimum 50%), coverage (minimum 50%) and organisation of genes within putative clusters to 1481 previously characterised SM biosynthesis clusters in the Minimum Information about a Biosynthetic Gene cluster (MIBiG) dataset (Medema et al. 2015; Fig. 1), GenBank (by BLASTP and TBLASTN; Altschul et al. 1990) and the literature.
Phylogenetic analysis of the bacterial core-gene set
Phylogenetic analysis of the bacterial core-gene set was undertaken using the up-to-date bacterial core gene (UBCG) set and pipeline for phylogenomic tree reconstruction programme, UBCG (ver. 3.0, see https://www.ezbiocloud.net/tools/ubcg, accessed September 2018; Na et al. 2018), configured with default settings (Jones, Taylor and Thorton model; Jones et al. 1992): where varying rates of evolution across sites were modelled using the ‘fixed number of rate categories’ (‘CAT’) model (Stamatakis 2006); codon-based alignment (i.e. where the output is nucleotide sequences, but the alignment is carried out using amino acid sequences); removal of alignment positions composed of more than 50% gap characters; 1000 resamples during tree construction phase. The UBCG pipeline was run with Prodigal for gene finding (ver. 2.6.3, see https://github.com/hyattpd/prodigal/releases/, accessed September 2018; Hyatt et al. 2010), hmmsearch for identification of UBCGs using HMMER (ver. 3.1b2, see http://eddylab.org/software/hmmer/hmmer-3.1b2.tar.gz, accessed September 2018; Eddy 2011), Multiple Alignment using Fast Fourier Transform (MAFFT) for alignments (ver. 7.402, see https://mafft.cbrc.jp/alignment/software/linux.html, accessed September 2018; Katoh and Standley 2013) and FastTree 2 (ver. 2.1.10, Double precision, number SSE3, see http://www.microbesonline.org/fasttree/, accessed September 2018; Price et al. 2010) to construct single gene trees and an unrooted maximum-likelihood (ML) tree from the concatenated alignment of the nucleotide sequences of 91 genes representing the bacterial core-gene set (length = 84 294 nucleotide positions; see Fig. S1, available as Supplementary material to this paper). The ML tree was subsequently rooted by applying the Minimum Ancestor Deviation function (ver. 1.1, https://www.mikrobio.uni-kiel.de/de/ag-dagan/ressourcen, accessed September 2018; Tria et al. 2017) with default settings, then visualised with the APE package (ver. 5.0, https://cran.r-project.org/web/packages/ape/, accessed September 2018; Paradis et al. 2004).
Phylogenetic distribution of SM biosynthesis gene clusters
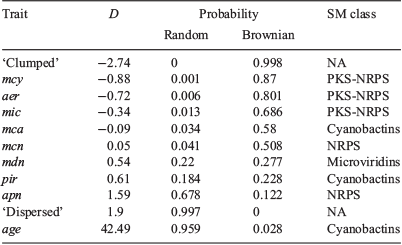
To characterise the phylogenetic distribution of SM gene clusters, Phylo.D (caper ver. 1.0.1, D. Orme, R. Freckleton, G. Thomas, T. Petzoldt, S. Fritz, N. Isaac, and W. Pearse, see https://CRAN.R-project.org/package=caper, accessed September 2018) was used to estimate the extent of phylogenetic clumping. Phylo.D (run with 10 000 permutations per trait model) calculates the D value (Fritz and Purvis 2010), a measure of phylogenetic signal in a binary trait, and tests the estimated D value for significant departure from both random association and the clumping expected under a Brownian evolution threshold model.
Genomic island search
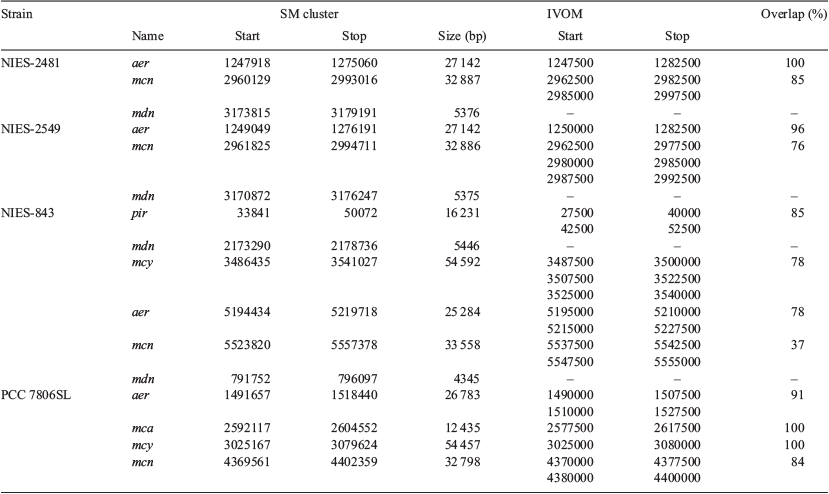
Putative genomic islands within the 27 M. aeruginosa genomes were identified using AlienHunter (ver. 1.7, see https://www.sanger.ac.uk/science/tools/alien-hunter, accessed September 2018) with default settings, which predicts putative horizontal gene transfer (HGT) events with the implementation of interpolated variable order motifs (IVOMs; Vernikos and Parkhill 2006). The IVOM approach exploits compositional biases at various levels (e.g. codon, dinucleotide and amino acid bias, structural constraints) and more reliably captures the local composition of a sequence than fixed-order methods. The relative position of SM biosynthesis clusters compared to IVOMs within the four closed M. aeruginosa genomes (NIES-2481, NIES-2549, NIES-843 and PCC 7806SL) was also examined.
Results
General genome features
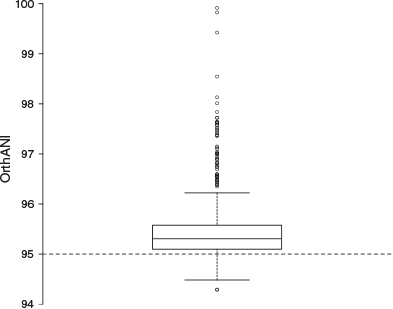
The genomes ranged in size from 4.3 to 5.9 Mb (mean = ~5 Mb) with GC contents ranging from 42.1 to 43.2% (mean = ~43%). They were assembled across 1 to 1363 scaffolds (mean = ~235) and contained 3839–5190 protein-coding regions (mean = ~4417; Table 1). In pairwise genome comparisons (Fig. S1), 85.2% of ANI values (which ranged from 94.3 to 99.9%; median 95.3%) were above the nominal prokaryote species threshold of 95% (Goris et al. 2007; Fig. 3).
|
SM cluster prediction
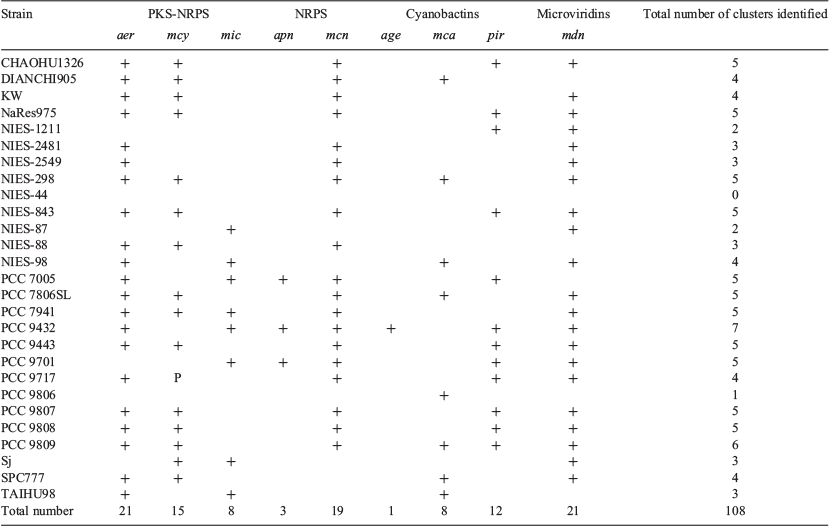
Homologues of nine known SM biosynthesis gene clusters were distributed among the 27 M. aeruginosa genomes, including three PK-NRP biosynthesis clusters (aeruginosin, aer; microcystin, mcy; and microginin, mic), two NRP biosynthesis clusters (anabaenopeptin, apn; and micropeptin, mcn), three cyanobactin biosynthesis clusters (aeruginosamide, age; microcyclamide, mca; and piricyclamide, pir) and a microviridin (mdn) biosynthesis cluster. The individual M. aeruginosa strains encoded 0–7 known SM biosynthesis clusters (mean = 4), which occupied ~0–3% of their total genomes (mean ≥2%). PCC 9432 encoded the greatest number of known clusters, whereas NIES-44 encoded none, although fragments of the pir cluster were found in the NIES-44 genome. The most commonly identified SM biosynthesis clusters were the aer and mdn clusters, which were identified in 21 different strains. In contrast, the age cluster was only found in a single strain (PCC 9432). The most common class of SM biosynthesis cluster identified among the 27 strains was the mixed PK-NRP class, which was identified at approximately twice the frequency of the NRP, cyanobactin and microviridin classes (Tables 2, S1).
Hybrid PK-NRP biosynthesis clusters
The complete microcystin (mcyA–J) biosynthesis gene cluster was identified in 15 M. aeruginosa strains, including the reference strain PCC 7806SL (MIBiG accession BGC000107). A partial microcystin synthetase cluster (mcyA–C) was identified in PCC 9717. Genes within the mcy cluster were highly conserved with inferred peptide sequence IDs ≥91% (mean = ~98%) compared with their homologous counterparts in strain PCC 7806SL.
The aeruginosin (aer) biosynthesis gene cluster was identified in 21 strains, including the reference strain NIES-98 (MIBiG accession BGC0000298). The consensus cluster within this group comprised aerA, aerB, aerD, aerE, aerF, aerG1, aerK, aerL, aerN and OxRed genes. A putative transposase gene was also identified just upstream of aerA. The full complement of aer genes (aerA, aerB, aerD, aerE, aerF, aerG1, aerG2, aerK, aerL, aerM, aerN) was only detected in three strains (NIES-98, PCC 7005 and TAIHU98). An additional open reading frame (ORF), namely aerJ, putatively encoding a halogenase, was detected in seven strains. The inferred peptide sequences of aer genes were mostly well conserved (mean = ~93% ID) compared with the NIES-98 sequences. However, the sequences of aerB and aerL were poorly conserved in some strains (≥58%; mean = ~76% ID).
The closest match to the microginin (mic) gene cluster in the MIBiG repository was the puwainaphycin cluster from Cylindrospermum alatosporum CCALA 988 (accession BGC0001125). Therefore, 27 M. aeruginosa genomes were screened for the mic cluster by BLASTP (Altschul et al. 1990) analysis with MicA, MicC, MicD and MicE from PCC 7941 used as query sequences (GenBank accession CCI09456–9). Homologues of all mic genes were identified in seven additional strains with an average inferred peptide sequence identity of ~90% compared with the reference sequences. Among these sequences, the PKS-NRPS micA and ABC transporter micE were the most conserved (≥90%; mean = ~95% ID), whereas the NRPSs micC and micD were less conserved (≥64%; mean = ~84% ID).
NRP biosynthesis clusters
The anabaenopeptin (apn) biosynthesis gene cluster was identified in three strains (PCC 7005, PCC 9432 and PCC 9701). All identified clusters encoded the full complement of apn genes (apnA, apnB, apnC, apnD, apnE). Sequence conservation was generally high compared with the Planktothrix agardhii NIVA-CYA 126/8 reference sequences (MIBiG accession BGC0000301; mean = ~88% ID). The NRPS genes apnA and apnB and the ABC-transporter gene apnE were particularly well conserved in terms of inferred peptide sequence (≥92%; mean = ~93% ID), whereas the NRPS genes apnC and apnD were less conserved (≥76%; mean = ~80% ID). As seen in P. agardhii, but in contrast with Anabaena sp. 90 (Rouhiainen et al. 2010), the M. aeruginosa apn gene clusters only had one copy of apnA.
The cyanopeptolin (mcn) biosynthesis gene cluster was identified in 19 strains. Only three of these clusters (NaRes975, PCC 7941 and PCC 9808) encoded the full complement of mcn genes (mcnA–G). However, most of the mcn clusters identified (17/19) encoded homologues of mcnB, mcnC, mcnE, mcnF and mcnG. Approximately half the mcn clusters identified (10/19) encoded the NRPS mcnA, whereas only four clusters encoded the halogenase mcnD. Interestingly, four of the strains that lacked mcnA (CHAOHU 1326, NIES-2481, NIES-2549 and NIES-298) had elongated (~2× compared with the reference) mcnB genes encoding an additional peptidyl carrier protein domain and an unusual FkbH domain. The inferred peptide sequences of these elongated mcnB homologues were poorly conserved (mean = ~65% ID) compared with the Microcystis sp. NIVA-CYA 172/5 reference sequence (MIBiG accession BGC0000332), but highly similar to each other. Sequence conservation among the remaining mcn genes was generally high (mean = ~92% ID).
Cyanobactin biosynthesis clusters
The microcyclamide (mca) biosynthesis gene cluster was identified in eight strains, including the reference strain PCC 7806SL (MIBiG accession BGC0000474). All clusters encoded the full complement of mca genes (mcaA–G), except TAIHU98, which lacked the subtilisin-like protein mcaA. Overall inferred peptide sequence conservation was high compared with the reference cluster (mean = ~95% ID). However, the sequence of the microcyclamide precursor gene mcaE was only moderately conserved (mean = ~82% ID).
An aeruginosamide (ageA, ageB, ageD, ageF1, ageF2, ageG) biosynthesis gene cluster was identified in only one strain, the reference strain PCC 9432 (MIBiG accession BGC0000483). The aer cluster has similar organisation and identity to the microcyclamide (mca) cluster, except it also encodes a methyltransferase gene ageF1.
Piricyclamide-like biosynthesis gene clusters were identified in 12 strains, including the reference strain PCC 7005 (MIBiG accession BGC0001167). Only one other strain (PCC 9807) encoded the full complement of pir genes (pirA, pirB, pirC, pirE2, pirE3, pirE4, pirF, pirG). All piricyclamide-like biosynthesis clusters contained homologues of the N- and C-terminal cyanobactin protease genes pirA and pirG and the hypothetical protein pirB. Most contained the hypothetical protein pirC, the prenyl transferase gene pirF and at least one cyanobactin precursor (pirE2, pirE3, pirE4). Overall inferred peptide sequence identity averaged ~86% compared with the reference cluster from PCC 7005. Sequence conservation was high among pirA, pirB, pirG and pirE4 genes (mean = ~94% ID), but lower among pirC, pirF, pirE2 and pirE3 genes (mean = ~75% ID).
Microviridin biosynthesis gene clusters
The microviridin (mdn) biosynthesis gene cluster was identified in 21 strains, including the reference strain NIES-298 (MIBiG accession BGC0000592). All strains encoded mdnB, mdnC, mdnD and mdnE, except PCC 7806SL, which lacked the acetyltransferase gene mdnD. Seven strains encoded the microviridin precursor gene mdnA. The remaining 14 strains encoded a gene with low inferred peptide sequence homology (mean = ~51%) to mdnA. This gene was located outside the mdn cluster in nine strains. The sequences of mdnB–E genes were highly conserved in all clusters (mean = ~97% ID). Fragments of the mdn cluster were also identified in PCC 9806 and TAIHU98.
Phylogenetic distribution of SM biosynthesis gene clusters
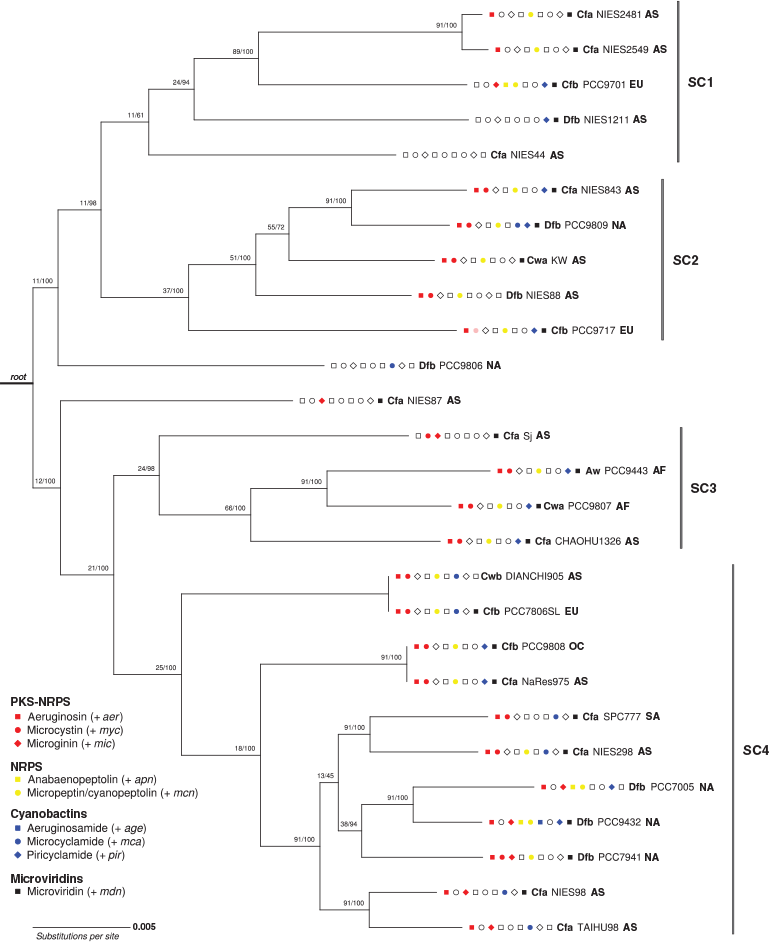
Maximum-likelihood phylogenetic analysis of the concatenated alignment of 91 bacterial core genes grouped the 27 M. aeruginosa genomes into 25 clades exhibiting a branching pattern broadly concordant with that found in the phylogenetic analysis of 10 strains by (Humbert et al. 2013), with generally high local support values (Fig. 4). The number of single gene trees supporting a given branch in the UBCG tree (designated the gene support index, GSI) ranged from 11 to 91, with lower GSI values observed in early branch splits. The 16S rRNA locus was also examined as a candidate single-locus phylogenetic marker, but because there was only a 0.5% sequence variation across aligned positions, this locus was not able to resolve closely related M. aeruginosa strains (data not shown).
|
Representative biosynthesis gene clusters from the four SM classes were detected in each of the major phylogenetic subclades (SC1–4). However, the distribution of individual SM biosynthesis clusters varied across the subclades. For example, all strains from SC1 lacked mcy, age and mca clusters; all strains from SC2 possessed the aer, mcy and mcn clusters, but lacked the mic, apn and age clusters; all strains from SC3 possessed mcy and mdn clusters, but lacked apn, age and mca clusters; and all strains from SC4 possessed the aer cluster (Fig. 4; Table 2).
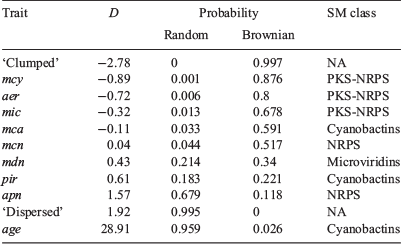
Phylo.D analysis revealed that the phylogenetic distribution of all PK-NRP biosynthesis clusters, particularly mcy, was clumped, the distribution of the apn cluster was random and the distribution of the age cluster (n = 1) was overdispersed. The phylogenetic patterning of the other gene clusters was less marked (Tables 3, 4).
|
|
Geoclimatic distribution of SM biosynthesis gene clusters
The 27 M. aeruginosa strains, from six different continents, were allocated to six different Köppen–Geiger climates: 12 to Cfa (humid subtropical), 7 to Dfb (warm-summer humid continental), 4 to Cfb (temperate oceanic), 2 to Cwa (monsoon-influenced humid subtropical), 1 to Aw (tropical wet) and 1 to Cwb (subtropical highland).
Representative gene clusters from the four SM classes were detected in each of the six continental groups, except the South American group (n = 1), which lacked the NRP class. However, the distribution of individual SM biosynthesis clusters varied across the geographical groups. For example, the African strains (n = 2) both possessed aer, mcy, mcn, pir and mdn gene clusters, but lacked mic, apn, age and mca clusters. The Asian strains (n = 15) all lacked apn and age clusters. The Australian strain (n = 1) possessed aer, mcy, mcn, pir and mdn gene clusters, but lacked mic, apn and age clusters. The European strains (n = 3) all possessed mcn and mdn clusters, but lacked the age cluster. When Köppen–Geiger climate zones (with n ≥ 2) were considered, all strains within the Cfa group (n = 12) lacked apn and age clusters; all strains within the Cfb group (n = 5) lacked the age cluster, but possessed the mcn and mdn clusters; and all strains within the Cwa group (n = 2) possessed the aer, mcy, mcn and mdn clusters, but lacked the mic, apn, age and mca clusters (Fig. 4; Table 2).
Genomic islands
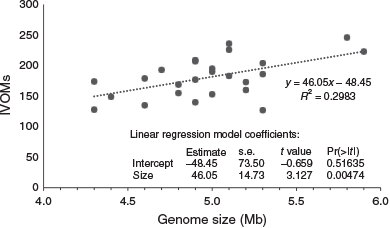
The number of IVOMs (Vernikos and Parkhill 2006) in the 27 M. aeruginosa genomes ranged from 127 to 246 (mean = 190; Table 1). A positive correlation between the number of IVOMs and genome size was observed (Fig. 5), but there was no correlation between the number of IVOMs and the number of SM biosynthesis clusters identified (data not shown). Examination of the relative position of SM biosynthesis gene clusters in the four closed M. aeruginosa genomes (NIES-2481, NIES-2549, NIES-843 and PCC 7806SL) revealed that the mcy, aer, mcn, mca and pir gene clusters completely or partially overlap with IVOMs (37–100%; mean = 84.2%). In contrast, the mdn cluster did not overlap with IVOMs (Table 5).
|
Discussion
Automated genome mining pipelines are powerful tools for identifying SM pathways in microorganisms, particularly when biochemical and genetic data are available for similar pathways in related organisms (Blin et al. 2017). In the present study, antiSMASH and BLAST analyses were used to screen 27 M. aeruginosa genomes for 1481 known SM pathways. Nine known SM biosynthesis gene clusters from four different metabolite classes were identified. However, the only cyanotoxin gene cluster identified within the 27 M. aeruginosa genomes was the mcy cluster, suggesting that the production of non-microcystin cyanotoxins by this taxon is absent or rare. This finding is in contrast with previous reports that the M. aeruginosa is also capable of producing anatoxin-a (strains TAC80, TAC87, TAC117 and TAC121; Park et al. 1993) and paralytic shellfish poison analogues (Sant’Anna et al. 2011).
Although the number of known SM pathways detected in each genome varied, the fact that SM biosynthesis gene clusters comprised on average 2% of the total genome size suggests that the corresponding compounds play an important ecological role in M. aeruginosa. The high frequency of aeruginosin, microviridin, cyanopeptolin and microcystin biosynthesis gene clusters further suggests that these compounds are particularly significant to the success of the species. However, the lack or complete absence of SM biosynthesis clusters in some strains (e.g. NIES-44) seems to contradict this. It is likely that the different SM cluster profiles observed in this study correspond to different M. aeruginosa ecotypes that have evolved to suit specific niche environments. Previous growth competition studies on toxic v. non-toxic M. aeruginosa strains support this theory, demonstrating that microcystin production, although not essential for survival, may be advantageous under certain growth conditions, such as nutrient limitation or high light (Kaebernick et al. 2000; Kardinaal et al. 2007; Zilliges et al. 2011).
Although gene sequence identity was generally high between homologous counterparts in each SM gene cluster, the composition of genes in each cluster sometimes varied. Gene identity and composition were particularly well conserved among microcystin, microginin, anabaenopeptin and microcyclamide pathways, suggesting that all mcy, mic, apn and mca genes play key roles in the biosynthesis or activity of their corresponding compounds, and major mutations are not tolerated. Previous studies have shown this to be the case. For example, mcyB- and mcyH-knockout mutants were unable to produce microcystin (Dittmann et al. 1997; Pearson et al. 2004), whereas mcyJ-knockout mutants produced toxin lacking the O-methylation on the C9 hydroxyl unit and had reduced inhibitory activity against protein phosphatases (Christiansen et al. 2003). Based on these examples, it is anticipated that strain PCC 9717, lacking mcyD–H, is unable to synthesise microcystin.
Interestingly, although microviridin biosynthesis genes were well conserved, homologues of the precursor peptide gene mdnA were poorly conserved in terms of sequence conservation and located outside the mdn gene cluster in some strains. The distal location of mdnA, although recognised previously (Ziemert et al. 2010; Humbert et al. 2013), is unusual given that most of SM biosynthesis genes are arranged in operons. Sequence variations in mdnA have recently been shown to underpin chemical diversity in the microviridin family (Gatte-Picchi et al. 2014). Furthermore, the two grasp ligases encoded by mdnB and mdnC could potentially modify a variety of other leader peptides scattered throughout the genome, as has been observed for the prochlorosin synthetase of Prochlorococcus MIT9313, which processes up to 29 different precursor peptides (Zhang et al. 2014). The absence of the acetyltransferase gene mdnD in PCC 7806SL suggests that novel microviridin variants lacking the N-acetylation could also be produced by this strain. Together, these results suggest that several different microviridins are produced by the M. aeruginosa group analysed in this study.
The identity and composition of core biosynthesis genes in the aeruginosin biosynthesis cluster was well conserved, but many strains lacked the halogenase gene aerJ and the NRPS genes aerG2 and aerM. The loss of aerJ suggests that these strains may produce non-chlorinated variants of aeruginosin. A previous study by Cadel-Six et al. (2008) has shown this to be the case for numerous M. aeruginosa strains, including several analysed in this study. The absence of the additional NRPS genes further suggests that some strains may produce aeruginosin variants with different amino acid backbones.
The composition of genes in the cyanopeptolin and piricyclamide biosynthesis clusters was highly variable, and no consensus gene sets could be established for these pathways. The loss of different genes in different strains suggests that a wide variety of cyanopeptolin- and piricyclamide-like compounds could be produced by M. aeruginosa. For example, strains lacking mcnD are likely to produce non-chlorinated cyanopeptolins, whereas strains lacking mcnD and mcnG are likely to produce micropeptins. The fusion of an FkbH domain to mcnB was observed in several cyanopeptolin biosynthesis gene clusters. This is interesting because most of the reported FkbH-like proteins are involved in polyketide biosynthesis, with cyanopetolin-1138 biosynthetic machinery as the only example of an FkbH-like protein found in a pure NRPS setting (Auerbach et al. 2018). The mosaic structure of these mcnB genes suggests that they may be products of horizontal gene transfer (HGT; Rounge et al. 2007).
Compositional and sequence diversity among the piricyclamide biosynthesis clusters was not surprising considering the size and complex evolutionary history of the cyanobactin family (Leikoski et al. 2013). However, the loss of multiple pir genes in some M. aeruginosa strains could be a sign of inactive pathways. For example, the absence of all but two piricyclamide genes, namely pirE3 and pirG, in the NIES-44 genome suggests that this strain is unable to produce cyanobactins.
Highly conserved genes within compositionally conserved SM biosynthesis clusters are good candidate targets for molecular diagnostic tests. PCR-based tests targeting core genes within the microcystin, nodularin, cylindrospermopsin and saxitoxin pathways have already proven to be reliable methods for detecting toxic cyanobacteria (Al-Tebrineh et al. 2010, 2011; Baker et al. 2013). Although mass spectrometry methods are still considered the ‘gold standard’ for detecting cyanotoxins, molecular methods are becoming more widely accepted and are often preferred by water quality managers and researchers because they are quicker, more economical and user friendly and provide evidence for toxigenic potential. Similar tests could be designed for the diagnosis of the other potentially harmful SM biosynthesis gene clusters identified in this study, but the choice of target genes may be limited in the case of the less conserved clusters, such as mcn and pir.
In addition to investigating the composition and conservation of SM biosynthesis gene clusters in M. aeruginosa, this study sought to determine whether phylogeny and geography or climate zone were good indicators of SM profiles. Although phylogeny appeared to be correlated with the distribution of certain SM biosynthesis clusters within certain subclades (particularly PK-NRP biosynthesis clusters), these relationships are not strong enough to allow reliable prediction of SM profiles based on phylogenetic markers alone. Similarly, although geoclimatic forces seem to affect SM gene cluster profiles in this group, further studies with larger sample sizes are required to establish the significance of these results.
The complex distribution of SM biosynthesis clusters in cyanobacteria has been mostly attributed to gene loss events, rather than HGT. For example, phylogenetic studies on mcy, aer and mcn gene clusters suggest that these pathways existed in an ancient common ancestor and were lost in subsequent lineages (Rantala et al. 2004; Rounge et al. 2007). Although the lateral transfer of whole clusters seems unlikely, HGT could explain some of the sequence variations observed in earlier studies and herein.
The 27 M. aeruginosa genomes analysed in this study contained a large number of IVOMs, suggesting that frequent HGT events occur within this species. Examination of the relative position of SM biosynthesis clusters in the four closed genomes revealed that the aer, mcy, mcn, mca and pir clusters overlap partially or completely with IVOMs. This suggests that HGT events may have led to the acquisition or recombination of genes within these clusters. These results are in agreement with previous phylogenetic studies that suggest HGT events led to DNA polymorphisms in the mcy and aer and cyanobactin biosynthesis clusters (Mikalsen et al. 2003; Tanabe et al. 2004; Ishida et al. 2009; Leikoski et al. 2009). The occurrence of transposase genes within, and proximal to, SM biosynthesis clusters, including aer (present study), mcy (Tillett et al. 2000), mcn (Nishizawa et al. 2011), mca (Ziemert et al. 2008b) and pir (Leikoski et al. 2013), lends further support to this theory.
Recombination events in cyanobacterial genomes can give rise to diverse SM variants. Knowing which variants are produced by a given strain can be important for both drug discovery and water management purposes because different variants often have different activities and toxicities. For example, non-sulfated variants of saxitoxin are ~10-fold more toxic than disulfated variants (Wiese et al. 2010). Similarly, the toxicity of microcystin-RR is approximately 10-fold higher than that of microcystin-LR (Sivonen and Jones 1999). Whole-genome approaches, capable of predicting SM profiles by the analysis of conserved domains within biosynthesis and tailoring enzymes, could foreseeably become routine features of water quality monitoring as rapid sequencing technologies become more accessible.
Conclusions
Cyanobacterial SMs exhibit a wide range of bioactivities, including the inhibition of eukaryotic protein phosphatases. These activities have the potential to disrupt aquatic ecosystems and affect water quality. This study has revealed the distribution, composition and conservation of 9 different SM biosynthesis gene clusters in 27 M. aeruginosa genomes, highlighting the potential chemical diversity inherent within this species. Furthermore, the combined results suggest that multiple factors, including geography and climate, gene loss and HGT, have played a role in shaping the evolution of these pathways. This research has also laid the foundations for future molecular screening tests for predicting SM profiles in uncharacterised, but potentially harmful, Microcystis cultures and bloom samples.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
Funding was received from Melbourne Water Corporation as part of the Nuisance and Harmful Algae Science–Practice Partnership and from the Australian Research Council (Grant LP130100311).
References
Al-Tebrineh, J., Mihali, T. K., Pomati, F., and Neilan, B. A. (2010). Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR. Applied and Environmental Microbiology 76, 7836–7842.| Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR.Crossref | GoogleScholarGoogle Scholar | 20935128PubMed |
Al-Tebrineh, J., Gehringer, M. M., Akcaalan, R., and Neilan, B. A. (2011). A new quantitative PCR assay for the detection of hepatotoxigenic cyanobacteria. Toxicon 57, 546–554.
| A new quantitative PCR assay for the detection of hepatotoxigenic cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 21194539PubMed |
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology 215, 403–410.
| Basic local alignment search tool.Crossref | GoogleScholarGoogle Scholar | 2231712PubMed |
Auerbach, D., Yan, F., Zhang, Y., and Müller, R. (2018). Characterization of an unusual glycerate esterification process in vioprolide biosynthesis. ACS Chemical Biology 13, 3123–3130.
| Characterization of an unusual glycerate esterification process in vioprolide biosynthesis.Crossref | GoogleScholarGoogle Scholar | 30286293PubMed |
Baker, L., Sendall, B. C., Gasser, R. B., Menjivar, T., Neilan, B. A., and Jex, A. R. (2013). Rapid, multiplex-tandem PCR assay for automated detection and differentiation of toxigenic cyanobacterial blooms. Molecular and Cellular Probes 27, 208–214.
| Rapid, multiplex-tandem PCR assay for automated detection and differentiation of toxigenic cyanobacterial blooms.Crossref | GoogleScholarGoogle Scholar | 23850895PubMed |
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., and Sayers, E. W. (2018). GenBank. Nucleic Acids Research 46, D41–D47.
| GenBank.Crossref | GoogleScholarGoogle Scholar | 29140468PubMed |
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., Suarez Duran, H. G., de los Santos, E. L. C., Kim, H. U., Nave, M., Dickschat, J. S., Mitchell, D. A., Shelest, E., Breitling, R., Takano, E., Lee, S. Y., Weber, T., and Medema, M. H. (2017). antiSMASH 4.0 – improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Research 45, W36–W41.
| antiSMASH 4.0 – improvements in chemistry prediction and gene cluster boundary identification.Crossref | GoogleScholarGoogle Scholar | 28460038PubMed |
Botes, D. P., Wessels, P. L., Kruger, H., Runnegar, M. T. C., Santikarn, S., Smith, R. J., Barna, J. C. J., and Williams, D. H. (1985). Structural studies on cyanoginosins-LR, -YR, -YA, and -YM, peptide toxins from Microcystis aeruginosa. Journal of the Chemical Society, Perkin Transactions 1, 2747–2748.
| Structural studies on cyanoginosins-LR, -YR, -YA, and -YM, peptide toxins from Microcystis aeruginosa.Crossref | GoogleScholarGoogle Scholar |
Cadel-Six, S., Dauga, C., Castets, A. M., Rippka, R., Bouchier, C., Tandeau de Marsac, N., and Welker, M. (2008). Halogenase genes in nonribosomal peptide synthetase gene clusters of Microcystis (cyanobacteria): sporadic distribution and evolution. Molecular Biology and Evolution 25, 2031–2041.
| Halogenase genes in nonribosomal peptide synthetase gene clusters of Microcystis (cyanobacteria): sporadic distribution and evolution.Crossref | GoogleScholarGoogle Scholar | 18614525PubMed |
Christiansen, G., Fastner, J., Erhard, M., Börner, T., and Dittmann, E. (2003). Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. Journal of Bacteriology 185, 564–572.
| Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation.Crossref | GoogleScholarGoogle Scholar | 12511503PubMed |
Christiansen, G., Philmus, B., Hemscheidt, T., and Kurmayer, R. (2011). Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and evolution of substrate promiscuity. Journal of Bacteriology 193, 3822–3831.
| Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and evolution of substrate promiscuity.Crossref | GoogleScholarGoogle Scholar | 21622740PubMed |
Dawson, R. M. (1998). The toxicology of microcystins. Toxicon 36, 953–962.
| The toxicology of microcystins.Crossref | GoogleScholarGoogle Scholar | 9690788PubMed |
Dittmann, E., Neilan, B. A., Erhard, M., von Döhren, H., and Börner, T. (1997). Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Molecular Microbiology 26, 779–787.
| Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806.Crossref | GoogleScholarGoogle Scholar | 9427407PubMed |
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Computational Biology 7, e1002195.
| Accelerated profile HMM searches.Crossref | GoogleScholarGoogle Scholar | 22039361PubMed |
Ersmark, K., Del Valle, J. R., and Hanessian, S. (2008). Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angewandte Chemie International Edition 47, 1202–1223.
| Chemistry and biology of the aeruginosin family of serine protease inhibitors.Crossref | GoogleScholarGoogle Scholar | 18076006PubMed |
Ferrão-Filho, A. da S., and Kozlowsky-Suzuki, B. (2011). Cyanotoxins: bioaccumulation and effects on aquatic animals. Marine Drugs 9, 2729–2772.
| Cyanotoxins: bioaccumulation and effects on aquatic animals.Crossref | GoogleScholarGoogle Scholar |
Fiore, M. F., Alvarenga, D. O., Varani, A. M., Hoff-Risseti, C., Crespim, E., Ramos, R. T., Silva, A., Schaker, P. D., Heck, K., Rigonato, J., and Schneider, M. P. (2013). Draft genome sequence of the Brazilian toxic bloom-forming cyanobacterium Microcystis aeruginosa strain SPC777. Genome Announcements 1, e00547-13.
| Draft genome sequence of the Brazilian toxic bloom-forming cyanobacterium Microcystis aeruginosa strain SPC777.Crossref | GoogleScholarGoogle Scholar | 23908289PubMed |
Frangeul, L., Quillardet, P., Castets, A. M., Humbert, J. F., Matthijs, H. C., Cortez, D., Tolonen, A., Zhang, C. C., Gribaldo, S., Kehr, J. C., Zilliges, Y., Ziemert, N., Becker, S., Talla, E., Latifi, A., Billault, A., Lepelletier, A., Dittmann, E., Bouchier, C., and de Marsac, N. T. (2008). Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9, 274.
| Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium.Crossref | GoogleScholarGoogle Scholar | 18534010PubMed |
Fritz, S. A., and Purvis, A. (2010). Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conservation Biology: the Journal of the Society for Conservation Biology 24, 1042–1051.
| Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits.Crossref | GoogleScholarGoogle Scholar |
Fujiki, H., and Suganuma, M. (2011). Tumor promoters-microcystin-LR, nodularin and TNF-α and human cancer development. Anti-cancer Agents in Medicinal Chemistry 11, 4–18.
| Tumor promoters-microcystin-LR, nodularin and TNF-α and human cancer development.Crossref | GoogleScholarGoogle Scholar | 21269254PubMed |
Gatte-Picchi, D., Weiz, A., Ishida, K., Hertweck, C., and Dittmann, E. (2014). Functional analysis of environmental DNA-derived microviridins provides new insights into the diversity of the tricyclic peptide family. Applied and Environmental Microbiology 80, 1380–1387.
| Functional analysis of environmental DNA-derived microviridins provides new insights into the diversity of the tricyclic peptide family.Crossref | GoogleScholarGoogle Scholar | 24334668PubMed |
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. International Journal of Systematic and Evolutionary Microbiology 57, 81–91.
| DNA–DNA hybridization values and their relationship to whole-genome sequence similarities.Crossref | GoogleScholarGoogle Scholar | 17220447PubMed |
Haft, D. H., DiCuccio, M., Badretdin, A., Brover, V., Chetvernin, V., O’Neill, K., Li, W., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, F., Marchler, G. H., Song, J. S., Thanki, N., Yamashita, R. A., Zheng, C., Thibaud-Nissen, F., Geer, L. Y., Marchler-Bauer, A., and Pruitt, K. D. (2018). RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Research 46, D851–D860.
| RefSeq: an update on prokaryotic genome annotation and curation.Crossref | GoogleScholarGoogle Scholar | 29112715PubMed |
Huang, I.-S., and Zimba, P. V. (2019). Cyanobacterial bioactive metabolites – a review of their chemistry and biology. Harmful Algae 83, 42–94.
| Cyanobacterial bioactive metabolites – a review of their chemistry and biology.Crossref | GoogleScholarGoogle Scholar | 31097255PubMed |
Humbert, J. F., Barbe, V., Latifi, A., Gugger, M., Calteau, A., Coursin, T., Lajus, A., Castelli, V., Oztas, S., Samson, G., Longin, C., Medigue, C., and de Marsac, N. T. (2013). A tribute to disorder in the genome of the bloom-forming freshwater cyanobacterium Microcystis aeruginosa. PLoS One 8, e70747.
| A tribute to disorder in the genome of the bloom-forming freshwater cyanobacterium Microcystis aeruginosa.Crossref | GoogleScholarGoogle Scholar | 23950996PubMed |
Hyatt, D., Chen, G.-L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119.
| Prodigal: prokaryotic gene recognition and translation initiation site identification.Crossref | GoogleScholarGoogle Scholar | 20211023PubMed |
Ishida, K., Welker, M., Christiansen, G., Cadel-Six, S., Bouchier, C., Dittmann, E., Hertweck, C., and Tandeau de Marsac, N. (2009). Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Applied and Environmental Microbiology 75, 2017–2026.
| Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 19201978PubMed |
Ishitsuka, M. O., Kusumi, T., Kakisawa, H., Kaya, K., and Watanabe, M. M. (1990). Microviridin. A novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis. Journal of the American Chemical Society 112, 8180–8182.
| Microviridin. A novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis.Crossref | GoogleScholarGoogle Scholar |
Janssen, E. M. L. (2019). Cyanobacterial peptides beyond microcystins – a review on co-occurrence, toxicity, and challenges for risk assessment. Water Research 151, 488–499.
| Cyanobacterial peptides beyond microcystins – a review on co-occurrence, toxicity, and challenges for risk assessment.Crossref | GoogleScholarGoogle Scholar |
Jeong, H., Chun, S. J., Srivastava, A., Cui, Y., Ko, S. R., Oh, H. M., and Ahn, C. Y. (2018). Genome sequences of two cyanobacterial strains, toxic green Microcystis aeruginosa KW (KCTC 18162P) and nontoxic brown Microcystis sp. strain MC19, under xenic culture conditions. Genome Announcements 6, e00378-18.
| Genome sequences of two cyanobacterial strains, toxic green Microcystis aeruginosa KW (KCTC 18162P) and nontoxic brown Microcystis sp. strain MC19, under xenic culture conditions.Crossref | GoogleScholarGoogle Scholar | 29930044PubMed |
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282.
| The rapid generation of mutation data matrices from protein sequences.Crossref | GoogleScholarGoogle Scholar | 1633570PubMed |
Jungblut, A. D., and Neilan, B. A. (2006). Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Archives of Microbiology 185, 107–114.
| Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 16402223PubMed |
Kaebernick, M., Neilan, B. A., Börner, T., and Dittmann, E. (2000). Light and the transcriptional response of the microcystin biosynthesis gene cluster. Applied and Environmental Microbiology 66, 3387–3392.
| Light and the transcriptional response of the microcystin biosynthesis gene cluster.Crossref | GoogleScholarGoogle Scholar | 10919796PubMed |
Kaneko, T., Nakajima, N., Okamoto, S., Suzuki, I., Tanabe, Y., Tamaoki, M., Nakamura, Y., Kasai, F., Watanabe, A., Kawashima, K., Kishida, Y., Ono, A., Shimizu, Y., Takahashi, C., Minami, C., Fujishiro, T., Kohara, M., Katoh, M., Nakazaki, N., Nakayama, S., Yamada, M., Tabata, S., and Watanabe, M. M. (2007). Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Research 14, 247–256.
| Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843.Crossref | GoogleScholarGoogle Scholar | 18192279PubMed |
Kardinaal, W. E. A., Tonk, L., Janse, I., Hol, S., Slot, P., Huisman, J., and Visser, P. M. (2007). Competition for light between toxic and nontoxic strains of the harmful cyanobacterium Microcystis. Applied and Environmental Microbiology 73, 2939–2946.
| Competition for light between toxic and nontoxic strains of the harmful cyanobacterium Microcystis.Crossref | GoogleScholarGoogle Scholar |
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30, 772–780.
| MAFFT multiple sequence alignment software version 7: Improvements in performance and usability.Crossref | GoogleScholarGoogle Scholar | 23329690PubMed |
Kottek, M., Grieser, J., Beck, C., Rudolf, B., and Rubel, F. (2006). World map of the Köppen–Geiger climate classification updated. Meteorologische Zeitschrift (Berlin) 15, 259–263.
| World map of the Köppen–Geiger climate classification updated.Crossref | GoogleScholarGoogle Scholar |
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. International Journal of Systematic and Evolutionary Microbiology 66, 1100–1103.
| OrthoANI: an improved algorithm and software for calculating average nucleotide identity.Crossref | GoogleScholarGoogle Scholar | 26585518PubMed |
Leikoski, N., Fewer, D. P., and Sivonen, K. (2009). Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Applied and Environmental Microbiology 75, 853–857.
| Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 19047393PubMed |
Leikoski, N., Fewer, D. P., Jokela, J., Alakoski, P., Wahlsten, M., and Sivonen, K. (2012). Analysis of an inactive cyanobactin biosynthetic gene cluster leads to discovery of new natural products from strains of the genus Microcystis. PLoS One 7, e43002.
| Analysis of an inactive cyanobactin biosynthetic gene cluster leads to discovery of new natural products from strains of the genus Microcystis.Crossref | GoogleScholarGoogle Scholar | 22952627PubMed |
Leikoski, N., Liu, L., Jokela, J., Wahlsten, M., Gugger, M., Calteau, A., Permi, P., Kerfeld, C. A., Sivonen, K., and Fewer, D. P. (2013). Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chemistry & Biology 20, 1033–1043.
| Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides.Crossref | GoogleScholarGoogle Scholar |
Lima, S. T., Alvarenga, D. O., Etchegaray, A., Fewer, D. P., Jokela, J., Varani, A. M., Sanz, M., Dörr, F. A., Pinto, E., Sivonen, K., and Fiore, M. F. (2017). Genetic organization of anabaenopeptin and spumigin biosynthetic gene clusters in the cyanobacterium Sphaerospermopsis torques-reginae itep-024. ACS Chemical Biology 12, 769–778.
| Genetic organization of anabaenopeptin and spumigin biosynthetic gene clusters in the cyanobacterium Sphaerospermopsis torques-reginae itep-024.Crossref | GoogleScholarGoogle Scholar | 28085246PubMed |
MacKintosh, C., Beattie, K. A., Klumpp, S., Cohen, P., and Codd, G. A. (1990). Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Letters 264, 187–192.
| Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants.Crossref | GoogleScholarGoogle Scholar | 2162782PubMed |
Martin, C., Oberer, L., Ino, T., König, W. A., Busch, M., and Weckesser, J. (1993). Cyanopeptolins, new depsipeptides from the cyanobacterium Microcystis sp. PCC 7806. The Journal of Antibiotics 46, 1550–1556.
| Cyanopeptolins, new depsipeptides from the cyanobacterium Microcystis sp. PCC 7806.Crossref | GoogleScholarGoogle Scholar | 8244882PubMed |
Medema, M. H., Kottmann, R., Yilmaz, P., Cummings, M., Biggins, J. B., Blin, K., de Bruijn, I., Chooi, Y. H., Claesen, J., Coates, R. C., Cruz-Morales, P., Duddela, S., Düsterhus, S., Edwards, D. J., Fewer, D. P., Garg, N., Geiger, C., Gomez-Escribano, J. P., Greule, A., Hadjithomas, M., Haines, A. S., Helfrich, E. J. N., Hillwig, M. L., Ishida, K., Jones, A. C., Jones, C. S., Jungmann, K., Kegler, C., Kim, H. U., Kötter, P., Krug, D., Masschelein, J., Melnik, A. V., Mantovani, S. M., Monroe, E. A., Moore, M., Moss, N., Nützmann, H.-W., Pan, G., Pati, A., Petras, D., Reen, F. J., Rosconi, F., Rui, Z., Tian, Z., Tobias, N. J., Tsunematsu, Y., Wiemann, P., Wyckoff, E., Yan, X., Yim, G., Yu, F., Xie, Y., Aigle, B., Apel, A. K., Balibar, C. J., Balskus, E. P., Barona-Gómez, F., Bechthold, A., Bode, H. B., Borriss, R., Brady, S. F., Brakhage, A. A., Caffrey, P., Cheng, Y.-Q., Clardy, J., Cox, R. J., De Mot, R., Donadio, S., Donia, M. S., van der Donk, W. A., Dorrestein, P. C., Doyle, S., Driessen, A. J. M., Ehling-Schulz, M., Entian, K.-D., Fischbach, M. A., Gerwick, L., Gerwick, W. H., Gross, H., Gust, B., Hertweck, C., Höfte, M., Jensen, S. E., Ju, J., Katz, L., Kaysser, L., Klassen, J. L., Keller, N. P., Kormanec, J., Kuipers, O. P., Kuzuyama, T., Kyrpides, N. C., Kwon, H.-J., Lautru, S., Lavigne, R., Lee, C. Y., Linquan, B., Liu, X., Liu, W., Luzhetskyy, A., Mahmud, T., Mast, Y., Méndez, C., Metsä-Ketelä, M., Micklefield, J., Mitchell, D. A., Moore, B. S., Moreira, L. M., Müller, R., Neilan, B. A., Nett, M., Nielsen, J., O’Gara, F., Oikawa, H., Osbourn, A., Osburne, M. S., Ostash, B., Payne, S. M., Pernodet, J.-L., Petricek, M., Piel, J., Ploux, O., Raaijmakers, J. M., Salas, J. A., Schmitt, E. K., Scott, B., Seipke, R. F., Shen, B., Sherman, D. H., Sivonen, K., Smanski, M. J., Sosio, M., Stegmann, E., Süssmuth, R. D., Tahlan, K., Thomas, C. M., Tang, Y., Truman, A. W., Viaud, M., Walton, J. D., Walsh, C. T., Weber, T., van Wezel, G. P., Wilkinson, B., Willey, J. M., Wohlleben, W., Wright, G. D., Ziemert, N., Zhang, C., Zotchev, S. B., Breitling, R., Takano, E., and Glöckner, F. O. (2015). Minimum information about a biosynthetic gene cluster. Nature Chemical Biology 11, 625–631.
| Minimum information about a biosynthetic gene cluster.Crossref | GoogleScholarGoogle Scholar | 26284661PubMed |
Mikalsen, B., Boison, G., Skulberg, O. M., Fastner, J., Davies, W., Gabrielsen, T. M., Rudi, K., and Jakobsen, K. S. (2003). Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. Journal of Bacteriology 185, 2774–2785.
| Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains.Crossref | GoogleScholarGoogle Scholar | 12700256PubMed |
Murakami, M., Sun, Q., Ishida, K., Matsuda, H., Okino, T., and Yamaguchi, K. (1997). Microviridins, elastase inhibitors from the cyanobacterium Nostoc minutum (NIES-26). Phytochemistry 45, 1197–1202.
| Microviridins, elastase inhibitors from the cyanobacterium Nostoc minutum (NIES-26).Crossref | GoogleScholarGoogle Scholar |
Na, S.-I., Kim, Y. O., Yoon, S.-H., Ha, S., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. Journal of Microbiology 56, 280–285.
| UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction.Crossref | GoogleScholarGoogle Scholar |
Nishizawa, T., Ueda, A., Nakano, T., Nishizawa, A., Miura, T., Asayama, M., Fujii, K., Harada, K., and Shirai, M. (2011). Characterization of the locus of genes encoding enzymes producing heptadepsipeptide micropeptin in the unicellular cyanobacterium Microcystis. Journal of Biochemistry 149, 475–485.
| Characterization of the locus of genes encoding enzymes producing heptadepsipeptide micropeptin in the unicellular cyanobacterium Microcystis.Crossref | GoogleScholarGoogle Scholar | 21212071PubMed |
Okano, K., Miyata, N., and Ozaki, Y. (2015). Whole genome sequence of the non-microcystin-producing Microcystis aeruginosa strain NIES-44. Genome Announcements 3, e00135-15.
| Whole genome sequence of the non-microcystin-producing Microcystis aeruginosa strain NIES-44.Crossref | GoogleScholarGoogle Scholar | 26227601PubMed |
Okino, T., Matsuda, H., Murakami, M., and Yamaguchi, K. (1993). Microginin, an angiotensin-converting enzyme inhibitor from the blue–green alga Microcystis aeruginosa. Tetrahedron Letters 34, 501–504.
| Microginin, an angiotensin-converting enzyme inhibitor from the blue–green alga Microcystis aeruginosa.Crossref | GoogleScholarGoogle Scholar |
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290.
| APE: analyses of phylogenetics and evolution in R language.Crossref | GoogleScholarGoogle Scholar | 14734327PubMed |
Parajuli, A., Kwak, D. H., Dalponte, L., Leikoski, N., Galica, T., Umeobika, U., Trembleau, L., Bent, A., Sivonen, K., Wahlsten, M., Wang, H., Rizzi, E., De Bellis, G., Naismith, J., Jaspars, M., Liu, X., Houssen, W., and Fewer, D. P. (2016). A unique tryptophan C-prenyltransferase from the Kawaguchipeptin biosynthetic pathway. Angewandte Chemie 55, 3596–3599.
| A unique tryptophan C-prenyltransferase from the Kawaguchipeptin biosynthetic pathway.Crossref | GoogleScholarGoogle Scholar | 26846478PubMed |
Park, H. D., Watanabe, M. F., Harada, K. I., Nagai, H., Suzuki, M., Watanabe, M., and Hayashi, H. (1993). Hepatotoxin (microcystin) and neurotoxin (anatoxin-a) contained in natural blooms and strains of cyanobacteria from Japanese freshwaters. Natural Toxins 1, 353–360.
| Hepatotoxin (microcystin) and neurotoxin (anatoxin-a) contained in natural blooms and strains of cyanobacteria from Japanese freshwaters.Crossref | GoogleScholarGoogle Scholar | 8167957PubMed |
Pearson, L. A., Hisbergues, M., Börner, T., Dittmann, E., and Neilan, B. A. (2004). Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Applied and Environmental Microbiology 70, 6370–6378.
| Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806.Crossref | GoogleScholarGoogle Scholar | 15528494PubMed |
Philmus, B., Christiansen, G., Yoshida, W. Y., and Hemscheidt, T. K. (2008). Post-translational modification in microviridin biosynthesis. ChemBioChem 9, 3066–3073.
| Post-translational modification in microviridin biosynthesis.Crossref | GoogleScholarGoogle Scholar | 19035375PubMed |
Portmann, C., Blom, J. F., Kaiser, M., Brun, R., Jüttner, F., and Gademann, K. (2008). Isolation of aerucyclamides C and D and structure revision of microcyclamide 7806A: heterocyclic ribosomal peptides from Microcystis aeruginosa PCC 7806 and their antiparasite evaluation. Journal of Natural Products 71, 1891–1896.
| Isolation of aerucyclamides C and D and structure revision of microcyclamide 7806A: heterocyclic ribosomal peptides from Microcystis aeruginosa PCC 7806 and their antiparasite evaluation.Crossref | GoogleScholarGoogle Scholar | 18973386PubMed |
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490.
| FastTree 2 – approximately maximum-likelihood trees for large alignments.Crossref | GoogleScholarGoogle Scholar | 20224823PubMed |
Rantala, A., Fewer, D. P., Hisbergues, M., Rouhiainen, L., Vaitomaa, J., Borner, T., and Sivonen, K. (2004). Phylogenetic evidence for the early evolution of microcystin synthesis. Proceedings of the National Academy of Sciences of the United States of America 101, 568–573.
| Phylogenetic evidence for the early evolution of microcystin synthesis.Crossref | GoogleScholarGoogle Scholar | 14701903PubMed |
Rhee, J. S., Choi, B. S., Han, J., Hwang, S. J., Choi, I. Y., and Lee, J. S. (2012). Draft genome database construction from four strains (NIES-298, FCY-26, -27, and -28) of the cyanobacterium Microcystis aeruginosa. Journal of Microbiology and Biotechnology 22, 1208–1213.
| Draft genome database construction from four strains (NIES-298, FCY-26, -27, and -28) of the cyanobacterium Microcystis aeruginosa.Crossref | GoogleScholarGoogle Scholar | 22814493PubMed |
Rohrlack, T., Christoffersen, K., Kaebernick, M., and Neilan, B. A. (2004). Cyanobacterial protease inhibitor microviridin J causes a lethal molting disruption in Daphnia pulicaria. Applied and Environmental Microbiology 70, 5047–5050.
| Cyanobacterial protease inhibitor microviridin J causes a lethal molting disruption in Daphnia pulicaria.Crossref | GoogleScholarGoogle Scholar | 15294849PubMed |
Rouhiainen, L., Vakkilainen, T., Siemer, B. L., Buikema, W., Haselkorn, R., and Sivonen, K. (2004). Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Applied and Environmental Microbiology 70, 686–692.
| Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90.Crossref | GoogleScholarGoogle Scholar | 14766543PubMed |
Rouhiainen, L., Jokela, J., Fewer, D. P., Urmann, M., and Sivonen, K. (2010). Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (cyanobacteria). Chemistry & Biology 17, 265–273.
| Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (cyanobacteria).Crossref | GoogleScholarGoogle Scholar |
Rounge, T. B., Rohrlack, T., Tooming-Klunderud, A., Kristensen, T., and Jakobsen, K. S. (2007). Comparison of cyanopeptolin genes in Planktothrix, Microcystis, and Anabaena Strains: evidence for independent evolution within each genus. Applied and Environmental Microbiology 73, 7322–7330.
| Comparison of cyanopeptolin genes in Planktothrix, Microcystis, and Anabaena Strains: evidence for independent evolution within each genus.Crossref | GoogleScholarGoogle Scholar | 17921284PubMed |
Rounge, T., Rohrlack, T., Nederbragt, A., Kristensen, T., and Jakobsen, K. (2009). A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genomics 10, 396.
| A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain.Crossref | GoogleScholarGoogle Scholar | 19706155PubMed |
Sandrini, G., Matthijs, H. C. P., Verspagen, J. M. H., Muyzer, G., and Huisman, J. (2014). Genetic diversity of inorganic carbon uptake systems causes variation in CO2 response of the cyanobacterium Microcystis. The ISME Journal 8, 589–600.
| Genetic diversity of inorganic carbon uptake systems causes variation in CO2 response of the cyanobacterium Microcystis.Crossref | GoogleScholarGoogle Scholar | 24132080PubMed |
Sant’Anna, C. L., de Carvalho, L. R., Fiore, M. F., Silva-Stenico, M. E., Lorenzi, A. S., Rios, F. R., Konno, K., Garcia, C., and Lagos, N. (2011). Highly toxic Microcystis aeruginosa strain, isolated from São Paulo, Brazil, produce hepatotoxins and paralytic shellfish poison neurotoxins. Neurotoxicity Research 19, 389–402.
| Highly toxic Microcystis aeruginosa strain, isolated from São Paulo, Brazil, produce hepatotoxins and paralytic shellfish poison neurotoxins.Crossref | GoogleScholarGoogle Scholar | 20376712PubMed |
Scherer, M., Bezold, D., and Gademann, K. (2016). Investigating the toxicity of the aeruginosin chlorosulfopeptides by chemical synthesis. Angewandte Chemie International Edition 55, 9427–9431.
| Investigating the toxicity of the aeruginosin chlorosulfopeptides by chemical synthesis.Crossref | GoogleScholarGoogle Scholar | 27332048PubMed |
Schreuder, H., Liesum, A., Lönze, P., Stump, H., Hoffmann, H., Schiell, M., Kurz, M., Toti, L., Bauer, A., Kallus, C., Klemke-Jahn, C., Czech, J., Kramer, D., Enke, H., Niedermeyer, T. H. J., Morrison, V., Kumar, V., and Brönstrup, M. (2016). Isolation, co-crystallization and structure-based characterization of anabaenopeptins as highly potent inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa). Scientific Reports 6, 32958.
| Isolation, co-crystallization and structure-based characterization of anabaenopeptins as highly potent inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa).Crossref | GoogleScholarGoogle Scholar | 27604544PubMed |
Sivonen, K., and Jones, G. (1999). Cyanobacterial toxins. In ‘Toxic Cyanobacteria in Water: a Guide to their Public Health Consequences, Monitoring and Management’. (Eds I. Chorus and J. Bartram.) pp. 41–111. (E and FN Spon: London, UK.)
Sivonen, K., Leikoski, N., Fewer, D. P., and Jokela, J. (2010). Cyanobactins – ribosomal cyclic peptides produced by cyanobacteria. Applied Microbiology and Biotechnology 86, 1213–1225.
| Cyanobactins – ribosomal cyclic peptides produced by cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 20195859PubMed |
Stamatakis, A. (2006). Phylogenetic models of rate heterogeneity: a high performance computing perspective (2006). In ‘Proceedings 20th IEEE International Parallel and Distributed Processing Symposium’, 25–29 April 2006, Rhodes Island, Greece. INSPEC Accession Number 8969655. (IEEE.)
Strangman, W. K., and Wright, J. L. C. (2016). Microginins 680, 646, and 612 – new chlorinated Ahoa-containing peptides from a strain of cultured Microcystis aeruginosa. Tetrahedron Letters 57, 1801.
| Microginins 680, 646, and 612 – new chlorinated Ahoa-containing peptides from a strain of cultured Microcystis aeruginosa.Crossref | GoogleScholarGoogle Scholar |
Tanabe, Y., Kaya, K., and Watanabe, M. M. (2004). Evidence for recombination in the microcystin synthetase (mcy) genes of toxic cyanobacteria Microcystis spp. Journal of Molecular Evolution 58, 633–641.
| Evidence for recombination in the microcystin synthetase (mcy) genes of toxic cyanobacteria Microcystis spp.Crossref | GoogleScholarGoogle Scholar | 15461420PubMed |
Tillett, D., Dittmann, E., Erhard, M., von Döhren, H., Börner, T., and Neilan, B. A. (2000). Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chemistry & Biology 7, 753–764.
| Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system.Crossref | GoogleScholarGoogle Scholar |
Tillett, D., Parker, D. L., and Neilan, B. A. (2001). Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Applied and Environmental Microbiology 67, 2810–2818.
| Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies.Crossref | GoogleScholarGoogle Scholar | 11375198PubMed |
Tooming-Klunderud, A., Rohrlack, T., Shalchian-Tabrizi, K., Kristensen, T., and Jakobsen, K. S. (2007). Structural analysis of a non-ribosomal halogenated cyclic peptide and its putative operon from Microcystis: implications for evolution of cyanopeptolins. Microbiology 153, 1382–1393.
| Structural analysis of a non-ribosomal halogenated cyclic peptide and its putative operon from Microcystis: implications for evolution of cyanopeptolins.Crossref | GoogleScholarGoogle Scholar | 17464052PubMed |
Tria, F. D. K., Landan, G., and Dagan, T. (2017). Phylogenetic rooting using minimal ancestor deviation. Nature Ecology & Evolution 1, 0193.
| Phylogenetic rooting using minimal ancestor deviation.Crossref | GoogleScholarGoogle Scholar |
Vernikos, G. S., and Parkhill, J. (2006). Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22, 2196–2203.
| Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands.Crossref | GoogleScholarGoogle Scholar | 16837528PubMed |
Weiz, A. R., Ishida, K., Makower, K., Ziemert, N., Hertweck, C., and Dittmann, E. (2011). Leader peptide and a membrane protein scaffold guide the biosynthesis of the tricyclic peptide microviridin. Chemistry & Biology 18, 1413–1421.
| Leader peptide and a membrane protein scaffold guide the biosynthesis of the tricyclic peptide microviridin.Crossref | GoogleScholarGoogle Scholar |
Wiese, M., D’Agostino, P. M., Mihali, T. K., Moffitt, M. C., and Neilan, B. A. (2010). Neurotoxic alkaloids: saxitoxin and its analogs. Marine Drugs 8, 2185–2211.
| Neurotoxic alkaloids: saxitoxin and its analogs.Crossref | GoogleScholarGoogle Scholar | 20714432PubMed |
Yamaguchi, H., Suzuki, S., Tanabe, Y., Osana, Y., Shimura, Y., Ishida, K., and Kawachi, M. (2015). Complete genome sequence of Microcystis aeruginosa NIES-2549, a bloom-forming cyanobacterium from Lake Kasumigaura, Japan. Genome Announcements 3, e00551-15.
| Complete genome sequence of Microcystis aeruginosa NIES-2549, a bloom-forming cyanobacterium from Lake Kasumigaura, Japan.Crossref | GoogleScholarGoogle Scholar | 26494683PubMed |
Yamaguchi, H., Suzuki, S., Osana, Y., and Kawachi, M. (2018). Complete genome sequence of Microcystis aeruginosa NIES-2481 and common genomic features of Group G M. aeruginosa. Journal of Genomics 6, 30–33.
| Complete genome sequence of Microcystis aeruginosa NIES-2481 and common genomic features of Group G M. aeruginosa.Crossref | GoogleScholarGoogle Scholar | 29576807PubMed |
Yang, C., Zhang, W., Ren, M., Song, L., Li, T., and Zhao, J. (2013). Whole-genome sequence of Microcystis aeruginosa TAIHU98, a nontoxic bloom-forming strain isolated from Taihu Lake, China. Genome Announcements 1, e00333-13.
| Whole-genome sequence of Microcystis aeruginosa TAIHU98, a nontoxic bloom-forming strain isolated from Taihu Lake, China.Crossref | GoogleScholarGoogle Scholar | 24092783PubMed |
Yang, C., Lin, F., Li, Q., Li, T., and Zhao, J. (2015). Comparative genomics reveals diversified CRISPR-Cas systems of globally distributed Microcystis aeruginosa, a freshwater bloom-forming cyanobacterium. Frontiers in Microbiology 6, 394.
| Comparative genomics reveals diversified CRISPR-Cas systems of globally distributed Microcystis aeruginosa, a freshwater bloom-forming cyanobacterium.Crossref | GoogleScholarGoogle Scholar | 26029174PubMed |
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek – International Journal of General and Molecular Microbiology 110, 1281–1286.
| A large-scale evaluation of algorithms to calculate average nucleotide identity.Crossref | GoogleScholarGoogle Scholar |
Zhang, Q., Yang, X., Wang, H., and van der Donk, W. A. (2014). High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chemical Biology 9, 2686–2694.
| High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis.Crossref | GoogleScholarGoogle Scholar | 25244001PubMed |
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C., and Dittmann, E. (2008a). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie 47, 7756–7759.
| Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria.Crossref | GoogleScholarGoogle Scholar | 18683268PubMed |
Ziemert, N., Ishida, K., Quillardet, P., Bouchier, C., Hertweck, C., de Marsac, N. T., and Dittmann, E. (2008b). Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Applied and Environmental Microbiology 74, 1791–1797.
| Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa.Crossref | GoogleScholarGoogle Scholar | 18245249PubMed |
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C., and Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and Environmental Microbiology 76, 3568–3574.
| Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides.Crossref | GoogleScholarGoogle Scholar | 20363789PubMed |
Zilliges, Y., Kehr, J. C., Meissner, S., Ishida, K., Mikkat, S., Hagemann, M., Kaplan, A., Börner, T., and Dittmann, E. (2011). The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6, e17615.
| The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions.Crossref | GoogleScholarGoogle Scholar | 21445264PubMed |