Genetic diversity and differentiation in south-western Australian bloodwoods (Corymbia section Calophyllae, Myrtaceae) with different ranges and abundance
Jane Sampson A , Sarah Tapper A , David Coates A , Margaret Hankinson A , Shelley McArthur A and Margaret Byrne A *
A *
A Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Locked Bag 104, Bentley Delivery Centre, Perth, WA 6983, Australia.
Australian Journal of Botany 70(2) 146-157 https://doi.org/10.1071/BT21081
Submitted: 4 July 2021 Accepted: 20 December 2021 Published: 16 February 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
An understanding of how variation is shared within and among closely related species is important for understanding evolutionary processes and managing biological diversity. We studied genetic structure in the three species occurring in south-western Australia that form the small and distinct monophyletic section Calophyllae of the genus Corymbia. We compared diversity in nuclear microsatellites and chloroplast DNA sequences in two species with patchy distributions, namely, Corymbia haematoxylon (Maiden) K.D. Hill & L.A.S. Johnson and Corymbia ficifolia (F. Muell.) K.D. Hill & L.A.S. Johnson, with that in the widespread congener, C. calophylla (Lindl.) K.D. Hill & L.A.S. Johnson. Consistent with predictions for the influence of range and abundance on genetic structure in the Australian flora, population differentiation was higher in the two restricted patchy species than in the widespread, semicontinuous C. calophylla. Genetic diversity in C. haematoxylon was similar to that in C. calophylla, but diversity was lower in the highly localised C. ficifolia, likely owing to genetic bottlenecks. All three species were distinguished by nuclear SSR variation, but C. haematoxylon and C. ficifolia each shared chloroplast haplotypes with C. calophylla from incomplete lineage sorting of ancestral variation and introgression. Limited evidence of recent hybridisation in two populations of C. haematoxylon was also present.
Keywords: bottleneck, differentiation, diversity, expansion, forest tree, hybridisation, inbreeding, lineage sorting, localised range, patchy abundance.
Introduction
Corymbia K.D.Hill & L.A.S.Johnson is a sister lineage to Eucalyptus and Angophora (Bayly et al. 2013) and one of the three ecologically and economically important sclerophyll genera commonly known as eucalypts. The southern lineage of Corymbia on the western side of the Australian continent is separated from other taxa in the genus by a large geographic disjunction. There are three closely related species that comprise the ‘south-western bloodwoods’ in section Calophyllae (Bayly et al. 2013), namely, Corymbia haematoxylon (Maiden) K.D.Hill & L.A.S.Johnson, Corymbia ficilfolia (F.Muell.) K.D.Hill & L.A.S.Johnson, and Corymbia calophylla (Lindl.) K.D.Hill & L.A.S.Johnson. Nicolle (2019) also included the taxon Corymbia chlorolampra K.D.Hill & L.A.S.Johnson in section Calophyllae; however, the name is not generally accepted for use in the Western Australian Flora.
Morphologically, C. haematoxylon, C. ficifolia and C. calophylla are distinct tree species that generally occupy different habitats in the mesic (6 001 500 mm of rainfall per annum) area of the south-western corner of Western Australia (Fig. 1). There is strong morphological and molecular evidence that section Calophyllae forms a monophyletic group (Parra-O et al. 2009; González-Orozco et al. 2016) with the nearest closest relative, Corymbia gummifera (Gaertn.) K.D.Hill & L.A.S.Johnson, being found in eastern Australia. A recent phylogenetic analysis of eucalypts (González-Orozco et al. 2016) based on nuclear (internal transcribed spacer, ITS, and external transcribed spacer, ETS) and chloroplast sequences (matK and psbA–trnH) proposed a temporal sequence of species divergence in section Calophyllae, beginning with divergence of C. haematoxylon, followed by C. calophylla and C. ficifolia. A detailed study of the evolutionary history of C. calophylla on the basis of chloroplast haplotype variation (Sampson et al. 2018) proposed an origin in the northern part of the distribution and a predominantly southward episodic spatial expansion from the early Pleistocene.
|
|
The widespread, semi-continuous distribution of C. calophylla extends over the forest and wetter woodland regions (800–1300 mm) of south-western Australia (Churchill 1968), with some isolated populations that extend north into the transitional rainfall zone (300–600 mm) on wetter patches of clay soils. In contrast, C. haematoxylon and C. ficifolia have restricted allopatric distributions within the range of C. calophylla. Corymbia haematoxylon occurs in sporadic patches on and near the crest of the Darling Escarpment, with a disjunct population ∼270 km north at Mount Lesueur. Hill and Johnson (1995) described the Mount Lesueur population as Corymbia chlorolampra K.D.Hill & L.A.S.Johnson, describing the taxon as a sharply distinguished northern variant, that is now relictual, rare and somewhat displaced by hybrids with C. calophylla. The distribution of C. haematoxylon is largely parapatric with C. calophylla; however, the species are sympatric in some populations. Corymbia ficifolia occurs in small, patchy populations in a very restricted area on the southern coast of Western Australia in the high-rainfall region and is occasionally sympatric with C. calophylla. Genetic analysis in C. calophylla found high nuclear diversity within populations and low differentiation among populations, indicating that the nuclear genetic structure of this widespread, semi-continuous species reflects significant associations with range and abundance (Sampson et al. 2018). This observation is consistent with the findings of a recent review of the Australian flora by Broadhurst et al. (2017). On the basis of these associations, species closely related to C. calophylla but with patchy abundance and regional or localised distributions would be expected to have greater nuclear genetic structure.
Here, we report a study of the distinct monophyletic south-western Australian section Calophyllae by using population genetic and phylogenetic analyses of cpDNA sequences and nSSR variation in C. haematoxylon and C. ficifolia, in combination with data reported for C. calophylla by Sampson et al. (2018). We examine diversity and differentiation to determine the contemporary and historical relationships among the species, and to compare the genetic structure of patchy, restricted species and semi-continuous, widespread species.
Materials and methods
Sampling and genotyping
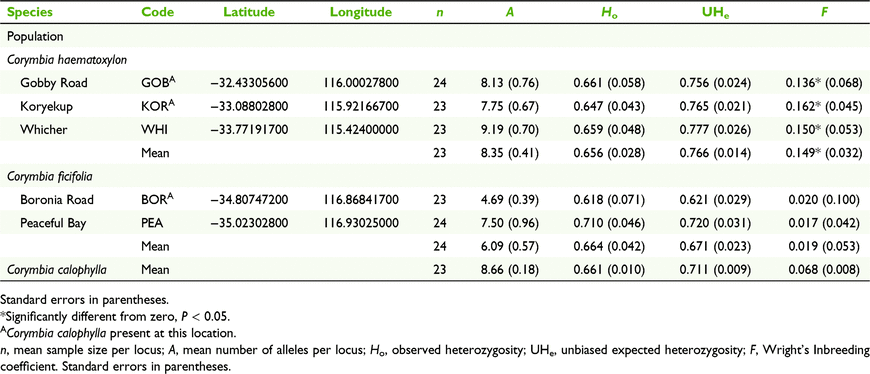
Leaves of individuals identified by morphology as C. haematoxylon or C. ficifolia were sampled from 24 well dispersed adult plants in each of three C. haematoxylon and two C. ficifolia populations (Table 1, Fig. 1). We were not able to sample the northern outlier population of C. haematoxylon because it had been recently burned. Genomic DNA was extracted from lysed, freeze-dried leaf material, following the methods in Byrne et al. (2016).
|
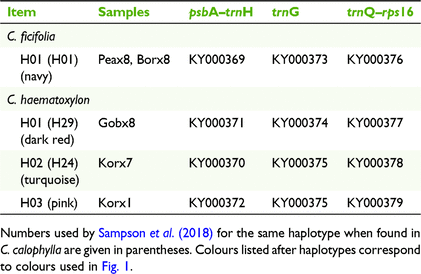
The chloroplast psbA–trnH and trnQ–rps16 intergenic spacer regions and the trnG intron were selected for amplification and sequencing in eight random samples from each of the five study populations. Sequence amplification and analysis were conducted according to Byrne and Hankinson (2012) and sequenced by Macrogen Inc. (Seoul, South Korea). SEQUENCHER (ver. 5.0, Genecodes Corp., Ann Arbor, MI, USA) was used to edit miscalls and to align and trim sequences. All three cpDNA regions were concatenated in MESQUITE (ver. 3.04, see http://www.mesquiteproject.org; Maddison and Maddison 2016) to a total sequence length of 2453 bp. One 21-bp inversion was uncovered in the psbA–trnH region. Following Whitlock et al. (2010), one configuration of the inversion was replaced with its reverse-complement and coded as a single transversion. Chloroplast haplotypes were identified using DNAsp (ver. 5.1.1, see http://www.ub.edu/dnasp/index_v5.html; Librado and Rozas 2009; Table 2).
|
Microsatellite loci developed for section Calophyllae (Sampson et al. 2018) were amplified using the Multiplex60 PCR program of the Qiagen Multiplex kit (Qiagen, Germany), separated on an Applied Biosystems 3730 capillary sequencer (Foster City, CA, USA), and 120 individuals (24 per population) were genotyped at 16 loci using GENEMAPPER (ver. 5.0, Applied Biosystems, Foster City, CA, USA). Tests for stutter bands and large allele dropout were conducted using MICROCHECKER (ver. 2.2.3, see http://www.nrp.ac.uk/nrp-strategic-alliances/elsa/software/microchecker/; Van Oosterhout et al. 2004). Tests of linkage disequilibrium among pairs of loci were performed with GENEPOP (ver. 4.2, see https://kimura.univ-montp2.fr/~rousset/Genepop.htm; Rousset 2008). The frequency of null alleles was estimated using Free NA (see https://www1.montpellier.inra.fr/CBGP/software/FreeNA/; Chapuis and Estoup 2007).
To enable comparisons of C. haematoxylon and C. ficifolia with the widespread congener C. calophylla, we obtained comparable data for C. calophylla from a previous study for cpDNA sequences from the same gene regions and nSSR data from the same loci (Sampson et al. 2018).
Chloroplast DNA diversity and divergence
We measured genetic diversity for cpDNA sequences in C. haematoxylon and C. ficifolia as nucleotide (π), haplotype (HD), and within-population haplotype (hs) diversity using ARLEQUIN (ver. 3.5.2.2, see http://cmpg.unibe.ch/software/arlequin35/; Excoffier and Lischer 2010). We estimated population genetic differentiation within species for cpDNA as GST and NST using PERMUT (ver. 2.0, see https://hal.inrae.fr/hal-02810373; Pons and Petit 1996). These measures are analogous to FST except that NST also takes the genetic distances between alleles (ordered analysis) into account as well as frequency. We estimated global and pairwise differentiation between species as FST using pooled data and ARLEQUIN (Excoffier and Lischer 2010). The partitioning of cpDNA genetic variation between species, among populations within species and within populations was examined by analysis of molecular variance (AMOVA) in ARLEQUIN (Excoffier and Lischer 2010) with significance tests being based on 1000 permutations based on distance matrices.
Tests for neutrality and population expansion were calculated with Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) in ARLEQUIN, and R2 (Ramos-Onsins and Rozas 2002), and F and D (Fu and Li 1993) in DNASP by using C. gummifera as an outgroup. To infer spatial and demographic history, we used mismatch distribution analyses in ARLEQUIN. Mismatch analyses test for deviation from the distribution of variation expected under spatial or demographic expansion models and, therefore, when P < 0.05, there is no support for expansion. Goodness-of-fit to models of spatial or demographic expansion were tested with Harpending’s raggedness index (HRag) and the sum of squared differences (SSD).
To examine the evolutionary relationships of chloroplast haplotypes in section Calophyllae, we constructed a median-joining maximum parsimony (MJMP) network in NETWORK (ver. 5.0, see https://www.fluxus-engineering.com/sharenet.htm; Bandelt et al. 1999). To estimate the divergence date of haplotypes, molecular dating and phylogeny reconstruction were simultaneously completed using a strict clock Bayesian analysis in BEAST (ver. 2.4.7, see https://www.beast2.org; Drummond and Rambaut 2007). All but one haplotype of C. haematoxylon and C. ficifolia were represented in the C. calophylla network (Sampson et al. 2018) and, therefore, so as to avoid repetition, we present only the analysis made using samples from C. haematoxylon and C. ficifolia, with C. gummifera as an outgroup. The substitution model was set to the GTR model as inferred as the best fit to the data by jModelTest (ver. 2.1.7, see https://github.com/ddarriba/jmodeltest2; Posada 2008). Divergence times were estimated under a strict clock model (i.e. with uniform rates across branches). Date estimates were constrained by the inclusion of a root calibration using the estimated time since most recent common ancestor (TMRCA) of C. haematoxylon, C. ficifolia and C. gummifera of 3.0 Ma. This calibration date was based on an unpublished dated version of the González-Orozco et al. (2016) eucalypt phylogeny that was calibrated by Andrew Thornhill (pers. comm.) by using the same eucalypt fossils as previously defined and used in Thornhill and Macphail (2012). In the absence of a 95% confidence interval for the TMRCA, dating was conducted using three different hypothetical confidence intervals applied to the root age calibration, namely, 2–4, 1–5 and 0–6 Ma, with four independent runs of 10 million generations performed for each of the three scenarios, sampling every 1000 generations. Convergence was assessed in Tracer (ver. 1.6, see https://github.com/beast-dev/tracer/releases/latest; Drummond and Rambaut 2007) and trees were combined using LogCombiner (ver. 1.6.2, see https://www.beast2.org) and TREEANNOTATOR (ver. 1.6.2, see https://www.beast2.org; Drummond and Rambaut 2007) was used to identify a maximum clade credibility tree.
Nuclear SSR diversity
We measured nSSR genetic variation for each species as mean multilocus parameters per population (number of alleles per locus, A; observed heterozygosity, Ho, unbiased expected heterozygosity, UHe; Wright’s inbreeding coefficient, F) by using GENALEX (ver. V6.501, see https://biology-assets.anu.edu.au/GenAlEx/Welcome.html; Peakall and Smouse 2012). We compared parameters between species using ANOVA and Fisher’s least significant difference (l.s.d.) post hoc test with data transformation for Ho and UHe.
We measured overall differentiation among populations within species (FST) by using FREENA with and without the excluding null alleles (ENA) method that corrects for null alleles, with 1000 bootstraps to generate 95% confidence intervals (Chapuis and Estoup 2007). We also estimated global and pairwise differentiation between populations and between species as FST, by using GENALEX with statistical testing by random permutations. If interspecific gene flow occurs between geographically close populations of two species, this may be reflected as a significant relationship between genetic and geographic distances for the species in parapatric or sympatric parts of their ranges. To test for this possibility in sympatric and parapatric populations of C. haematoxylon (GOB, KOR, WHI) and C calophylla (SER, KER, PEE, EAT, LEN, MEE), we used a Mantel procedure in GENALEX to calculate a correlation between log10 pairwise geographic distances and linearised pairwise genetic distances. Similar tests were not undertaken for C. ficifolia with C. calophylla because of the small number of populations.
Nuclear DNA structure
We used both phenetic and Bayesian analyses to examine genetic structure and whether species boundaries based on morphology corresponded with genetic differentiation. Phenetic analyses in PHYLIP (ver. 3.69, see https://evolution.genetics.washington.edu/phylip.html; Felsenstein 1989) were used to construct an unrooted neighbour-joining (NJ) tree based on CS chord genetic distance calculated in MSA: MicroSatellite Analyzer (ver. 4.05, see https://www.softpedia.com/get/Science-CAD/MSAnalyzer.shtml; Dieringer and Schlötterer 2003), with clustering patterns validated with 1000 bootstraps. We used Bayesian methods implemented in the program STRUCTURE (ver. 2.3.4, see https://web.stanford.edu/group/pritchardlab/structure.html; Pritchard et al. 2000) to identify genetic clusters (K) and their distribution in individuals. To assess the optimum number of clusters and the presence of subclusters in STRUCTURE analyses, we used the ΔK statistic of Evanno et al. (2005) estimated in CLUMPAK (see http://clumpak.tau.ac.il; Kopelman et al. 2015), which tends to identify the uppermost level of structure. Two or more optimal K may be found if samples are taken, as they were in this study, from hierarchically structured samples (section Calophyllae, species, population; Evanno et al. 2005; Wang 2017). Following the recommendations of (Gilbert et al. 2012) to promote reproducibility of analyses using STRUCTURE, we ran hierarchical analyses with the combined species as the highest level. This showed two clusters (q1, q2) that corresponded almost entirely to C. calophylla (97.3% of individuals with q1 ≥ 0.95), and a second cluster (q2 ≥ 0.95) containing both C. haematoxylon and C. ficifolia (Fig. 2a). We analysed C. haematoxylon and C. ficifolia as the lower level of the hierarchy. We ran 20 replicates with a burn-in of 100 000 with 300 000 iterations for Markov-chain Monte Carlo parameters for K = 1–20 or K = 1–10 possible clusters for upper or lower levels of the hierarchy respectively.
|
|
In a recent review of studies using STRUCTURE, Wang (2017) concluded that when the sample sizes from groups are highly unbalanced, as was the case in this study, STRUCTURE tends to merge populations represented by smaller samples. In this situation, Wang (2017) recommends use of a specific parameter combination to avoid misidentification of structure and to estimate admixture. We adopted these recommendations and used the parameters of no prior knowledge, the alternative ancestry prior of separate alphas for each population, an initial ALPHA value of 0.1, and the correlated allele frequency models.
Results
Chloroplast diversity and divergence
Haplotypes from each species are denoted by subscripts (e.g. H01H, C. haematoxylon; H01F, C. ficifolia; H01C, C. calophylla). No cpDNA diversity was found in C. ficifolia because it had only one haplotype. The single C. ficifolia haplotype (H01F; Table 2) was also the most common and one of the two more widely distributed haplotypes found in C. calophylla by Sampson et al. (2018) (H01C; Fig. 1). Chloroplast DNA diversity was low in C. haematoxylon, (π = 0.009, s.d. = 0.005; HD = 0.721, s.d. = 0.036; hS = 0.083, s.d. = 0.083) with four haplotypes being detected, but differentiation among populations was high (GST = 0.917, s.d. = 0.083, NST = 0.976, s.d. = 0.032), with no haplotypes shared among populations. There was one haplotype specific to C. haematoxylon (H03H), and three haplotypes (H01H, H02H, H04H) that were shared with C. calophylla (H29C, H24C, H15C; Sampson et al. 2018). The shared haplotypes were found in populations within 50 km of each other (Fig. 1).
In the haplotype network, the C. ficifolia haplotype and three of the four C. haematoxylon haplotypes were located at one end of the network with weakly diverged haplotypes from C. calophylla (H01F, H02H, H03H, H04H; Fig. 3a). In contrast, the divergent H01H haplotype, which was also found in C. calophylla (H29C), was located at the opposite end of the network among highly divergent haplotypes of C. calophylla.
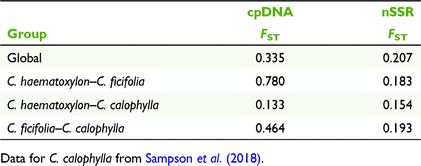
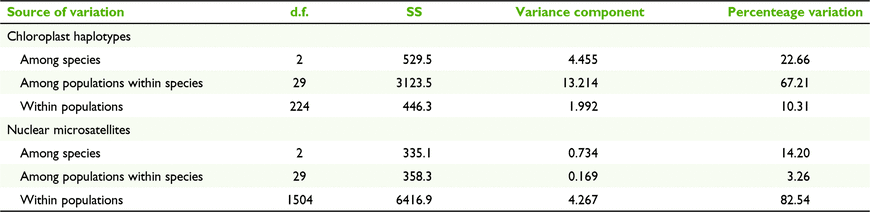
Overall, cpDNA genetic differentiation among taxa was moderate (FST = 0.335; Table 3), but C. ficifolia was more highly differentiated from C. haematoxylon (FST = 0.780) and C. calophylla (FST = 0.464) than C. haematoxylon was from C. calophylla (FST = 0.133). AMOVA analyses of the combined species showed more cpDNA genetic diversity among populations within species (67.21%; Table 4) than among species (22.66%).
|
|
We found evidence of spatial expansion in chloroplast DNA sequence data from C. haematoxylon as deviation from neutral expectations [(SSD) = 0.112, P > 0.05 (HRag) = 0.342, P > 0.05)]. However, there was no significant evidence to support demographic expansion (Tajima’s D = 2.504; Ramos-Onsins and Rozas R2 = 0.159, Fu’s FS = 20.929, Fu and Li’s D = 1.624, Fu and Li’s F = 2.327). Mismatch analyses did not conform to a model of demographic expansion (SSD = 0.178, P < 0.05; HRag = 0.343, P < 0.05).
In the BEAST (ver. 2.4.7) phylogenetic reconstruction analysis using the 0–6-Ma CI (Fig. 3b), divergence dates for haplotypes shared with C. calophylla were similar to those obtained by Sampson et al. (2018), although confidence intervals were larger owing to the smaller sample size. In this analysis, the H01H (=H29C) haplotype, placed at one end of the haplotype network (Fig. 3a), diverged in the early Pleistocene c. 2.152 Ma. All other haplotypes, found at the opposite end of the network with weakly diverged C. calophylla haplotypes, were estimated to have diverged later (c. 0.413 Ma). The unique C. haematoxylon haplotype (H03H) diverged most recently (c.0.122 Ma). Trees and dates derived from the 0–6-, 1–5- and 2–4-Ma CI applied to the root age calibration were similar.
Nuclear diversity and structure
We identified 18 species-specific alleles in C. haematoxylon and seven in C. ficifolia among 347 alleles found in section Calophyllae. No evidence of stutter or large allele dropout was detected for nSSR loci in C. haematoxylon and C. ficifolia. The overall frequency of missing data was low (2.28% missing single-locus genotypes), with a very low number of single-locus genotypes missing per individual (mean = 0.023). Two individuals were excluded from STRUCTURE analyses because of the frequency of missing single-locus genotypes (>50%), which can result in poor assignment. The frequency of significant composite genotypic disequilibrium among loci was high in one C. haematoxylon population (WHI) and very high in one C. ficifolia population (BOR); 28 instances of a possible 360 in C. haematoxylon where 18 were expected by chance (P < 0.05; 1XGOB, 1XKOR, 26XWHI), and 73 instances of a possible 240 in C. ficifolia where 12 were expected by chance (P < 0.05; 72XBOR, 1XPEA). Disequilibrium was concentrated in one population in each species and is unlikely to indicate chromosomal linkage. We detected 24 and 7 frequencies of null alleles significantly greater than 0.05 in C. haematoxylon and C. ficifolia respectively (data not shown). Comparison of FST (95% CI) estimates with and without ENA adjustment (reported here) showed that null alleles did not cause significant bias and, therefore, loci were not excluded from analyses.
Nuclear microsatellite diversity in C. ficifolia was moderate (A = 6.09, Ho = 0.664, UHe = 0.671; Table 1) and in C. haematoxylon it was high (A = 8.35, Ho = 0.656, UHe = 0.766), similar to that in C. calophylla (A = 8.66, Ho = 0.661, UHe = 0.711; Sampson et al. 2018). When all three species were compared, diversity was lowest in C. ficifolia, with A and UHe values significantly lower than in both C. calophylla and C. haematoxylon (F2,29 = 16.80, P < 0.01; F2,29 = 13.34, P < 0.01). Inbreeding coefficients were significantly positive for all populations of C. haematoxylon (F = 0.136–0.142; Table 1), but significant inbreeding was not detected in C. ficifolia.
All species were delimited by patterns of nSSR variation as shown by moderately high interspecific FST (Table 3), a greater proportion of diversity among species than among populations (Table 4), and by individuals and populations grouping as species in the STRUCTURE and NJ tree analyses (Fig. 2b, c). Genetic differentiation of populations within species was significantly higher in C. ficifolia (FST = 0.119, CI 0.086–0.152) than in C. haematoxylon (FST = 0.064, CI 0.048–0.081) or C. calophylla (FST = 0.032, CI 0.029–0.037; Sampson et al. 2018), and also higher in C. haematoxylon than in C. calophylla. Within the geographic range of C. haematoxylon, there was no significant association of pairwise geographic and genetic distances between C. haematoxylon and C. calophylla populations (r2 = −0.017, P > 0.05). Differentiation of species based on nSSRs was lowest between C. haematoxylon and C. calophylla (FST = 0.154; Table 3). Differentiation of C. ficifolia and C. haematoxylon, and of C. ficifolia and C. calophylla was higher, although similar. In contrast to cpDNA, more nSSR variation was found among species (14.20%; Table 4) than among populations within species (3.26%).
Species were clearly separated in the consensus NJ tree of CS chord genetic distance, with populations clustering as species with strong (>99%) bootstrap support and with no other significant groupings (Fig. 2c). In the STRUCTURE analysis, the three species were identified using two levels of hierarchical analyses (Fig. 2a for the higher level, and Supplementary Material Fig. S1a for the lower level). The lower level is not presented separately here because it is largely replicated in Fig. 2b showing all species. At the highest level (Fig. 2a), the optimal number of clusters identified was K = 2 by using ΔK (see Supplementary Material Fig. S1b), with high similarity of the program runs (h′ = 0.999). At K = 2, one cluster (q1) comprised predominantly C. calophylla individuals (97.3%, q1 ≥ 0.95) and a second cluster (q2) comprised all C. haematoxylon and C. ficifolia individuals (100%, q2 ≥ 0.95). At the lower level of analysis (Fig. S1a), the q2 cluster was subdivided into two subclusters by optimal ΔK (Supplementary Material Fig. S1c; h′ = 0.999), corresponding largely to C. haematoxylon and C. ficifolia. Admixture of the two subclusters above 5% was found in 1.4 and 10.4% of individuals respectively. Overall, the three species could be identified with high accuracy using two levels of hierarchical analysis. This pattern was also supported by the clustering pattern of the entire dataset (Fig. 2b), for which there was a minor change in the rate of ΔK at 3 (Supplementary Material Fig. S1b). A further peak at K = 5 showed three clusters corresponding to the species, with additional clusters indicating substructure within C. calophylla (see Sampson et al. 2018 for a description of this substructure).
When the entire dataset was anaysed at K = 3, admixture levels were low with 8.5, 2.1 and 0.8% of individuals in C. haematoxylon, C. ficifolia and C. calophylla respectively showing total admixture of ≥10%. Although low overall, C. haematoxylon showed the most admixture with 17.5% of WHI and 8.4% of GOB, with individuals showing ≥10% admixture from C. calophylla. The only C. ficifolia individual with admixture ≥10% was from PEA population and had ∼5% admixture from both parapatric C. calophylla and allopatric C. haematoxylon clusters. Two individuals in the KER population of C. calophylla that is sympatric with C. haematoxylon showed combined admixture to C. haematoxylon and C. ficifolia of more than 10%, and one individual in the MOW population showed admixture from C. ficifolia.
Discussion
Different patterns of diversity and differentiation were evident among the three Corymbia species of south-western Australia that have differences in range and abundance. Patterns of higher diversity in regional or widespread v. highly localised species, and greater differentiation among patchy v. semi-continuous species were consistent with the predictions of population genetic theory and the associations identified in a meta-analysis of the Australian flora (Broadhurst et al. 2017). Species were clearly differentiated in the nuclear genome but chloroplast haplotypes were shared across species. Incomplete lineage sorting, that is the retention of genetic variation from common ancestors, may be the most parsimonious explanation for shared haplotypes between C. ficifolia and C. calophylla, whereas both incomplete lineage sorting and ancient hybridisation have probably influenced the pattern of shared haplotypes in C. haematoxylon and C. calophylla. Admixture in nuclear variation in a few individuals also suggests some more recent hybridisation between C. calophylla and C. haematoxylon.
Relationships among species
All three species were separated by nuclear variation; however, although cpDNA sequence variation clearly delimited C. haematoxylon from C. ficifolia, neither of these two species were delimited from C. calophylla. The presence of shared haplotypes in cpDNA despite clear distinction of species on the basis of nuclear variation has been noted previously in other eucalypts (McKinnon et al. 2010), as well as in other angiosperms (Kikuchi et al. 2010; Ley and Hardy 2010; Wang et al. 2011). Shared haplotypes can arise as a result of retention of variation from a common ancestor because of the slower rate of lineage sorting in the maternally inherited chloroplast genome (Currat et al. 2008). It can also indicate past hybridisation in areas of past or present sympatry owing to chloroplast capture or seed dispersal (Dixon et al. 2007; Kikuchi et al. 2010).
The lack of shared haplotypes and clear differentiation in the nuclear genome between C. haematoxylon and C. ficifolia is likely to reflect complete lineage sorting in cpDNA, a lack of gene flow between allopatric populations of C. haematoxylon and C. ficifolia, and the timing of divergence of C. haematoxylon and C. ficifolia from a common ancestor. This would be consistent with the phylogeny proposed by González-Orozco et al. (2016) in which C. haematoxylon diverged first, followed by C. calophylla and, most recently, C. ficifolia.
For C. ficifolia, the southern location, low diversity and strong nSSR delineation of C. ficifolia from C. calophylla, together with a shared recently diverged haplotype, suggest that incomplete lineage sorting may be the more parsimonious explanation for shared variation between these species. Corymbia ficifolia is found in the wetter southern extremity of the distribution of C. calophylla. In the expansion scenario proposed by Sampson et al. (2018), the range of C. calophylla is proposed to have expanded southward, beginning c.0.426 million years ago, into the area where C. ficifolia now occurs, following the southward progress of increasing aridity. The shared recently diverged haplotype in C. ficifolia and divergence in the late Pleistocene are consistent with the proposed recent divergence of C. ficifolia from C. calophylla on the basis of the phylogeny of González-Orozco et al. (2016).
The relationship of C. haematoxylon and C. calophylla is probably more complex. The distribution of shared chloroplast haplotypes reflects some influence of the broad geographic distribution of populations, but haplotypes are not shared by the geographically closest populations, suggesting historical introgression and incomplete lineage sorting rather than recent hybridisation through seed-mediated gene flow. A geographic pattern of cpDNA variation has previously been interpreted as evidence of historical introgression in other Eucalyptus and Corymbia species (McKinnon et al. 2004; Pollock et al. 2013; Healey et al. 2018). Similarly, shared ancestral polymorphism is inferred when cpDNA haplotype sharing occurs among haplotypes internal to network rather than at the tips (Schaal and Leverich 2001) and for populations in close geographic proximity (Muir and Schlötterer 2005). This is the general pattern found in C. haematoxylon and C. calophylla for three of the four haplotypes. The other haplotype is highly divergent (separated by c.1.5 million years ago from the three haplotypes) tip haplotype that suggests introgression rather than incomplete lineage sorting, although that haplotype is not common in C. caloplhylla.
Current hybridisation has been observed in many eucalypts (e.g. Field et al. 2011; Bradbury et al. 2016; Robins et al. 2021) and also in species of Corymbia (Shepherd et al. 2008; Ochieng et al. 2010). Even though hybridisation between C. haematoxylon and C. calophylla, and between C. ficifolia and C. calophylla, has been noted anecdotally in localised situations, we did not seek to test this explicitly and rather sought to determine species-level relationships separate from any localised recent hybridisation. As expected, we did not see evidence of extensive hybridisation among the species because we observed a pattern of high differentiation and identified structure among species in the nuclear genome, and limited instances of admixture unrelated to geographic proximity. This is contrary to expectations when hybridisation is extensive where one might expect to find low differentiation and structure in nSSRs among the genetically distinct species, a significant association between geographical proximity and relatedness, and extensive admixture of individuals in geographically proximal populations of the different species (Edwards et al. 2008). The levels of genetic differentiation among species (FST = 0.154–0.193) are as expected for closely related taxa of eucalypts (Byrne 2008; Bradbury et al. 2021), indicating little introgression at the species level. Some admixture was evident in a few individuals in the WHI population of C. haematoxylon, suggesting a small level of recent hybridisation with C. calophylla in this population. Admixture identified in two individuals in the KER population of C. calophylla was from both C. haematoxylon and C. ficifolia, suggesting secondary genomic admixture rather than recent gene flow because this population is 275 km distant from the range of C. ficifolia. Other populations of C. calophylla within the distribution of C. haematoxylon did not show evidence of recent hybridisation.
Some historic and recent hybridisation may also be an explanation for the relatively lower level of differentiation between C. haematoxlyon and C. calophylla compared with C. ficifolia and C. calophylla, which is not consistent with the phylogenetic relationships and species divergence proposed by González-Orozco et al. (2016), in which C. haematoxylon diverged earlier and C. ficifolia later.
Species’ range and abundance and patterns of diversity
Many reviews of nuclear genetic variation have found that genetic structure is influenced by the interaction of mating systems, life-history traits, chromosomal variation, population distribution, and other ecological traits related to gene flow (Loveless and Hamrick 1984; Gitzendanner and Soltis 2000; Nybom 2004; Duminil et al. 2007; Ellstrand 2014). In a review focussed on the Australian flora, Broadhurst et al. (2017) found that the most important attributes influencing nuclear DNA differentiation were range disjunctions and abundance (patchy v. semi-continuous), whereas range size (localised v. regional or widespread) and abundance (localised or patchy v. semi-continuous) had greater influence on diversity. There are no significant range disjunctions in any of these three Corymbia species, but comparison of the patterns of differentiation reflected the predicted influence of abundance in the semi-continuously distributed C. calophylla compared with the patchily distributed C. haematoxylon and C. ficifolia. As predicted by population genetic theory, nuclear population differentiation was higher in C. haematoxylon and C. ficifolia than in C. calophylla. This can be attributed to lower gene flow among patchily distributed populations than among semi-continuous populations, because lower gene flow reduces the immigration of new variants and increases genetic divergence among populations. Identification of the influence of abundance and distribution is somewhat unexpected because gene flow through pollen dispersal can be extensive in eucalypts over long distances in open woodlands and in fragmented landscapes (Byrne et al. 2008; Sampson and Byrne 2008; Mimura et al. 2009), whereas it is generally localised in more continuous forest populations (Barbour et al. 2008; Jones et al. 2008). Seed-mediated dispersal is likely to be low in all three species because seeds are primarily gravity dispersed in eucalypts (Booth 2017) and thus unaffected by abundance or population distribution.
Comparisons within section Calophyllae also illustrated the influence of range and abundance on genetic diversity as well as differentiation. Abundance can influence diversity because patchily distributed populations are often smaller than semi-continuous populations, making them more prone to genetic drift and inbreeding that can reduce genetic variation (Loveless and Hamrick 1984; Ellstrand and Elam 1993). Wider-ranging and more abundant species should be buffered against loss of genetic diversity by their larger effective population size. The lower levels of diversity in the localised C. ficifolia than in both the regional C. haematoxylon and the widespread C. calophylla suggest that a threshold effect from range size has a stronger effect than abundance in these species. We did find evidence of significant inbreeding in populations of C. haematoxylon, although levels were low and not associated with reduced diversity. Overall, Wright’s F-values indicated mixed mating systems in all three species. This type of mating system is commonly found in eucalypts and considered to contribute to maintaining diversity (Byrne 2008; Barrett and Harder 2017), and we found no evidence that the observed significant inbreeding led to loss of diversity in patchy v. semi-continuous species, suggesting that the influence of range on diversity may be more important in these species.
Species such as C. ficifolia that are distributed as small, patchy populations within highly localised ranges, might be expected to be more prone to loss of diversity through drift and stochastic processes such as genetic bottlenecks. We found evidence of a bottleneck in one population of C. ficifolia (BOR) as a very high frequency of composite genotypic diseqilibrium, resulting in significantly lower diversity but without inbreeding. Bottlenecks might occur following recurrent severe fires that characterise the south-western landscape (Pickett 1997; Prideaux et al. 2010) and there was evidence of recent fires in BOR at the time of collection. The wider range and larger effective population size in C. haematoxylon probably buffers this species against the loss of diversity apparent in the more localised C. ficifolia.
The relatively low haplotype diversity we found in the patchy populations of regional C. haematoxylon and localised C. ficifolia when compared with high diversity in widespread C. calophylla was expected because the chloroplast genome generally maintains lower diversity than does the nuclear genome and is more influenced by abundance because of its lower effective population size. However, as yet, there are few comparative studies of closely related species with different distribution and abundance. In Western Australia, where patchy distributions are common, low levels of chloroplast nucleotide and haplotype diversity have also been reported by several studies of species with regional or localised patchy distributions (Kunzea pulchella, Tapper et al. 2014; Hakea oldfieldii, Sampson et al. 2015; Acacia atkinsiana, Levy et al. 2016), supporting this association.
Conclusions
Analysis of genetic relationships among the three species of Corymbia in south-western Australia confirmed distinction of the species, with greater differentiation between the geographically disjunct C. haematoxylon and C. ficifolia, and closer relationships between each of these with the widespread C. calophylla. As expected in closely related eucalypt species, some incomplete lineage sorting, introgression and ancient hybridisation was evident because cpDNA haplotypes in the two restricted species were shared with the widespread species. There was some evidence of limited recent hybridisation with admixture in a few individuals in a population of C. haematoxylon and in a population of C. calophylla within the distribution of C. haematoxylon.
The genetic structure in C. haematoxylon and C. ficifolia reflects the influence of geographic range and abundance on nuclear DNA diversity and differentiation. Greater differentiation within C. haematoxylon and C. ficifolia than within C. calophylla is probably due to a lower gene flow between small and patchy populations, despite the potential for extensive gene flow through pollen dispersal in eucalypt taxa. Lower diversity in C. ficifolia is likely to reflect the vulnerability of highly localised small populations to loss of diversity through stochastic events.
Supplementary material
Supplementary material is available online.
Data availability
The data that support this study will be shared on request to the corresponding author.
Conflicts of interest
Margaret Byrne is an Associate Editor of Australian Journal of Botany but did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Journal of Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
This work was funded by the Department of Biodiversity, Conservation and Attractions.
Acknowledgements
We thank Andrew Thornhill for providing node dates for divergence estimates and advice on BEAST analyses.
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48.| Median-joining networks for inferring intraspecific phylogenies.Crossref | GoogleScholarGoogle Scholar | 10331250PubMed |
Barbour RC, Otahal Y, Vaillancourt RE, Potts BM (2008) Assessing the risk of pollen-mediated gene flow from exotic Eucalyptus globulus plantations into native eucalypt populations of Australia. Biological Conservation 141, 896–907.
| Assessing the risk of pollen-mediated gene flow from exotic Eucalyptus globulus plantations into native eucalypt populations of Australia.Crossref | GoogleScholarGoogle Scholar |
Barrett SCH, Harder LD (2017) The ecology of mating and its evolutionary consequences in seed plants. Annual Review of Ecology, Evolution, and Systematics 48, 135–157.
| The ecology of mating and its evolutionary consequences in seed plants.Crossref | GoogleScholarGoogle Scholar |
Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, Bossinger G, Merchant A, Udovicic F, Woodrow IE, Tibbits J (2013) Chloroplast genome analysis of Australian eucalypts – Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics and Evolution 69, 704–716.
| Chloroplast genome analysis of Australian eucalypts – Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 23876290PubMed |
Booth TH (2017) Going nowhere fast: a review of seed dispersal in eucalypts. Australian Journal of Botany 65, 401–410.
| Going nowhere fast: a review of seed dispersal in eucalypts.Crossref | GoogleScholarGoogle Scholar |
Bradbury D, Grayling PM, MacDonald B, Hankinson M, Byrne M (2016) Clonality, interspecific hybridisation and inbreeding in a rare mallee eucalypt, Eucalyptus absita (Myrtaceae), and implications for conservation. Conservation Genetics 17, 193–205.
| Clonality, interspecific hybridisation and inbreeding in a rare mallee eucalypt, Eucalyptus absita (Myrtaceae), and implications for conservation.Crossref | GoogleScholarGoogle Scholar |
Bradbury D, Binks RM, Byrne M (2021) Genomic data inform conservation of rare tree species: clonality, diversity and hybridity in Eucalyptus series in a global biodiversity hotspot. Biodiversity and Conservation 30, 619–641.
| Genomic data inform conservation of rare tree species: clonality, diversity and hybridity in Eucalyptus series in a global biodiversity hotspot.Crossref | GoogleScholarGoogle Scholar |
Broadhurst L, Breed M, Lowe A, Bragg J, Catullo R, Coates D, Encinas-Viso F, Gellie N, James E, Krauss S, Potts B, Rossetto M, Shepherd M, Byrne M (2017) Genetic diversity and structure of the Australian flora. Diversity and Distributions 23, 41–52.
| Genetic diversity and structure of the Australian flora.Crossref | GoogleScholarGoogle Scholar |
Byrne M (2008) Eucalypt phylogeny, diversity and evolution. In ‘Plant genome: biodiversity and evolution. 1E: phanerogam – angiosperm’. (Eds AK Sharma, A Sharma) pp. 303–346. (Science Publishers: Enfield, NH, USA)
Byrne M, Hankinson M (2012) Testing the variability of chloroplast sequences for plant phylogeography. Molecular Ecology Resources 60, 569–574.
| Testing the variability of chloroplast sequences for plant phylogeography.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Elliott CP, Yates CJ, Coates DJ (2008) Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conservation Genetics 9, 97–105.
| Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne M, Coates DJ, Macdonald BM, Hankinson M, McArthur SM, van Leeuwen S (2016) High nuclear genetic differentiation, but low chloroplast diversity in a rare species, Aluta quadrata (Myrtaceae), with a disjunct distribution in the Pilbara, Western Australia. Australian Journal of Botany 64, 687–695.
| High nuclear genetic differentiation, but low chloroplast diversity in a rare species, Aluta quadrata (Myrtaceae), with a disjunct distribution in the Pilbara, Western Australia.Crossref | GoogleScholarGoogle Scholar |
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24, 621–631.
| Microsatellite null alleles and estimation of population differentiation.Crossref | GoogleScholarGoogle Scholar | 17150975PubMed |
Churchill DM (1968) The distribution and prehistory of Eucalyptus diversicolor F.Muell., E. marginata Donn ex SM., and E. calophylla R.Br. in relation to rainfall. Australian Journal of Botany 16, 125–151.
| The distribution and prehistory of Eucalyptus diversicolor F.Muell., E. marginata Donn ex SM., and E. calophylla R.Br. in relation to rainfall.Crossref | GoogleScholarGoogle Scholar |
Currat M, Ruedi M, Petit RJ, Excoffier L (2008) The hidden side of invasions: massive introgression by local genes. Evolution 62, 1908–1920.
| The hidden side of invasions: massive introgression by local genes.Crossref | GoogleScholarGoogle Scholar | 18452573PubMed |
Dieringer D, Schlötterer C (2003) MICROSATELLITE ANALYSER: a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes 3, 167–169.
| MICROSATELLITE ANALYSER: a platform independent analysis tool for large microsatellite data sets.Crossref | GoogleScholarGoogle Scholar |
Dixon CJ, Schönswetter P, Schneeweiss GM (2007) Traces of ancient range shifts in a mountain plant group (Androsace halleri complex, Primulaceae). Molecular Ecology 16, 3890–3901.
| Traces of ancient range shifts in a mountain plant group (Androsace halleri complex, Primulaceae).Crossref | GoogleScholarGoogle Scholar | 17850552PubMed |
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7, 214
| BEAST: Bayesian evolutionary analysis by sampling trees.Crossref | GoogleScholarGoogle Scholar | 17996036PubMed |
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits? The American Naturalist 169, 662–672.
| Can population genetic structure be predicted from life-history traits?Crossref | GoogleScholarGoogle Scholar | 17427136PubMed |
Edwards CE, Soltis DE, Soltis PS (2008) Using patterns of genetic structure based on microsatellite loci to test hypotheses of current hybridization, ancient hybridization and incomplete lineage sorting in Conradina (Lamiaceae). Molecular Ecology 17, 5157–5174.
| Using patterns of genetic structure based on microsatellite loci to test hypotheses of current hybridization, ancient hybridization and incomplete lineage sorting in Conradina (Lamiaceae).Crossref | GoogleScholarGoogle Scholar | 19120994PubMed |
Ellstrand NC (2014) Is gene flow the most important evolutionary force in plants? American Journal of Botany 101, 737–753.
| Is gene flow the most important evolutionary force in plants?Crossref | GoogleScholarGoogle Scholar | 24752890PubMed |
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24, 217–242.
| Population genetic consequences of small population size: implications for plant conservation.Crossref | GoogleScholarGoogle Scholar |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study.Crossref | GoogleScholarGoogle Scholar | 15969739PubMed |
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567.
| Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows.Crossref | GoogleScholarGoogle Scholar | 21565059PubMed |
Felsenstein J (1989) PHYLIP – phylogeny inference package (version 3.2). Cladistics 5, 164–166.
| PHYLIP – phylogeny inference package (version 3.2).Crossref | GoogleScholarGoogle Scholar |
Field DL, Ayre DJ, Whelan RJ, Young AG (2011) Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida. Heredity 106, 841–853.
| Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida.Crossref | GoogleScholarGoogle Scholar | 21063438PubMed |
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
| Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection.Crossref | GoogleScholarGoogle Scholar | 9335623PubMed |
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133, 693–709.
| Statistical tests of neutrality of mutations.Crossref | GoogleScholarGoogle Scholar | 8454210PubMed |
Gilbert KJ, Andrew RL, Bock DG, Franklin MT, Kane NC, Moore JS, Moyers BT, Renaut S, Rennison DJ, Veen T, Vines TH (2012) Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program structure. Molecular Ecology 21, 4925–4930.
| Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program structure.Crossref | GoogleScholarGoogle Scholar | 22998190PubMed |
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87, 783–792.
| Patterns of genetic variation in rare and widespread plant congeners.Crossref | GoogleScholarGoogle Scholar | 10860909PubMed |
González-Orozco CE, Pollock LJ, Thornhill AH, Mishler BD, Knerr N, Laffan SW, Miller JT, Rosauer DF, Faith DP, Nipperess DA, Kujala H, Linke S, Butt N, Külheim C, Crisp MD, Gruber B (2016) Phylogenetic approaches reveal biodiversity threats under climate change. Nature Climate Change 6, 1110–1114.
| Phylogenetic approaches reveal biodiversity threats under climate change.Crossref | GoogleScholarGoogle Scholar |
Healey A, Lee DJ, Furtado A, Henry RJ (2018) Evidence of inter-sectional chloroplast capture in Corymbia among sections Torellianae and Maculatae. Australian Journal of Botany 66, 369–378.
| Evidence of inter-sectional chloroplast capture in Corymbia among sections Torellianae and Maculatae.Crossref | GoogleScholarGoogle Scholar |
Hill K, Johnson L (1995) Systematic studies in the eucalypts 7. A revision of the bloodwoods, genus Corymbia (Myrtaceae). Telopea 6, 185–504.
| Systematic studies in the eucalypts 7. A revision of the bloodwoods, genus Corymbia (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Jones ME, Shepherd M, Henry R, Delves A (2008) Pollen flow in Eucalyptus grandis determined by paternity analysis using microsatellite markers. Tree Genetics & Genomes 4, 37–47.
| Pollen flow in Eucalyptus grandis determined by paternity analysis using microsatellite markers.Crossref | GoogleScholarGoogle Scholar |
Kikuchi R, Jae-Hong P, Takahashi H, Maki M (2010) Disjunct distribution of chloroplast DNA haplotypes in the understory perennial Veratrum album ssp. oxysepalum (Melanthiaceae) in Japan as a result of ancient introgression. New Phytologist 188, 879–891.
| Disjunct distribution of chloroplast DNA haplotypes in the understory perennial Veratrum album ssp. oxysepalum (Melanthiaceae) in Japan as a result of ancient introgression.Crossref | GoogleScholarGoogle Scholar |
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15, 1179–1191.
| Clumpak: a program for identifying clustering modes and packaging population structure inferences across K.Crossref | GoogleScholarGoogle Scholar | 25684545PubMed |
Levy E, Byrne M, Coates DJ, Macdonald BM, McArthur S, van Leeuwen S (2016) Contrasting influences of geographic range and distribution of populations on patterns of genetic diversity in two sympatric Pilbara acacias. PLoS ONE 11, e0163995
| Contrasting influences of geographic range and distribution of populations on patterns of genetic diversity in two sympatric Pilbara acacias.Crossref | GoogleScholarGoogle Scholar | 27768703PubMed |
Ley AC, Hardy OJ (2010) Species delimitation in the Central African herbs Haumania (Marantaceae) using georeferenced nuclear and chloroplastic DNA sequences. Molecular Phylogenetics and Evolution 57, 859–867.
| Species delimitation in the Central African herbs Haumania (Marantaceae) using georeferenced nuclear and chloroplastic DNA sequences.Crossref | GoogleScholarGoogle Scholar | 20813193PubMed |
Librado P, Rozas J (2009) DnaSP v.5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
| DnaSP v.5: a software for comprehensive analysis of DNA polymorphism data.Crossref | GoogleScholarGoogle Scholar | 19346325PubMed |
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15, 65–95.
| Ecological determinants of genetic structure in plant populations.Crossref | GoogleScholarGoogle Scholar |
Maddison WP, Maddison DR (2016) Mesquite: a modular system for evolutionary analysis. Available at http://mesquiteproject.org.
McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM (2004) Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359, 275–284.
| Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts.Crossref | GoogleScholarGoogle Scholar | 15101583PubMed |
McKinnon GE, Smith JJ, Potts BM (2010) Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Molecular Ecology 19, 1367–1380.
| Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species.Crossref | GoogleScholarGoogle Scholar | 20298471PubMed |
Mimura M, Barbour RC, Potts BM, Vaillancourt RE, Watanabe KN (2009) Comparison of contemporary mating patterns in continuous and fragmented Eucalyptus globulus native forests. Molecular Ecology 18, 4180–4192.
| Comparison of contemporary mating patterns in continuous and fragmented Eucalyptus globulus native forests.Crossref | GoogleScholarGoogle Scholar | 19769693PubMed |
Muir G, Schlötterer C (2005) Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.). Molecular Ecology 14, 549–561.
| Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.).Crossref | GoogleScholarGoogle Scholar | 15660945PubMed |
Nicolle D (2019) Classification of the eucalypts (Angophora, Corymbia and Eucalyptus) Version 4. Available at http://www.dn.com.au/Classification-Of-The-Eucalypts.
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13, 1143–1155.
| Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants.Crossref | GoogleScholarGoogle Scholar | 15078452PubMed |
Ochieng JW, Shepherd M, Baverstock PR, Nikles G, Lee DJ, Henry RJ (2010) Two sympatric spotted gum species are molecularly homogeneous. Conservation Genetics 11, 45–56.
| Two sympatric spotted gum species are molecularly homogeneous.Crossref | GoogleScholarGoogle Scholar |
Parra-O C, Bayly MJ, Drinnan A, Udovicic F, Ladiges P (2009) Phylogeny, major clades and infrageneric classification of Corymbia (Myrtaceae), based on nuclear ribosomal DNA and morphology. Australian Systematic Botany 22, 384–399.
| Phylogeny, major clades and infrageneric classification of Corymbia (Myrtaceae), based on nuclear ribosomal DNA and morphology.Crossref | GoogleScholarGoogle Scholar |
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539.
| GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update.Crossref | GoogleScholarGoogle Scholar | 22820204PubMed |
Pickett EJ (1997) ‘A Late Pleistocene and Holocene vegetation history of three lacustrine sequences from the Swan Coastal Plain.’ (The University of Western Australia: Perth, WA, Australia)
Pollock LJ, Bayly MJ, Nevill PG, Vesk PA (2013) Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia. Journal of Biogeography 40, 155–167.
| Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245.
| Measuring and testing genetic differentiation with ordered versus unordered alleles.Crossref | GoogleScholarGoogle Scholar | 8913764PubMed |
Posada D (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25, 1253–1256.
| jModelTest: phylogenetic model averaging.Crossref | GoogleScholarGoogle Scholar | 18397919PubMed |
Prideaux GJ, Gully GA, Couzens AMC, Ayliffe LK, Jankowski NR, Jacobs Z, Roberts RG, Hellstrom JC, Gagan MK, Hatcher LM (2010) Timing and dynamics of Late Pleistocene mammal extinctions in southwestern Australia. Proceedings of the National Academy of Sciences of the United States of America 107, 22157–22162.
| Timing and dynamics of Late Pleistocene mammal extinctions in southwestern Australia.Crossref | GoogleScholarGoogle Scholar | 21127262PubMed |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
| Inference of population structure using multilocus genotype data.Crossref | GoogleScholarGoogle Scholar | 10835412PubMed |
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution 19, 2092–2100.
| Statistical properties of new neutrality tests against population growth.Crossref | GoogleScholarGoogle Scholar | 12446801PubMed |
Robins TP, Binks RM, Byrne M, Hopper SD (2021) Landscape and taxon age are associated with differing patterns of hybridization in two Eucalyptus (Myrtaceae) subgenera. Annals of Botany 127, 49–62.
| Landscape and taxon age are associated with differing patterns of hybridization in two Eucalyptus (Myrtaceae) subgenera.Crossref | GoogleScholarGoogle Scholar | 32914170PubMed |
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8, 103–106.
| GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux.Crossref | GoogleScholarGoogle Scholar | 21585727PubMed |
Sampson JF, Byrne M (2008) Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Molecular Ecology 17, 2769–2781.
| Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba.Crossref | GoogleScholarGoogle Scholar | 18444981PubMed |
Sampson JF, Hankinson M, Mcarthur S, Tapper S, Langley M, Gibson N, Yates C, Byrne M (2015) Long-term ‘islands’ in the landscape: low gene flow, effective population size and genetic divergence in the shrub Hakea oldfieldii (Proteaceae). Botanical Journal of the Linnean Society 179, 319–334.
| Long-term ‘islands’ in the landscape: low gene flow, effective population size and genetic divergence in the shrub Hakea oldfieldii (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Sampson J, Tapper S, Coates D, Hankinson M, Mcarthur S, Byrne M (2018) Persistence with episodic range expansion from the early Pleistocene: the distribution of genetic variation in the forest tree Corymbia calophylla (Myrtaceae) in south-western Australia. Biological Journal of the Linnean Society. Linnean Society of London 123, 545–560.
| Persistence with episodic range expansion from the early Pleistocene: the distribution of genetic variation in the forest tree Corymbia calophylla (Myrtaceae) in south-western Australia.Crossref | GoogleScholarGoogle Scholar |
Schaal BA, Leverich WJ (2001) Plant population biology and systematics. Taxon 50, 679–695.
| Plant population biology and systematics.Crossref | GoogleScholarGoogle Scholar |
Shepherd M, Kasem S, Ablett G, Ochieng J, Crawford A (2008) Genetic structuring in the spotted gum complex (genus Corymbia, section Politaria). Australian Systematic Botany 21, 15–25.
| Genetic structuring in the spotted gum complex (genus Corymbia, section Politaria).Crossref | GoogleScholarGoogle Scholar |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
| Statistical method for testing the neutral mutation hypothesis by DNA polymorphism.Crossref | GoogleScholarGoogle Scholar | 2513255PubMed |
Tapper SL, Byrne M, Yates CJ, Keppel G, Hopper SD, Van Niel K, Schut AGT, Mucina L, Wardell-Johnson GW (2014) Isolated with persistence or dynamically connected? Genetic patterns in a common granite outcrop endemic. Diversity and Distributions 20, 987–1001.
| Isolated with persistence or dynamically connected? Genetic patterns in a common granite outcrop endemic.Crossref | GoogleScholarGoogle Scholar |
Thornhill AH, Macphail M (2012) Fossil myrtaceous pollen as evidence for the evolutionary history of Myrtaceae: a review of fossil Myrtaceidites species. Review of Palaeobotany and Palynology 176–177, 1–23.
| Fossil myrtaceous pollen as evidence for the evolutionary history of Myrtaceae: a review of fossil Myrtaceidites species.Crossref | GoogleScholarGoogle Scholar |
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4, 535–538.
| MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data.Crossref | GoogleScholarGoogle Scholar |
Wang J (2017) The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Molecular Ecology Resources 17, 981–990.
| The computer program structure for assigning individuals to populations: easy to use but easier to misuse.Crossref | GoogleScholarGoogle Scholar | 28028941PubMed |
Wang J, Wu Y, Ren G, Guo Q, Liu J, Lascoux M (2011) Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa. PLoS ONE 6, e26530
| Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa.Crossref | GoogleScholarGoogle Scholar | 22028897PubMed |
Whitlock BA, Hale AM, Groff PA (2010) Intraspecific inversions pose a challenge for the trnH–psbA plant DNA barcode. PLoS ONE 5, e11533
| Intraspecific inversions pose a challenge for the trnH–psbA plant DNA barcode.Crossref | GoogleScholarGoogle Scholar | 20644717PubMed |


