Phage research in ‘organ-on-chip’ devices
Wai Hoe Chin A B and Jeremy J Barr A CA School of Biological Sciences, Monash University, 17 Rainforest Walk, Clayton, Vic. 3800, Australia. Tel: +61 3 9905 5486.
B Email: wai.chin@monash.edu
C Email: jeremy.barr@monash.edu
Microbiology Australia 40(1) 28-32 https://doi.org/10.1071/MA19006
Published: 21 February 2019
The use of ‘organ-on-chip’ devices in microbiology research presents enormous opportunities for fundamental and translational research1–4. Yet these approaches have not been widely embraced by the microbiology field. This is particularly evident with bacteriophage (phage) research applications. Traditionally phage research has been an early adopter of experimental techniques and approaches5, having catalysed research in biotechnology, environmental biology, sequencing, and synthetic biology. Here we discuss some of the opportunities that organ-on-chip devices present to both phage and microbiology research, and provide a ‘how to’ guide for researchers interested in utilising this approach.
‘Organ-on-chips’ are micro-engineered biomimetic devices that replicate key functions, activities and physiological responses of entire living organs6. The approach has been used to develop beating hearts7, simulate breathing lungs8, sustain a gut microbiome3,9 and even develop interconnected neurons of the brain10. Devices are typically micro-fabricated to contain channels that are lined with cultured human cells, which mimic organ-specific architecture and functions in vitro6. The device structure varies depending on the organ of interest. For instance, the gut-on-chip can comprise of a single4 or double channel structure9, with channel dimensions varying between 500–1000 µm wide and 150–250 µm high. The single-channel gut-on-chip forms the simplest structure, being enclosed by a glass slide upon which a layer of gut epithelial cells is grown. In comparison, the double-channel gut-on-chip is constructed by joining two single-channel devices together with a thin porous membrane separating the two channels. The membrane supports the gut cell layer within the top channel while the bottom channel represents the vascular system of the gut.
The fabrication, operation and experimentation of organ-on-chip devices typically require the convergence of numerous fields including engineering, cell biology and microbiology; presenting a high technical barrier for research applications. Yet overcoming these challenges allows us to probe the interactions between phages, their bacterial hosts and ‘life-like’ organs to answer therapeutic, ecological, and fundamental questions. For example, a mucus-producing lung-on-chip model was used to describe phage adherence to mucus layer, thereby forming a non-host-derived barrier against bacterial infection4. Other studies have demonstrated the maintenance of a gut microbiome and Coxsackie virus infection using a gut-on-chip model3,9; approaches that can be modified to investigate gut phage-bacteria interactions. In essence, the organ-on-chip provides researchers the benefit of in vitro amenability while experimenting with phages under biologically relevant conditions.
The organ-on-chip in four steps
Step 1: designing the organ-on-chip mould
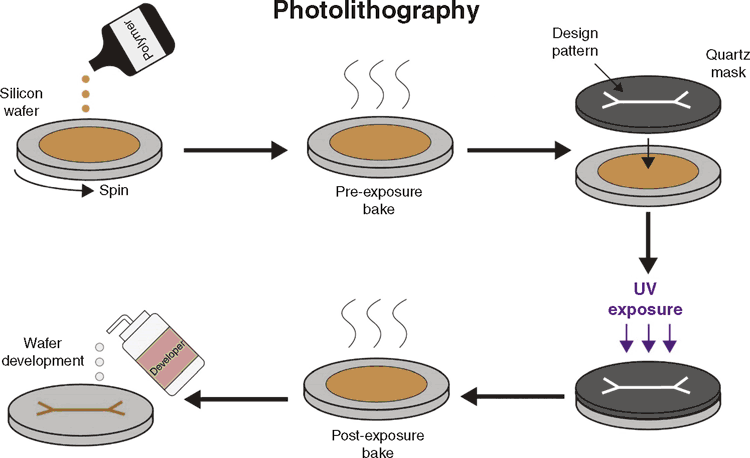
The first step to creating an organ-on-chip is to fabricate a mould. Two commonly used options are photolithography and 3D-printing. Photolithography (Figure 1) is commonly used in engineering fields, but is technically challenging; requiring specialist equipment and reagents. However, this technique is virtually limitless in creating complex designs at the nanoscale11. The technique starts with depositing a photosensitive polymer on a substrate. By controlling ultraviolet (UV) light exposure on the substrate, the polymer will polymerise to the desired feature pattern, which is subsequently developed by washing away soluble unpolymerised regions.
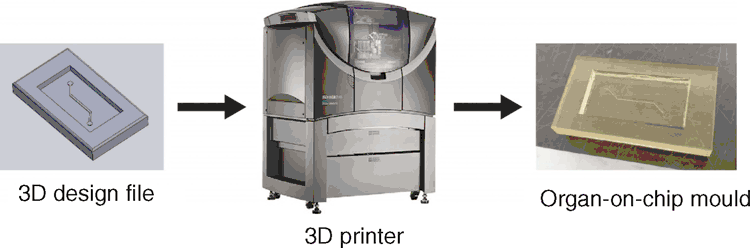
Alternatively, 3D-printing (Figure 2) offers a much quicker, easier, and cheaper route to fabricate organ-on-chip moulds. However, unlike photolithography, 3D printing has a much lower printing resolution, typically in the micrometre scale12. Nonetheless, the accessibility and speed that 3D-printing offers enable researchers to quickly create simple organ-on-chip moulds for subsequent manufacturing, set-up, and experimentation11.
Step 2: making the organ-on-chip
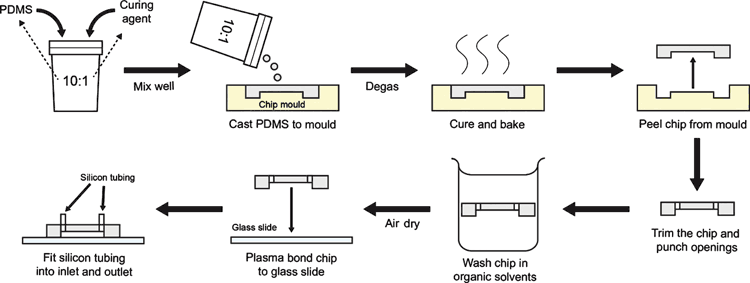
Once a mould is obtained, a variety of materials can be used to manufacture organ-on-chip devices. However, none has matched polydimethylsiloxane (PDMS) for its advantages in biocompatibility, permeability to gases, optical transparency and material flexibility13. In addition to its advantages in biological applications, fabricating with PDMS is fairly straightforward (Figure 3) and does not require special expertise. The only specialist equipment required is a plasma cleaner to bond the PDMS device onto a substrate (typically a glass slide or another PDMS base). However, labs without access to this equipment can utilise a portable plasma ‘torch’ for bonding organ-on-chips (Corona SB, Elveflow Microfluidics). Alternatively, researchers can purchase ready-made devices that are immediately amenable to cell culture, such as the LiverChip® (CN Bio Innovations, United Kingdom) or Intestine Bio-Kit (Emulate Bio, USA). For further details on organ-on-chip fabrication methods, consult references6,11,14.
Step 3: recreating the ‘organ’ in the organ-on-chip
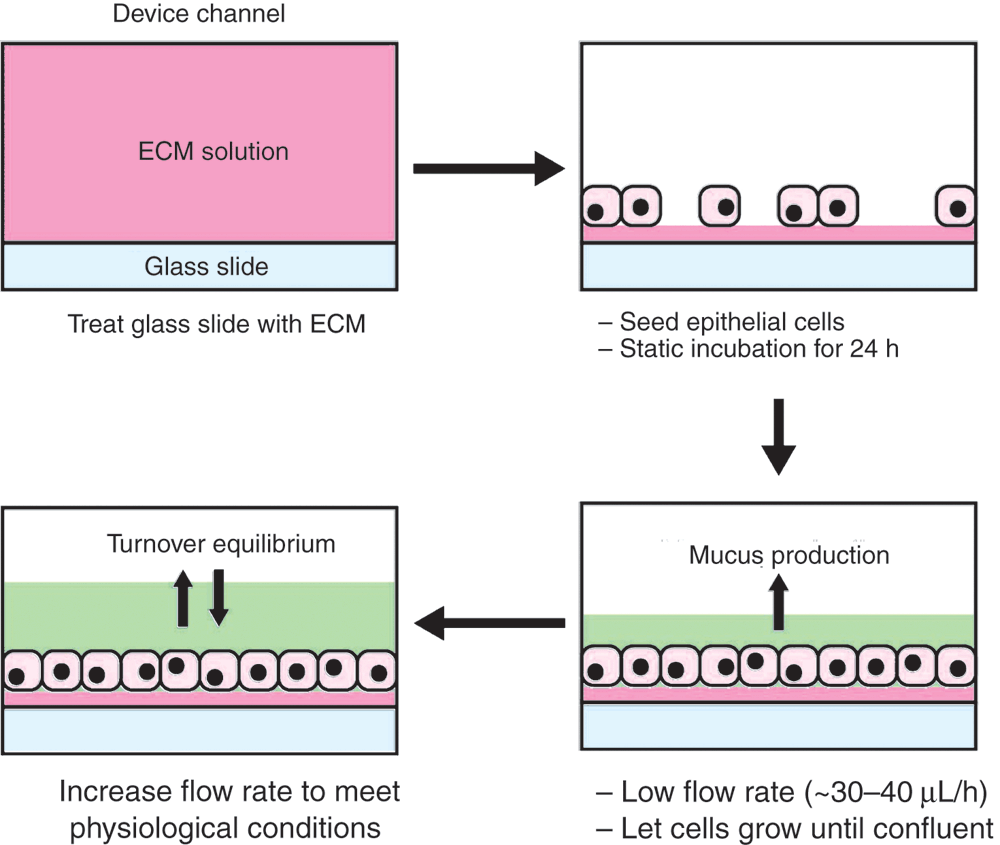
Any given organ is functionally and architecturally complex. Therefore, we must be mindful that organ-on-chips serve to approximate these complexities by ‘building the organ’ using tissues or cells in culture. Nonetheless, with a fair amount of creativity and innovation, these approximations can recapitulate key functions and fundamental architecture of an organ unit. Recreating the functioning organ-on-chip relies on tissue culture work that is no different to traditional cell culture in flasks (Figure 4). Researchers need only to scale their techniques to efficiently handle tissue culture at the microfluid-level – a simple act of replacing serological pipettes with micropipettes.
|
Difficulties in transitioning culture cells from flasks into the organ-on-chip are often encountered, but can be overcome with a few simple solutions. Toxicity from uncured PDMS in the devices can potentially cause cell death, but is easily eliminated through organic solvent washes16. Determining the optimal cell seeding density will vary depending on the device and cell line used and often requires troubleshooting. Cell layer maintenance within the device requires a continual flow rate that does not impose excessive shear stress to the cells. Again, this will depend on the cell line used as some cell lines, such as endothelial lines, are more robust in withstanding high shear stress17. Consulting publications that have used similar cell lines and devices will provide a ballpark figure to start troubleshooting.
Step 4: operating the organ-on-chip
As outlined in step 3, cell growth and maintenance within the organ-on-chip is dependent on constant perfusion with culture media. Syringe pumps and pressure-driven systems are two widely adopted approaches to perfuse organ-on-chip devices, each with their advantages and limitations. Setting up syringe pumps is simpler and requires less tubing, but has limited flow control and sample inoculation options. Conversely, pressure-driven systems are computerised setups made up of multiple components to regulate air pressure that will drive fluid flow from a media reservoir into the device. Connecting these components requires various adaptors and considerable tubing length, but offer increased flexibility for device control and inoculation. Furthermore, the computer interface in these systems offers fast response times and can incorporate flow sensor feedback loops that provide superior fluid flow stability compared to syringe pumps18.
Moving forward: phage research in organ-on-chips
Traditionally, investigations of phage-bacteria interactions have been confined to in vitro broth culture. While these studies have proven instrumental for our understanding of phage biology, they neglect the complex environment and interactions seen in vivo. Recently, animal models have demonstrated the surprising diversity and stability of the phageome19, and tissue culture-based in vitro studies have shown surprising interactions between phage and eukaryotic cells and tissues4,20–22. Organ-on-chip systems offer a unique way to study phage interactions within life-like systems that are cheap, accessible, and experimentally amenable.
Phage therapy approaches utilising organ-on-chip
Phages are known for their antimicrobial properties and are currently being pursued as an alternative to antibiotics in treating bacterial infections. Today, animal models are still the ‘bread-and-butter’ for preclinical testing of therapeutics, including the therapeutic validation of phages. However, animal models are costly, labour-intensive, and ethically questionable9. There are further concerns regarding the suitability of animal infection models to recapitulate human pathological conditions. Organ-on-chip models provide a middle ground between traditional static cell cultures and animal models for preclinical testing. A recent example was the use of a gut-on-chip to reproduce Coxsackie virus infection of a highly differentiated human villus intestinal epithelium, which reproduced cytopathic effects3. The use of organ-on-chip devices for phage therapy approaches offers large potentials, including the validation of antimicrobial capacity within an organ of interest, pharmacokinetic and pharmacodynamics studies, and tracking the emergence of phage resistance.
Gut-on-chip: moving gut phageome and microbiome studies from faeces to mucus
The human gut is home to a diverse repertoire of microbial species. This gut microbiome is comprised of trillions of microbial cells that influence our health, well-being and even psychological behaviour23. Numerically, the gut viruses, of which phages account for ~90%, are as abundant, if not more, than their microbial counterparts24. However, very little is known regarding the nature of phage-bacteria interactions within the gut. This is primarily due to the difficulty in studying and sampling the gut environment directly. Faecal samples are often used as a proxy to direct sampling, yet the faecal microbial communities differ significantly from intestinal mucosa25. Gut-on-chip devices address these limitations by providing a life-like environment for phage-bacteria experimental studies (Figure 4). This relatively simple set-up mimics essential aspects of the in vivo gut, namely the mucus layer, luminal flow, and spatial elements of the cell layer. Using gut-on-chip devices, it was demonstrated that phages were able to adhere to gut-produced mucus layer and as a result, exhibit enhanced antimicrobial activity within the mucus layer, providing a layer of non-host-derived immunity4,20. A microbiome gut-on-chip approach demonstrated the recapitulation of pathogenic microbially induced inflammation and the correction of these effects through probiotic and antibiotic therapies26. Finally, recent cell culture studies demonstrated that phages targeting the gut pathogen Clostridium difficile had increased antimicrobial affects when in co-culture with human gut cell lines22. These studies illustrate the potential of phage and microbiology studies within organ-on-chip devices.
Phage-bacteria ecology and evolution using organ-on-chip
To date, most evolutionary and ecological hypotheses attempting to explain phage-bacteria diversity in nature are confined to test-tube experiments and mathematical models. However, these are limited by the complexity of the experimental environment and assumptions of the models tested. Comparatively, the organ-on-chip approach allows for experimental investigations of these hypotheses under life-like conditions, adding increased complexity and biological relevance. Building off recent organ-on-chip microbiome devices4,26, researchers are now able to study emergent microbial properties, such as co-evolutionary phage-host dynamics, experimental evolution of microbial communities, and investigations of gut phage-bacteria ecology. These devices are further amenable to the introduction of genetically modified phages and bacteria, including the insertion of fluorescence markers for real-time visualisation27 or antibiotic or CRISPR locus for quantification of target populations28,29. The collective evolutionary and ecological results obtained may validate models and further explain gut microbiome diversity.
Conclusion
Phages have been at the forefront of many biological advances. Today, not only are they impacting the medical field through therapeutic applications, but also continually fueling fundamental research, such as evolutionary biology and ecology. However, experimental phage research has been mostly confined in vitro and in silico. To that, we propose organ-on-chips as an experimental approach to further propel phage and microbiology research. The amenability of organ-on-chips allows researchers to conduct various phage and microbiological studies within life-like conditions; without the cost associated with animal models. Despite requiring high interdisciplinary knowledge, the organ-on-chip remains accessible to non-engineers through collaborations or simpler alternatives in setting up the platform.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was performed in part at the Melbourne Centre for Nanofabrication (MCN) in the Victorian Node of the Australian National Fabrication Facility (ANFF). This work was supported by a Monash University PhD Scholarship (awarded to WHC), an ARC DECRA Fellowship (DE170100525; awarded to JJB), an NHMRC New Investigator grant (1156588; awarded to JJB), and a Perpetual Trustees Australia award (2018HIG00007; awarded to JJB).
References
[1] Jalili-Firoozinezhad, S. et al. (2018) Complex human gut microbiome cultured in anaerobic human intestine chips. bioRxiv , 421404.| Complex human gut microbiome cultured in anaerobic human intestine chips.Crossref | GoogleScholarGoogle Scholar |
[2] Bein, A. et al. (2018) Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterol. Hepatol. 5, 659–668.
| Microfluidic organ-on-a-chip models of human intestine.Crossref | GoogleScholarGoogle Scholar | 29713674PubMed |
[3] Villenave, R. et al. (2017) Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One 12, e0169412.
| Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro.Crossref | GoogleScholarGoogle Scholar | 28146569PubMed |
[4] Barr, J.J. et al. (2015) Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 112, 13675–13680.
| Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters.Crossref | GoogleScholarGoogle Scholar | 26483471PubMed |
[5] Rohwer, F. and Segall, A.M. (2015) In retrospect: a century of phage lessons. Nature 528, 46–48.
| In retrospect: a century of phage lessons.Crossref | GoogleScholarGoogle Scholar | 26632584PubMed |
[6] Huh, D. et al. (2013) Microfabrication of human organs-on-chips. Nat. Protoc. 8, 2135–2157.
| Microfabrication of human organs-on-chips.Crossref | GoogleScholarGoogle Scholar | 24113786PubMed |
[7] Grosberg, A. et al. (2011) Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip 11, 4165.
| Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip.Crossref | GoogleScholarGoogle Scholar | 22072288PubMed |
[8] Huh, D. et al. (2010) Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668.
| Reconstituting organ-level lung functions on a chip.Crossref | GoogleScholarGoogle Scholar | 20576885PubMed |
[9] Kim, H.J. et al. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174.
| Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow.Crossref | GoogleScholarGoogle Scholar | 22434367PubMed |
[10] Park, J. et al. (2015) Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 15, 141–150.
| Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease.Crossref | GoogleScholarGoogle Scholar | 25317977PubMed |
[11] Leester-Schädel, M. et al. (2016) Fabrication of microfluidic devices. In Microsystems for Pharmatechnology. pp. 23–57. Springer International Publishing, Switzerland.
[12] Gross, B.C. et al. (2014) Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Anal. Chem. 86, 3240–3253.
| Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences.Crossref | GoogleScholarGoogle Scholar | 24432804PubMed |
[13] Beißner, N. et al. (2016) Organ on chip. In Microsystems for Pharmatechnology. pp. 299–339. Springer International Publishing, Switzerland.
[14] Faustino, V. et al. (2016) Biomedical microfluidic devices by using low-cost fabrication techniques: a review. J. Biomech. 49, 2280–2292.
| Biomedical microfluidic devices by using low-cost fabrication techniques: a review.Crossref | GoogleScholarGoogle Scholar | 26671220PubMed |
[15] Ahadian, S. et al. (2018) Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv. Healthc. Mater. 7, 1700506.
| Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies.Crossref | GoogleScholarGoogle Scholar | 30134074PubMed |
[16] Regehr, K.J. et al. (2009) Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 9, 2132–2139.
| Biological implications of polydimethylsiloxane-based microfluidic cell culture.Crossref | GoogleScholarGoogle Scholar | 19606288PubMed |
[17] Kim, L. et al. (2007) A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip 7, 681–694.
| A practical guide to microfluidic perfusion culture of adherent mammalian cells.Crossref | GoogleScholarGoogle Scholar | 17538709PubMed |
[18] Zeng, W. et al. (2015) Characterization of syringe-pump-driven induced pressure fluctuations in elastic microchannels. Lab Chip 15, 1110–1115.
| Characterization of syringe-pump-driven induced pressure fluctuations in elastic microchannels.Crossref | GoogleScholarGoogle Scholar | 25537266PubMed |
[19] Reyes, A. et al. (2015) Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. USA 112, 11941–11946.
| Gut DNA viromes of Malawian twins discordant for severe acute malnutrition.Crossref | GoogleScholarGoogle Scholar | 26351661PubMed |
[20] Barr, J.J. et al. (2013) Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 110, 10771–10776.
| Bacteriophage adhering to mucus provide a non-host-derived immunity.Crossref | GoogleScholarGoogle Scholar | 23690590PubMed |
[21] Nguyen, S. et al. (2017) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio 8, e01874-17.
| Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers.Crossref | GoogleScholarGoogle Scholar | 29162715PubMed |
[22] Shan, J. et al. (2018) Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 8, 5091.
| Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells.Crossref | GoogleScholarGoogle Scholar | 29572482PubMed |
[23] Lozupone, C.A. et al. (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230.
| Diversity, stability and resilience of the human gut microbiota.Crossref | GoogleScholarGoogle Scholar | 22972295PubMed |
[24] Scarpellini, E. et al. (2015) The human gut microbiota and virome: potential therapeutic implications. Dig. Liver Dis. 47, 1007–1012.
| The human gut microbiota and virome: potential therapeutic implications.Crossref | GoogleScholarGoogle Scholar | 26257129PubMed |
[25] Ingala, M.R. et al. (2018) Comparing microbiome sampling methods in a wild mammal: fecal and intestinal samples record different signals of host ecology, evolution. Front. Microbiol. 9, 803.
| Comparing microbiome sampling methods in a wild mammal: fecal and intestinal samples record different signals of host ecology, evolution.Crossref | GoogleScholarGoogle Scholar | 29765359PubMed |
[26] Kim, H.J. et al. (2016) Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 113, E7–E15.
| Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip.Crossref | GoogleScholarGoogle Scholar | 26668389PubMed |
[27] Trinh, J.T. et al. (2017) Cell fate decisions emerge as phages cooperate or compete inside their host. Nat. Commun. 8, 14341.
| Cell fate decisions emerge as phages cooperate or compete inside their host.Crossref | GoogleScholarGoogle Scholar | 28165024PubMed |
[28] Borges, A.L. et al. (2018) Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell 174, 917–925.e10.
| Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity.Crossref | GoogleScholarGoogle Scholar | 30033364PubMed |
[29] Chaudhry, W.N. et al. (2018) Leaky resistance and the conditions for the existence of lytic bacteriophage. PLoS Biol. 16, e2005971.
| Leaky resistance and the conditions for the existence of lytic bacteriophage.Crossref | GoogleScholarGoogle Scholar | 30114198PubMed |
Biographies
Wai Hoe Chin, BSc (Hons), graduated with a Bachelors in Biomedical Sciences with first class honours from The University of Edinburgh. He is currently a first-year PhD student in Monash University under supervision by Dr Jeremy Barr. His research adopts the gut-on-chip platform to investigate phage-bacteria co-evolution and ecology within the gut.
Jeremy J Barr, BBiot (Hons), PhD, is a Lecturer and ARC DECRA Fellow at the School of Biological Sciences, Monash University. He leads a research group investigating the tripartite interactions between bacteriophages and their bacterial and human hosts (https://thebarrlab.org).