Long-lifetime green-emitting Tb3+ complexes for bacterial staining
Weronika Rochowiak A , Ewa Kasprzycka A C , Israel P. Assunção A D , Ulrich Kynast A * and Marina Lezhnina B *
A * and Marina Lezhnina B *
A Muenster University of Applied Sciences, Institute for Optical Technologies, Stegerwaldstr. 39, 48565 Steinfurt, Germany.
B Quantum Analysis GmbH, Mendelstraße 17, 48149 Münster, Germany.
C Present address: Faculty of Chemistry, University of Wroclaw, 14 F Joliot-Curie Street, 50-383 Wroclaw, Poland.
D Present address: Instituto Federal de Ciência e Tecnologia (IFSP), Campus Suzano, Avenue Mogi das Cruzes, 1501 Parque Suzano, Suzano, SP, Brazil.
Handling Editor: George Koutsantonis
Australian Journal of Chemistry 75(9) 754-759 https://doi.org/10.1071/CH21315
Submitted: 1 December 2021 Accepted: 18 February 2022 Published: 18 May 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
The present report describes a new approach to stain bacteria by means of rare earth complexes. We demonstrate with selected Gram-negative and positive bacteria (Escherichia coli, Micrococcus luteus, Bacillus megaterium) that these microbes can be stained efficiently with derivatives of N-phenylanthranilic acid, flufenamic acid in particular, and Tb3+ ions. Hence, the inherent advantages of rare earth complexes, e.g. strong optical absorption (>50 000 L × M−1 × cm−1) due to the antenna effect, large Stokes’ shifts (~10 000 cm−1) and very long emission decay times (millisecond range), and, not least, enhanced photostability can be fully exploited in fluorescence microscopy and spectroscopy of the bacteria; foreseeably, these findings will also be useful in flow cytometry and ELISA techniques.
Keywords: bacteria, fluorescence microscopy, lifetime, luminescence, rare earth complexes, staining.
Introduction
In the context of microbes, rare earth ions have scarcely been studied. Only a few reports are devoted to ‘direct’ rare earth stains for bacteria and spores,[1–4] the term ‘direct bacterial staining’ refering to simple procedures as known for ‘classical’ organic cell dyes like Sybr Green (SG) or propidium iodide (PI).[5] Often, these essentially just require the incubation of the bacterial suspensions with the dye. In contrast to that and larger in number, rare earth-based techniques predominantly employ ‘indirect’ staining techniques, i.e. aim at liberated bacterial DNA, other secreted biomolecules, or dedicated antibodies, which have been shown to be useful in several bioanalytical and immunological methods, like polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), fluorescence flow cytometry and lateral flow assays.[6–14] However, these indirect methods require tedious and time-consuming preliminary preparations of, for example, antibodies, fairly costly equipment and highly skilled personnel.
The intrinsic optical properties of Tb3+ ions, the focus here, arise from the nature of the f-orbitals: on one hand, their screening from the chemical environment advantageously leads to very narrow f–f excitation and emission lines, while the quantum mechanically forbidden nature of f–f transitions effect long decay times (ms) of the excited f-states, but also to disadvantageously weak absorption intensities on the other hand. To cope with the poor f–f absorptivity viz excitability in the bio-analytically relevant spectral range above 300 nm, the ‘antenna-effect’ is used to sensitize Tb3+. Numerous articles and reviews outline the mechanisms involved and their benefits for bio-analytics, just three of which, covering mechanisms, biomedical applications and some economical aspects are quoted here.[15–17] In brief, a suitable ligand with allowed, strong and comparably broad absorptions in the relevant spectral range forms a complex with the Tb3+ ion; after excitation of the ligand (1S0 → 1S*), it undergoes inter-system crossing into an associated triplet state (1S* → 3T, enabled by spin–orbit coupling), from which the energy is transferred radiationlessly to energetically close Tb3+ states. Finally, characteristic Tb3+ emissions will typically occur from the excited 5D4 state of Tb3+. The process is limited by the ligands’ triplet energies, which have to be located at least 2000 cm−1 above the emitting 5D4-level (20 500 cm−1) to prevent backtransfer into the ligand’s 3T level.[18,19] A most important complex described in the past is tri-anionic tris(pyridine-2,6-dicarboxylato)terbium, [Tb((C5H4N)(COO)2)3]3− (also referred to as [Tb(dipicolinate)3]3− or [Tb(dpa)3]3−),[20] especially as a stain for endospores,[1,2] though their excitation unfortunately lies in the deep UV below 300 nm.
The abovementioned long decay times can most favourably be used to discriminate the emission signal from other dyes, and the highly problematic autofluorescence in matrices of biological origin in particular (by time-gated luminescence).[21–23] The feasibility of using long-lived red-emitting microbe stains based on Eu diketonate complexes has recently been evaluated.[4] Furthermore, the narrow-line emissions of Tb3+, as well as most other rare earth ions of interest, can be of extreme value where filters have to be used to separate excitation and emission. Combined with the broad excitation bands of the complexes just below 400 nm, i.e. Stokes shifts of more than 150 nm, superb microbial luminescence monitors become available for the photoluminescent applications listed above.
Last but not least, we envisage that rare earth stains hold the promise of superior photostability over purely organic dyes like SG, photobleaching being an omnipresent bottleneck in fluorescence microscopy and flow cytometry.[24–26]
Experimental
Materials
The ligand, with a purity of >98%, was purchased from TCI chemicals (Belgium). For the experiments, ethanolic solutions were prepared and the desired TbCl3 ethanolic solutions were prepared from a 100 mM stock solution. The pure complex Tb(fluf)3 (fluf = flufenamate) was prepared as described earlier.[27]
Spectroscopy
Emission and excitation spectra as well as luminescence decay of the dispersions were measured on an Edinburgh FS5 spectrofluorometer (180 W Xe lamp, 5 W microsecond Xe flash lamp, cooled and stabilized photomultiplier R928P) at room temperature. The excitation and emission spectra were recorded after centrifugation and re-dispersion of the bacteria in buffer, yielding approximately 2.4 × 108 bacteria mL–1.
Microscopy
Microscope images were taken with a Leica DMi8 fluorescence microscope equipped with filtercubes (Tb3+: excitation 365 nm, emission 545 nm) and a CoolLED pE-4000 LED light source. The pictures of SG staining were obtained using an FITC filtercube (excitation 460–500 nm, emission 512–542 nm) and for PI staining, a TXR filtercube (excitation 540–580 nm, emission 592–668 nm) was used. The pictures were taken using 100× and 63× magnification objectives; the irradiance intensity was set to 30%. The camera exposure time was 130 ms.
Bacteria
The bacterial strains in this work (DSM 1116, Escherichia coli; DSM 20030, Micrococcus luteus; DSM 90, Bacillus megaterium) were purchased from DSMZ (Leibniz Institute DSMZ; German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany). The bacterial cultures were taken from tryptic soy agar plates and inoculated into tryptic soy broth (caso bouillon, Merck KGaA, Darmstadt, Germany) to grow overnight. Then the bacteria were centrifuged and redispersed in 50 mM HEPES buffer (pH 7.5) of above 99.5% purity purchased from Sigma–Aldrich.
The approximate numbers of bacteria in samples were calculated using the optical density of the dispersion at 600 nm. The density was measured on a Specord 200 Plus UV-visible spectrophotometer (Analytik Jena) using 1 cm acrylic cuvettes.
Bacterial dispersions in HEPES buffer (2.4 × 107 cells mL–1) were first incubated with 10 µL HFluf (30 mM ethanolic solution, 30 min) followed by treatment with 10 µL TbCl3 (10 mM ethanolic solution, 30 min).
Staining with the whole complex was done by incubating bacterial dispersions with 10 µL of 30 mM complex dissolved in ethanol for 1–2 h.
Results and discussion
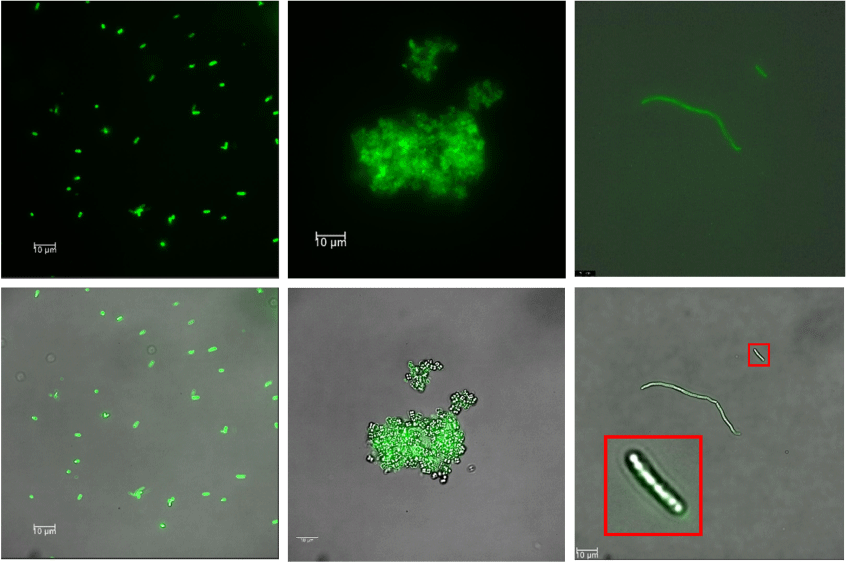
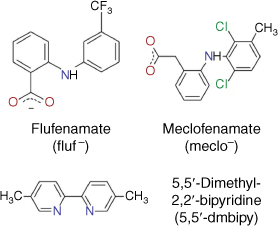
We recently described a new series of green-emitting Tb3+ complexes based on derivatives of N-phenylanthranilic acid (fenamic acid), with efficiencies up to 33% and decay times of 873 µs for Tb(meclofenamate)3(H2O)2 for example.[27] The efficiency of the complexes could be greatly enhanced by co-coordination with an ancillary 5,5′-dimethyl-2,2-bipyridine ligand (5,5'-dmbipy) to yield a quantum efficiency of 60% at a decay time of 1270 µs, this for the complex Tb(flufenamate)3(5,5′-dmbipy) (Fig. 1),[28] which showed the largest gain over the non-co-coordinated complexes. This gain via co-coordination nurtured our hope that the numerous functional groups on or in bacteria would be able to provide an analogous efficiency boost. Indeed, Tb(flufenamate)3H2O (Tb(fluf)3) proved to be the best complex for microbial staining and is the focus of the following discussion. To include the different membrane construction of Gram-negative (thickness of cell wall typically less than 10 nm, lipopolysaccharide surface) and Gram-positive (thickness of cell wall up to 80 nm, containing teichoic acids) bacteria, Escherichia coli (Gram-negative), Micrococcus luteus and Bacillus megaterium (both Gram-positive) were chosen; B. megaterium was selected as the largest of these bacteria and is known to readily form spores on stress.[29]
|
Generally, a two-step procedure was pursued: a first step involving incubation with flufenamate (fluf−), followed by the addition of Tb3+ solution as the second step. The fluorescence microscopy images and their overlay with the bright-field images are shown in Fig. 2. Obviously, all bacteria could be stained to give satisfying and bright fluorescence images, staining of E. coli working the best owing to its thin cell wall. M. luteus is known to form smaller aggregates (tetrads), which seem to agglomerate even further in the presence of Tb3+; B. megaterium takes up the least stain, this most likely being related to sporulation, as apparent in Fig. 1 (inset right image and Supplementary Fig. S1).
On plating the fluf−/Tb3+ incubated dispersions on agar plates, all samples formed colonies after 3 days. However, live-dead staining with SG (green emission in live bacteria) and PI (red emission in dead bacteria) revealed an increasing number of dead microbes, as indicated by the dominance of the PI emission from E. coli and B. megaterium (Supplementary Fig. S2). It should be pointed out, though, that care has to be taken in evaluating the fluorescence images: SG emission and PI absorption exhibit a large spectral overlap and energy transfer fluorescence resonance energy transfer (FRET); hence, the red PI emission can easily be overemphasized and falsify the picture. In the case of B. megaterium, no live bacteria were found after staining; however, as pointed out above, this may be related to sporulation.
We also tested bacterial viability by incubating all three species for 0.5, 2 and 24 h in the buffered fluf− solution alone with subsequent plating on agar plates; after 3 days, no difference with blanks (bacteria in buffer only) was noted. Additionally, live–dead stains (SG and PI) support the conclusion that the addition of fluf− in the first step leaves the bacteria generally intact.
Furthermore, we evaluated the cytotoxicity of Tb3+ alone by first incubating the bacteria in buffered Tb3+ solution. Subsequently, they were plated on agar plates, which revealed growth of all three, although somewhat inhibited in comparison with plated blanks. In parallell, we used SG and PI staining right after the Tb3+ incubation. Supplementary Fig. S3 shows the presence of predominantly live bacteria, except for E. coli, for which a larger number appears to be dead, although the same consideration with regard to SG-PI-FRET as above holds true here as well.
The reverse order of incubation (Tb3+ followed by addition of fluf−) led to very similar results with respect to staining intensities in the fluorescence images. Generally, however, the degree of agglomeration is somewhat higher if Tb3+ incubation is conducted first, which can be ascribed to the fact, well known from colloid chemistry, that trivalent cations are strong coagulants for negatively charged particles.
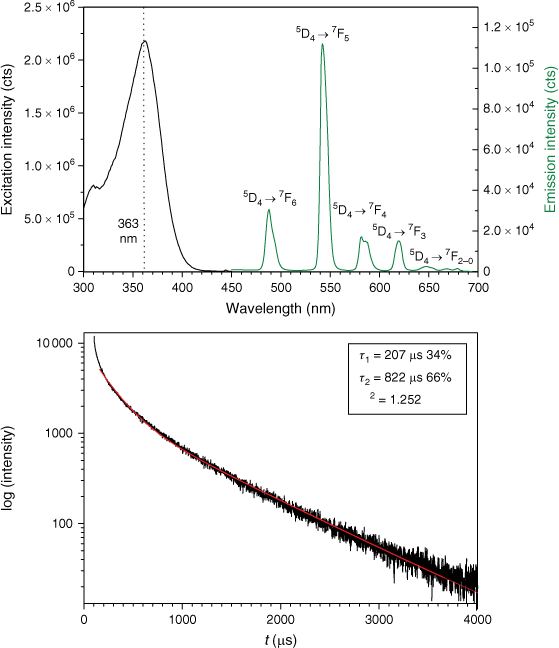
By the aid of fluorescence microscopy, it is furthermore possible to show that the stains are contained within the cells rather than on the surface alone: cross-section brightness profiles are shown in Fig. 3 for E. coli (for other species, see Supplementary Fig. S1). We furthermore took excitation and emission spectra of dispersions of the bacteria and determined the lifetimes of the ingested complexes; again the spectra for E. coli displayed in Fig. 4 serve as an example (see Supplementary Fig. S4 for the other species). The excitation maxima are found at ~365 nm, which perfectly matches the emission of commercial high-power UV-LEDs, such that fairly low-cost monitoring is accessible; of course, our advanced fluorescence microscope also profits from this coincidence.
|
We should point out here that, other than suggested by the bright fluorescence image on first sight (Fig. 2, centre row), M. luteus unexpectedly had the lowest spectroscopic excitation and emission signals. At the same time, the luminescence images showed that the complex is also strongly allocated in between the microbes. The emission intensity was clearly the lowest for M. luteus; it took several incubation attempts to obtain sufficiently emissive bacteria for recording the lifetimes. We have to concede that we had significant problems with the reproducible growth of M. luteus in previous investigations. The reason for this is not completely clear, but may be related to the fact that M. luteus lacks a glykokalyx capsule at the outer cell wall, which may promote the uptake of the growth-inhibiting complex on one hand. On the other hand, with respect to luminescence, the yellow carotenoid sarcinaxanthin (λmax = 450 nm), inevitably present in M. luteus, readily interferes via its triplet state with the Tb3+ ion’s 5D4 f state and decreases its emission. Whether M. luteus partly metabolizes the complex, or the capsule is needed for more efficient endocytosis of the complexes is an open question still to be tackled.
The decay times τ2 of E. coli, B. megaterium and M. luteus are considerably higher than for the free complex Tb(fluf)3(H2O), especially considering that the free complex was measured as a solid. The true nature of the intracellular complex, e.g. neutral [Tb(fluf)3], monocationic [Tb(fluf)2]+ or dicationic [Tb(fluf)]2+, could not be verified; evidently, however, the complexes are unambiguously linking to coordinating functional groups of the bacteria. The fact that the decays were best fitted as bi-exponentials yielding a comparably short decay τ1 is tentatively ascribed to species adhering to the surface of the bacteria, where they are exposed to quenching through the aqueous environment. Near-surface allocated complexes would obviously be in accord with the brightness profiles as well (Table 1).

|
Somewhat to our surprise, a one-step staining procedure with the complex itself for the Gram-negative E. coli (see Supplementary Fig. S5) also led to acceptable results, although at appreciably lower brightness levels. In contrast, the Gram-positive bacterial cells did not respond to the whole complex as such; instead, the complex precipitated from the dispersion in the vicinity of the bacteria. This observation again corroborates the role of the thinner cell walls of the Gram-negative bacteria. It is conceivable that via this pathway a simple alternative Gram stain can be devised.
Last but not least, Tb(fluf)3 in the bacteria proved to be far more stable photochemically than SG (half-life 69 vs 17 s), which motivates us to conduct further systematic studies on this issue.
Conclusions
We have demonstrated for the first time that green-emitting Tb complexes with highly desired excitation in the spectral range above 350 nm can be used as efficient luminescent stains for bacteria, Gram-negative bacteria (E. coli) yielding greater brightness in the fluorescence images than Gram-positives, which can be attributed to the thickness of the cell walls. The predominant luminescence lifetimes of up to approximately 800 µs in the aqueous bacteria dispersions, by far in excess of the free complexes, show the synergistic interaction with functional groups of the bacteria. The long lifetimes will be an indispensable asset, exploitable in time-gating analyses such as luminescent flow cytometry and time gated microscopy. Furthermore, although yet to be detailed for other organic dyes, the photostability of the bacterial complexes was shown to outshine SG as a prominent example for an organic dye.
Data availability
The data that support this study are available in the article and accompanying online supplementary material.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
WR and UK greatly appreciate the funding granted by the European Union and the State of North Rhine-Westphalia (EFRE OP NRW 2014-2020; Optics Center-R&D). EK, IPA and ML are grateful for support by the German Ministry of Education and Research (BMBF – Bundesministerium für Bildung und Forschung, FKZ 13N14363).
Supplementary material
Supplementary material is available online consisting of supplementary Figs S1–S5 with additional microscopic images, excitation and emission spectra, decay curves and brightness profiles.
References
[1] PM Pellegrino, NF Fell, DL Rosen, JB Gillespie, Anal Chem 1998, 70, 1755.| Crossref | GoogleScholarGoogle Scholar | 21651270PubMed |
[2] DL Rosen, C Sharpless, LB McGown, Anal Chem 1997, 69, 1082.
| Crossref | GoogleScholarGoogle Scholar |
[3] M Högmander, CJ Paul, S Chan, E Hokkanen, V Eskonen, T Pahikkala, S Pihlasalo, Anal Chem 2017, 89, 3208.
| Crossref | GoogleScholarGoogle Scholar | 28194955PubMed |
[4] MM Lezhnina, W Rochowiak, W Göhde, R Kuczius, U Kynast, J Biophotonics 2020, 13, e202000068.
| Crossref | GoogleScholarGoogle Scholar | 32500670PubMed |
[5] Sabnis RW. Handbook of biological dyes and stains: synthesis and industrial applications. John Wiley & Sons; 2010.
[6] V Hagren, P von Lode, A Syrjälä, T Soukka, T Lövgren, H Kojola, J Nurmi, Anal Biochem 2008, 374, 411.
| Crossref | GoogleScholarGoogle Scholar | 18191467PubMed |
[7] GA Dasch, S Halle, AL Bourgeois, J Clin Microbiol 1979, 9, 38.
| Crossref | GoogleScholarGoogle Scholar | 107185PubMed |
[8] Jin D. Background-free Cytometry Using Rare Earth Complex Bioprobes in: Darzynkiewicz Z, Holden E, Orfao A, Telford W, Wlodkowic D, editors. Methods in Cell Biology. Academic Press; 2011. Vol. 102, pp. 479–513.
[9] D Jin, JA Piper, RC Leif, S Yang, BC Ferrari, J Yuan, G Wang, LM Vallarino, JW Williams, J Biomed Opt 2009, 14, 024023.
| Crossref | GoogleScholarGoogle Scholar | 19405753PubMed |
[10] J Gordon, G Michel, Clin Chem 2008, 54, 1250.
| Crossref | GoogleScholarGoogle Scholar | 18593968PubMed |
[11] JY Ham, J Jung, B-G Hwang, W-J Kim, Y-S Kim, E-J Kim, M-Y Cho, M-S Hwang, DI Won, JS Suh, Ann Lab Med 2015, 35, 50.
| 25553280PubMed |
[12] J Nurmi, T Wikman, M Karp, T Lövgren, Anal Chem 2002, 74, 3525.
| Crossref | GoogleScholarGoogle Scholar | 12139064PubMed |
[13] P Zhang, X Liu, C Wang, Y Zhao, F Hua, C Li, R Yang, L Zhou, PLos One 2014, 9, e105305.
| Crossref | GoogleScholarGoogle Scholar | 25144726PubMed |
[14] Matsumoto K. In: Bünzli J-CG, Pecharsky VK, editors. Handbook on the Physics and Chemistry of Rare Earths. Elsevier; 2020; Vol. 57, pp. 119–191.
[15] J-CG Bünzli, Coord Chem Rev 2015, 293-294, 19.
| Crossref | GoogleScholarGoogle Scholar |
[16] PR Selvin, Annu Rev Biophys Biomol Struct 2002, 31, 275.
| Crossref | GoogleScholarGoogle Scholar | 11988471PubMed |
[17] J-CG Bünzli, J Lumin 2016, 170, 866.
| Crossref | GoogleScholarGoogle Scholar |
[18] M Latva, H Takalo, V-M Mukkala, C Matachescu, JC Rodríguez-Ubis, J Kankare, J Lumin 1997, 75, 149.
| Crossref | GoogleScholarGoogle Scholar |
[19] Bünzli J-CG. Rare Earth Luminescent Centers in Organic and Biochemical Compounds in: Hull R, Parisi J, Osgood RM, Warlimont H, Liu G, Jacquier B, editors. Spectroscopic Properties of Rare Earths in Optical Materials. Berlin, Heidelberg: Springer; 2005. pp. 462–499.
[20] A-S Chauvin, F Gumy, D Imbert, J-CG Bünzli, Spectrosc Lett 2004, 37, 517.
| Crossref | GoogleScholarGoogle Scholar |
[21] KY Zhang, Q Yu, H Wei, S Liu, Q Zhao, W Huang, Chem Rev 2018, 118, 1770.
| Crossref | GoogleScholarGoogle Scholar | 29393632PubMed |
[22] N Sayyadi, RE Connally, TS Lawson, J Yuan, NH Packer, JA Piper, Molecules 2019, 24, 2083.
| Crossref | GoogleScholarGoogle Scholar |
[23] Garcia-Fernandez E, Pernagallo S, González-Vera JA, Ruedas-Rama MJ, Díaz-Mochón JJ, Orte A. In: Pedras B, editor. Fluorescence in Industry. Cham: Springer International Publishing; 2019. pp. 213–267.
[24] K Tajiri, H Kishi, T Ozawa, T Sugiyama, A Muraguchi, Cytometry Part A 2009, 75A, 282.
| Crossref | GoogleScholarGoogle Scholar |
[25] P Stiefel, S Schmidt-Emrich, K Maniura-Weber, Q Ren, BMC Microbiol 2015, 15, 36.
| Crossref | GoogleScholarGoogle Scholar | 25881030PubMed |
[26] F Ou, C McGoverin, S Swift, F Vanholsbeeck, Anal Bioanal Chem 2019, 411, 3653.
| Crossref | GoogleScholarGoogle Scholar | 31049617PubMed |
[27] E Kasprzycka, IP Assunção, M Bredol, M Lezhnina, UH Kynast, J Lumin 2021, 232, 117818.
| Crossref | GoogleScholarGoogle Scholar |
[28] IP Assunção, M Bredol, E Kasprzycka, UH Kynast, M Lezhnina, Inorg Chim Acta 2021, 515, 120071.
| Crossref | GoogleScholarGoogle Scholar |
[29] RA Greene, RA Slepecky, J Bacteriol 1972, 111, 557.
| Crossref | GoogleScholarGoogle Scholar | 4626503PubMed |