Exploring HIV latency using transcription profiling
Sushama Telwatte A B C and Steven A Yukl A BA Department of Medicine, Veteran Affairs Medical Center, 4150 Clement Street, San Francisco, CA, United States
B University of California, San Francisco (UCSF), San Francisco, CA, United States
C Tel: +1 415 221 4810, Email: Sushama.telwatte@ucsf.edu
Microbiology Australia 38(3) 137-139 https://doi.org/10.1071/MA17050
Published: 9 August 2017
The major barrier to a cure for HIV is the existence of reservoirs consisting predominantly of latently infected CD4+ T cells, which do not produce virus constitutively but can be induced to produce infectious virus on activation. HIV latency research has largely focused on peripheral blood, yet most HIV-infected cells reside in tissues, especially the gut, where differences in drug penetration, cell types, and immune responses may impact mechanisms of persistence. Exploring the differences between the gut and the blood in transcriptional blocks may reveal fundamental insights into mechanisms that contribute to HIV latency. Our novel transcriptional profiling assays enable us to determine where blocks to HIV transcription occur in various tissues and the magnitude of their contribution. These assays could also be adapted to investigate latency established by other retroviridae or even DNA viruses such as herpesviridae with a view to pinpointing mechanisms underlying latency in vivo and ultimately contribute to designing a cure.
Probing HIV in the gut
HIV remains a major pandemic, with more than 36 million people affected worldwide. Over 1.1 million people in the US are currently living with HIV. In Australia, increased awareness and high profile health promotion campaigns have been unsuccessful in reducing the number of new infections, which have remained steady over the last few years1. Despite the success of combination antiretroviral therapy (ART) in suppressing HIV-1 replication, ART is not curative and residual virus continues to cause immune activation, organ damage, and reduction in life expectancy2,3. HIV-1 evades ART and immune responses through latent infection of CD4+ T cells4–7. Since these latently infected cells do not produce HIV proteins, they escape viral cytopathic effects and evade detection by the immune system. Latent HIV has been primarily found in long-lived memory CD4+ T cells, which can survive for decades and expand the viral reservoir by cell proliferation8–11. These latently infected cells are considered to be the main barrier to HIV eradication4 and their reactivation in vivo likely contributes to sustained immune activation observed during suppressive ART12.
Although much HIV latency research utilises in vitro models or cells from peripheral blood, prior work has highlighted differences between the gut and blood in the phenotype of infected T and non-T cells12,13. Furthermore, gut and blood compartments differ in levels of T cell activation and its relationship with HIV transcription12. Considering that the gut harbors up to 85% of all lymphoid tissue and over 90% of all lymphocytes14,15, it is imperative to investigate how mechanisms of HIV persistence and latency differ between gut and blood in vivo. To this end, we are employing a cutting-edge ‘transcription profiling’ approach, which features a novel panel of highly conserved, sensitive, quantitative reverse transcription droplet digital PCR (RT-ddPCR) assays. This approach quantifies the levels of HIV transcripts that suggest different mechanisms of transcriptional blockade and/or progression though various stages of HIV transcription. The levels and ratios of different HIV transcripts can be used to determine the degree to which different mechanisms contribute to reversible inhibition of HIV gene expression, and hence latency, in cells from HIV-infected individuals.
Exploiting transcriptional features of HIV
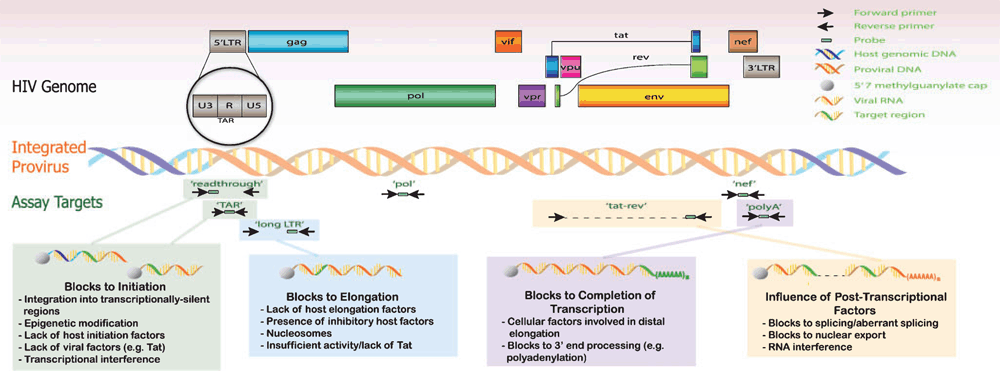
The compact genome of HIV features major coding regions, including: 1) gag, pol and env, common to all retroviruses, which encode essential structural proteins (such as envelope, matrix and capsid) and critical enzymes, including protease (catalyses cleavage), reverse transcriptase (reverse transcribes RNA genome into double-stranded DNA) and integrase (mediates integration into host genome); 2) regulatory genes (tat and rev); and 3) accessory genes (vif, vpr, vpu and nef) (Figure 1). A major putative mechanism driving HIV latency is transcriptional interference (TI), caused by ongoing transcription of host genes in cis that inhibit the assembly of the RNA polymerase complex on the HIV promoter region, the 5’-long terminal repeat (LTR)16–19. ‘Read-through’ transcripts (Figure 1) are suggestive of TI since they include the U3-U5 region that distinguishes them from canonical HIV transcripts. Other mechanisms that can lead to a block to HIV transcription initiation include epigenetic modification, a lack of host initiation factors18,20, suboptimal activity of the viral transcription factor Tat21 and integration into transcriptionally silent regions of the genome18,22,23. The degree of transcriptional initiation can be assessed by detection of transcripts containing the ‘transactivation response’ (TAR) element, which is the RNA target of Tat protein and is present in all HIV transcripts (Figure 1). Our ‘TAR’ assay has been specifically designed to maximise the detection of short, prematurely terminated transcripts with an efficiency equal to longer transcripts24,25 by incorporating an additional polyadenylation step that generates an accessible priming site for reverse transcription. This strategy offers a considerable advantage over other assays, which can detect only 4% of true short transcripts and thus significantly underestimate the abundance of these transcripts24.
Other proposed mechanisms of HIV latency include downstream blocks to elongation due to the lack of host elongation factors, the presence of inhibitory factors, nucleosome conformation and insufficient Tat activity21,26–28. Such mechanisms can be evaluated by targets for downstream sequences (such as ‘Long LTR’) that indicate elongation past the TAR loop. To assess how efficiently transcription proceeds through pol to the 3’end, transcripts containing pol and nef target sequence are also detected by our panel of assays. Levels of polyadenylated HIV RNA (‘PolyA’), indicative of transcription completion29, are detected using primers that span the LTR (U3) and polyA tail. Polyadenylated transcripts can act as surrogate markers for HIV protein. Similarly, multiply spliced HIV RNA (‘Tat-Rev’), heralding the completion of splicing, can serve as a surrogate for productive infection30. The levels of each distinct transcript and the ratios between them can be used to quantify the degree to which HIV transcription is inhibited in vivo by TI or blocks to transcriptional initiation, elongation, completion and splicing.
The novelty of this approach lies in the ability to simultaneously investigate multiple mechanisms of transcriptional blocks in vivo. Combined with RT-ddPCR, which enables absolute cDNA quantification24, this approach provides a considerable advantage over previous work that mostly focuses on one mechanism of latency at a time and has typically utilised in vitro models of latency, which may not recapitulate what happens in vivo.18–20,22,23,26–28. Unlike previously employed strategies, specific blocks to transcription and the magnitude of their impact on the prevailing levels of HIV RNA can be simultaneously assessed by determining the expression of these processive transcripts. These data can then inform strategies to target latency reversal. Using matched tissues from ART-suppressed HIV-infected individuals, this transcription profiling approach is beginning to reveal differences between blood and gut in the blocks to HIV transcription, which is of particular interest due to the difficulty in accessing tissue samples and the subsequent paucity of data examining HIV latency in the gut. This work, which contributes to elucidating the molecular mechanisms that govern HIV latency, may lead to new therapies aimed at curing HIV.
Acknowledgements
This project was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institutes of Health (R01DK108349-01).
References
[1] The Kirby Institute. (2014) HIV, viral hepatitis and sexually transmissible infections in Australia Annual Surveillance Report 2014. The Kirby Institute, UNSW, Sydney, NSW, Australia.[2] Brenchley, J.M. and Douek, D.C. (2008) HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1, 23–30.
| HIV infection and the gastrointestinal immune system.Crossref | GoogleScholarGoogle Scholar |
[3] Marchetti, G. et al. (2013) Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev. 26, 2–18.
| Microbial translocation in the pathogenesis of HIV infection and AIDS.Crossref | GoogleScholarGoogle Scholar |
[4] Chun, T.W. et al. (1997) Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94, 13193–13197.
| Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy.Crossref | GoogleScholarGoogle Scholar |
[5] Chun, T.W. et al. (1997) Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188.
| Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection.Crossref | GoogleScholarGoogle Scholar |
[6] Finzi, D. et al. (1997) Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300.
| Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy.Crossref | GoogleScholarGoogle Scholar |
[7] Wong, J.K. et al. (1997) Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295.
| Recovery of replication-competent HIV despite prolonged suppression of plasma viremia.Crossref | GoogleScholarGoogle Scholar |
[8] Finzi, D. et al. (1999) Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5, 512–517.
| Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy.Crossref | GoogleScholarGoogle Scholar |
[9] Siliciano, J.D. et al. (2003) Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728.
| Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells.Crossref | GoogleScholarGoogle Scholar |
[10] Strain, M.C. et al. (2003) Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100, 4819–4824.
| Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence.Crossref | GoogleScholarGoogle Scholar |
[11] Zhang, L. et al. (1999) Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340, 1605–1613.
| Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy.Crossref | GoogleScholarGoogle Scholar |
[12] Wong, J.K. and Yukl, S.A. (2016) Tissue reservoirs of HIV. Curr. Opin. HIV AIDS 11, 362–370.
| Tissue reservoirs of HIV.Crossref | GoogleScholarGoogle Scholar |
[13] Yukl, S.A. et al. (2013) The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J. Infect. Dis. 208, 1212–1220.
| The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence.Crossref | GoogleScholarGoogle Scholar |
[14] Cerf-Bensussan, N. and Guy-Grand, D. (1991) Intestinal intraepithelial lymphocytes. Gastroenterol. Clin. North Am. 20, 549–576.
[15] Smit-McBride, Z. et al. (1998) Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4+ T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72, 6646–6656.
[16] Jordan, A. et al. (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20, 1726–1738.
| The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation.Crossref | GoogleScholarGoogle Scholar |
[17] Pion, M. et al. (2003) Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J. Virol. 77, 4025–4032.
| Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation.Crossref | GoogleScholarGoogle Scholar |
[18] Han, Y. et al. (2004) Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78, 6122–6133.
| Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes.Crossref | GoogleScholarGoogle Scholar |
[19] Lenasi, T. et al. (2008) Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4, 123–133.
| Transcriptional interference antagonizes proviral gene expression to promote HIV latency.Crossref | GoogleScholarGoogle Scholar |
[20] Ghose, R. et al. (2001) Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75, 11336–11343.
| Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines.Crossref | GoogleScholarGoogle Scholar |
[21] Yukl, S. et al. (2009) Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387, 98–108.
| Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity.Crossref | GoogleScholarGoogle Scholar |
[22] Winslow, B.J. et al. (1993) HIV-1 latency due to the site of proviral integration. Virology 196, 849–854.
| HIV-1 latency due to the site of proviral integration.Crossref | GoogleScholarGoogle Scholar |
[23] Schröder, A.R. et al. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529.
| HIV-1 integration in the human genome favors active genes and local hotspots.Crossref | GoogleScholarGoogle Scholar |
[24] Kaiser, P. et al. (2017) Assays for precise quantification of total (including short) and elongated HIV-1 transcripts. J. Virol. Methods 242, 1–8.
| Assays for precise quantification of total (including short) and elongated HIV-1 transcripts.Crossref | GoogleScholarGoogle Scholar |
[25] Yukl, S. et al. (2016) Investigating the mechanisms that control HIV transcription and latency in vivo. In CROI 2016, Boston, MA.
[26] Lassen, K.G. et al. (2004) Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 78, 9105–9114.
| Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo.Crossref | GoogleScholarGoogle Scholar |
[27] Emiliani, S. et al. (1996) A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 93, 6377–6381.
| A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency.Crossref | GoogleScholarGoogle Scholar |
[28] Lassen, K. et al. (2004) The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10, 525–531.
| The multifactorial nature of HIV-1 latency.Crossref | GoogleScholarGoogle Scholar |
[29] Bullen, C.K. et al. (2014) New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429.
| New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo.Crossref | GoogleScholarGoogle Scholar |
[30] Fischer, M. et al. (2004) Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J. Infect. Dis. 190, 1979–1988.
| Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef.Crossref | GoogleScholarGoogle Scholar |
Biographies
Sushama Telwatte is a Postdoctoral Scholar undertaking postdoctoral studies in the laboratory of Associate Professor Steven A Yukl at the Veteran Affairs Medical Center in San Francisco. Her work focuses on uncovering the mechanisms underlying HIV persistence and latency in the gut and blood in vivo. Sushama completed her PhD under the supervision of Associate Professor Gilda Tachedjian at the Burnet Institute/Monash University, investigating the role of synonymous mutations in HIV-1 selected during drug therapy. Sushama was awarded the ASM BD Award (Victorian Branch) and presented her PhD work at the ASM 2015 meeting.
Dr Steven Yukl is an Associate Professor of Medicine at the University of California, San Francisco (UCSF) and a staff physician at the San Francisco Veterans Affairs Medical Center. His research focuses on the mechanisms that allow HIV to persist despite immune defenses and antiretrovirals, thereby preventing HIV cure. Dr Yukl has been involved in basic, translational, and clinical research on HIV persistence since 2005, with specialisation in HIV latency, gut tissue reservoirs, cellular reservoirs, and development of ultrasensitive assays to measure HIV persistence.