Multi-gene insights into the taxonomy and conservation of Tasmania’s galaxiid fishes
Mark Adams A B , Michael P. Hammer C , Peter J. Unmack D , Tarmo A. Raadik E , Charlotte Jense F and Christopher P. Burridge F *
F *
A Evolutionary Biology Unit, South Australian Museum, North Terrace, Adelaide, SA 5000, Australia.
B School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia.
C Museum & Art Gallery of the Northern Territory, Darwin, NT 0810, Australia.
D Centre for Applied Water Science, Institute for Applied Ecology, University of Canberra, Canberra, ACT 2617, Australia.
E Arthur Rylah Institute for Environmental Research, Department of Environment, Land, Water and Planning, Heidelberg, Vic. 3084, Australia.
F Discipline of Biological Sciences, University of Tasmania, Sandy Bay, Tas. 7001, Australia.
Marine and Freshwater Research 74(13) 1113-1128 https://doi.org/10.1071/MF22263
Submitted: 6 December 2022 Accepted: 6 July 2023 Published: 28 July 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Galaxiids are a widespread, southern hemisphere, radiation of mostly obligate freshwater fishes. Tasmania houses a diversity of endemic species of Galaxias and Paragalaxias. Of these, many are at risk of extinction, being landlocked, range-restricted and subject to anthropogenic threats, placing a high-conservation priority on the region.
Aim: Our aim was to synthesise historic and published molecular datasets to provide the sound systematic framework needed to underpin future conservation and taxonomic efforts for Tasmanian galaxiids.
Methods: Novel and published nuclear (allozyme) and matrilineal (cytb) datasets were generated and integrated for every putative Tasmanian galaxiid species lacking a comparable multi-gene assessment.
Key results: The Tasmanian galaxiids are phylogenetically diverse, with molecular data generally supporting the accepted taxonomy, but with potential species-level diversity noted within an alpine radiation of the Galaxias truttaceus complex and further support for synonymy of G. niger within G. brevipinnis.
Conclusions: This study highlights the value of multi-locus studies in both validating species-level taxonomy and resolving taxonomic ambiguities and conservation priorities within Tasmania’s galaxiids.
Implications: Our integrated genetic analyses provide a framework to underpin more in-depth genomic approaches to assess additional cryptic diversity and conservation planning, such as genetic rescue and ex situ population security.
Keywords: allozymes, conservation genetics, cytonuclear discordance, Galaxiidae, landlocked population, mtDNA, Pedder Galaxias, species delimitation.
Introduction
Despite their habitat occupying less than 1% of the earth’s surface, freshwater fishes represent around one quarter of all described vertebrate species, making them more speciose than all other vertebrate groups, including marine fishes (Dudgeon et al. 2006; Lintermans et al. 2020; World Wildlife Fund 2020). At the same time, due to a broad range of human-induced challenges, freshwater ecosystems worldwide are arguably also the most threatened of all biomes (Dudgeon et al. 2006; Reid et al. 2019). In consequence, nearly one-third of the planet’s freshwater fishes are under the threat of extinction (World Wildlife Fund 2020).
With no fully documented extinctions and only one species listed as extinct in the wild (Department of Climate Change, Energy, the Environment and Water 2022), it might seem at first glance that Australia’s freshwater fishes are faring better than most other Australian vertebrates (60 species listed as extinct; Department of Climate Change, Energy, the Environment and Water 2022) and freshwater fishes elsewhere (Duncan and Lockwood 2001). However, the reality is quite different, and much more in keeping with the well documented degradation of freshwater ecosystems in southern and eastern Australia and consequent major declines in most freshwater fishes that live therein (Arthington et al. 1983; Lintermans 2007; Hammer et al. 2013a; Morgan et al. 2014; Faulks et al. 2017; Lintermans et al. 2020). The true situation could also be concealed by underestimation of Australia’s freshwater fish diversity.
For a continent of its size and latitudinal range, Australia has comparatively few genera and species of freshwater fish (Lundberg et al. 2000; Allen et al. 2002). Some researchers have largely attributed this taxonomic austerity to unfavourable geographic or climatic comparisons relating to aridity, isolation, topography, habitat and rainfall (Merrick and Schmida 1984; Berra 1998; Allen et al. 2002). However, others have pointed to a key additional consideration, a comparative lack of taxonomic effort (Lundberg et al. 2000; Leis et al. 2007; Adams et al. 2013; Hammer et al. 2013b). This latter view has been strongly supported by the increasing availability of multi-locus genetic studies (i.e. not stand-alone DNA barcoding), which have routinely found compelling evidence for additional candidate species in most genera surveyed, e.g. Cairnsichthys (Hammer et al. 2018), Gadopsis (Hammer et al. 2014; Unmack et al. 2017), Galaxias (Adams et al. 2014), Galaxiella (Unmack et al. 2012), Glossamia (Cook et al. 2017), Glossogobius (Hammer et al. 2021a), Hypseleotris (Unmack et al. 2019), Melanotaenia (Unmack 2016; Hammer et al. 2019a), Mogurnda (Cook et al. 2011; Adams et al. 2013), Nannoperca (Unmack et al. 2011, 2013), Philypnodon (Hammer et al. 2019b), Pseudogobius (Hammer et al. 2021b), Retropinna (Hammer et al. 2007; Unmack et al. 2022), Syncomistes (Shelley et al. 2018) and Tandanus (Jerry 2008).
Although only surveying a small fraction of Australia’s freshwater fish fauna, these studies have already added more than 50 candidate species to the national inventory. Of these, many are now formally recognised (Welsh et al. 2014, 2017; Raadik 2014; Coleman et al. 2015; Shelley et al. 2017; Hammer et al. 2018, 2019a; Larson and Hammer 2021; Hoese and Hammer 2021; Thacker et al. 2022), some have become widely accepted as valid ‘sp. nov.’ (Raadik 2019a, 2019b, 2019c, 2019d; Lintermans et al. 2020), and the rest occupy a ‘twilight zone’ of taxonomic anonymity, awaiting attention from the nation’s small, over-stretched and underfunded ichthyological community (Leis et al. 2007).
Another great strength of multi-locus molecular datasets is that they can either help validate the species status of already-described species or expose doubts about their taxonomic distinctiveness (Richardson et al. 1986; Georges and Adams 1996). Examples of the former abound in the above-cited studies, while the latter outcome has already been demonstrated in several genera, including Chlamydogobius (Mossop et al. 2015), Craterocephalus (Adams et al. 2011), Milyeringa (Page et al. 2018), and Retropinna (Hammer et al. 2007). Three of these four cases involve ‘species’ with a restricted geographic range (i.e. a predisposing factor in extinction risk), with the genetic data inferring that they may instead represent a peripheral subpopulation of a more wide-ranging, valid species and thus not merit the enhanced conservation attention often afforded to short-range endemics (Harvey et al. 2011). Finally, molecular markers can also highlight inconsistencies with natural distributional patterns that can signal potential human-mediated dispersal or range extension (translocation), especially for smaller species that often lack detailed historic baseline distribution data (Waters et al. 2002; Hammer et al. 2013b). Multi-locus studies of Australian freshwater fishes are particularly valuable in each of these contexts, given their often-imperilled status.
In all, 40 of the 55 Australian fishes listed as endangered or vulnerable live in freshwater, and 10 of these are Tasmanian galaxiids (Department of Climate Change, Energy, the Environment and Water 2022). Furthermore, Tasmanian galaxiids represent a significant component of what is Australia’s most speciose and imperilled group of freshwater fishes (the family Galaxiidae; Bray 2018; Lintermans et al. 2020). Tasmanian galaxiids are therefore a prime group for taxonomic or conservation-focused genetic investigation, having a high proportion of endemic and threatened species and with biological and geographic attributes that set them apart from most other Australian freshwater fishes. Most species are naturally range-restricted, occur in specific habitats (e.g. flowing water, alpine lakes) and complete their life as obligate freshwater species (landlocked), thus rendering them susceptible to anthropogenic change (Hardie et al. 2006). Many galaxiids have suffered heavily from interaction with introduced piscivores (various salmonids), along with other general catchment modification and degradation (Crowl et al. 1992; Hardie et al. 2006). Hydroelectric development has both created human-mediated dispersal opportunities (e.g. canals, headwater connections: Waters et al. 2002; Chilcott et al. 2013) and impounded facultatively diadromous populations, thus allowing their greater penetration inland and enabling competition with non-diadromous species (Humphries 1990; Chilcott et al. 2013). Furthermore, galaxiids are often used as bait by recreational anglers and, hence, bait-bucket transfer may have influenced their current range (Lintermans 2004) and perhaps even created artificial hybrid zones.
In this study, we present the results of an allozyme ‘overview’ of the Tasmanian galaxiid fishes, buttressed with extended analyses of two already-published allozyme datasets on several members of that radiation, and all cross-referenced using a mitochondrial DNA (mtDNA) gene tree combining both previously published and newly generated sequence data. Together, these multi-locus data provide a range of important taxonomic and conservation perspectives on 15 of the 17 described Tasmanian galaxiid species (Table 1; historic distribution maps for each species are presented in Fig. 1), with comprehensive nuclear and matrilineal datasets already available to confirm species status in the remaining two species (Lovettia sealii and Galaxiella pusilla, Table 1). The synthesis of allozyme and mtDNA datasets has already proved pivotal in the delineation and discovery of ~18 additional endemic Galaxias species on mainland south-eastern Australia, all previously considered to be part of a single widespread Galaxias olidus sensu lato (McDowall and Frankenberg 1981; Adams et al. 2014; Raadik 2014; Lintermans et al. 2020).
| Species | Geographic distribution | EPBC | IUCN | Range-wide nuclear genetic data |
|---|---|---|---|---|
| Galaxias auratus, L | Tasmania | Endangered | EN | Morgan et al. (2016) |
| Galaxias brevipinnis, D | South-eastern Australia and New Zealand | LC | ||
| Galaxias fontanus | Tasmania | Endangered | EN | |
| Galaxias johnstoni | Tasmania | Endangered | EN | |
| Galaxias maculatus | Southern Australia and southern hemisphere | LC | ||
| Galaxias niger, L | Tasmania | – | ||
| Galaxias parvus | Tasmania | Vulnerable | VU | |
| Galaxias pedderensis, L | Tasmania | Extinct in the wild | EN | |
| Galaxias tanycephalus, L | Tasmania | Vulnerable | CR | Morgan et al. (2016) |
| Galaxias truttaceus, D | Southern Australia | WA population endangered | LC | Morgan et al. (2016) |
| Galaxiella pusilla | Tasmania and south-eastern Victoria | Vulnerable | EN | Coleman et al. (2010), Unmack et al. (2012) |
| Lovettia sealii, D | Tasmania and Victoria | LC | Schmidt et al. (2014) | |
| Neochanna cleaveri, D | South-eastern Australia | EN | Whiterod et al. (2020) | |
| Paragalaxias dissimilis, L | Tasmania | Vulnerable | EN | |
| Paragalaxias eleotroides, L | Tasmania | Vulnerable | EN | |
| Paragalaxias julianus, L | Tasmania | EN | ||
| Paragalaxias mesotes, L | Tasmania | Endangered | EN |
Also shown are any published studies that present range-wide multi-locus data to assess taxonomic validity. Conservation status: EPBC, Environment Protection and Biodiversity Conservation Act 1999 national listing; IUCN, International Union for Conservation of Nature Red List of threatened species reviewed in 2019.
D, diadromous; L, landlocked or lacustrine species; CR, critically endangered; EN, endangered; VU, vulnerable; LC, least concern.
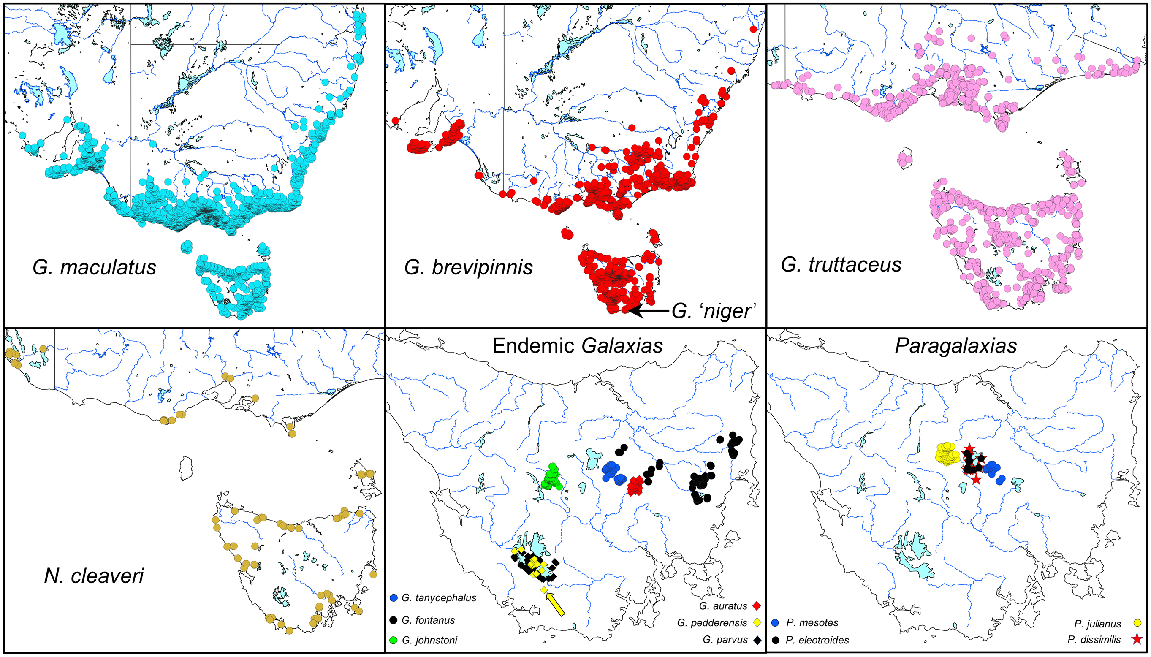
Historic distribution maps (south-eastern Australia) for all the Tasmanian galaxiids surveyed in this study. Source data from the ‘Atlas of Living Australia’ (see https://www.ala.org.au/, accessed 2 February 2023). There have been considerable but difficult-to-quantify reductions in the distribution or abundance of several endemic Tasmanian Galaxias and Paragalaxias species in recent times, most notably for G. pedderensis, which now occurs only as a translocated population (identified by the yellow arrow).
Methods
Timeline of allozyme analyses
The original allozyme overview study was undertaken in 1985, to identify areas of taxonomic uncertainty, obvious phylogenetic and phylogeographic patterns and a suite of markers for follow-up assessments of population structure in selected species (L. sealii and Galaxias maculatus; Pavuk 1997). As such, it focused on small numbers of individuals per site and taxon, screened for as many allozyme markers as were established in the SA Museum’s allozyme laboratory at that time. Whole fish were collected and snap frozen, often with the assistance of staff from the Tasmanian Inland Fisheries Commission, before being shipped to the SA Museum for inclusion in their frozen-tissue collection (the Australian Biological Tissues Collection; ABTC). These collections were planned to include exemplars of all species of Galaxias (10 species), Paragalaxias (4 species) and Neochanna (Neochanna cleaveri at that time was included in the genus Galaxias; Waters and White 1997), plus a single population of the mainland galaxiid G. olidus sensu stricto (i.e. referable to correct taxon on the basis of the taxonomic revision of Raadik 2014). It should be noted that Galaxias pedderensis and Galaxias niger are morphologically similar to Galaxias brevipinnis, and specimens were largely assigned to these two species on the basis of collection locality. Furthermore, although many (but not all) researchers have regarded G. niger as likely synonymous with G. brevipinnis (McDowall and Fulton 1996; Hardie et al. 2006; Raadik 2014), we have conservatively applied the name to the original 1985 collection from its type locality.
A subsequent allozyme study was then conducted in 1988, using additional frozen material collected and dispatched earlier that year. The aim of this study was to explore population structure and species integrity in the Galaxias truttaceus species group, including G. truttaceus plus its two sister species G. tanycephalus and G. auratus. This study focused on the polymorphic markers identified for these three species in the initial overview study, plus a handful of additional enzymes that are generally likely to be polymorphic. Ultimately, this second screen of 20 polymorphic loci, along with companion mtDNA restriction fragment-length polymorphism (RFLP) data, were published as two discrete studies, one on the widespread G. truttaceus (Ovenden and White 1990), and the other on its two lake-restricted congeners (Ovenden et al. 1993). Of relevance here is that the raw genotypes for all individuals screened were generated in our laboratory, and so were able to be integrated with those obtained in the final study (see below).
We next conducted one final allozyme study in 2012, focused on assessing the genetic distinctiveness of the threatened Western Australian population of G. truttaceus (thought prior to the study to be a subspecies). By this time, the ABTC collection of frozen tissues had been enhanced to include additional Tasmanian and some mainland populations of G. truttaceus. As a consequence, this final allozyme screen surveyed all populations for which frozen tissues were available (12 coastal and two landlocked sites for G. truttaceus) and again included both G. tanycephalus and G. auratus. The resultant published study (Morgan et al. 2016) included a companion mtDNA tree, but excluded the genetic data for the eastern landlocked populations; this latter decision reflected our desire to focus on the broader Australian perspective and subsequently explore the distinctive genetic profiles of these landlocked populations in a future study.
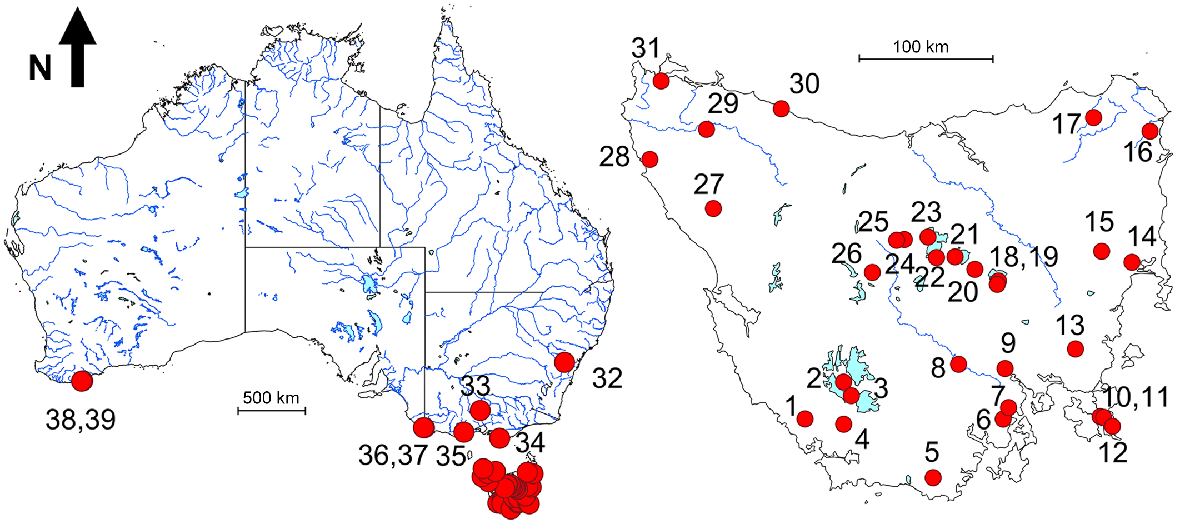
Herein, we present two analyses of all existing allozyme data for the G. truttaceus species group, one for the full 2012 allozyme dataset and the other incorporating the allozyme profiles of all sites surveyed for the 20 polymorphic loci common across all three allozyme studies (excluding sites with n = 1). Sampling details and sample sizes for all allozyme studies and the ‘2022’ analysis (i.e. the integrated dataset of 20 loci for the 1985, 1988 and 2012 studies) are presented in Table 2 and the geographic arrangement of these sites are displayed in Fig. 2.
| Site | Locality | Species | 1985 | 1988 | 2012 | 2022 | cytb | Latitude | Longitude | Tissue code |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Giblin River, Tas. | G. brevipinnis | 1 | −43.083 | 145.860 | UTas:Gbr014 | ||||
| 2 | Lake Pedder, Tas. [L] | G. parvus | 3 | 3 | −42.832 | 146.122 | White#2 | |||
| G. pedderensis | 3 | 2 | White#1 | |||||||
| 3 | Creek near Lake Pedder, Tas. [L] | G. brevipinnis | 2 | −42.925 | 146.175 | UTas:LP01+ | ||||
| 4 | Crossing River, Tas. | G. brevipinnis | 1 | −43.119 | 146.123 | UTas:Gbr010 | ||||
| 5 | Reservoir Lakes, Tas. [L] | G. niger | 3 | 3 | −43.483 | 146.731 | White#27 (1985) | |||
| G. brevipinnis | 2 | UTas:RL (2014) | ||||||||
| 6 | Snug Falls, Tas. | G. brevipinnis | 1 | −43.084 | 147.207 | UTas:SnugF05 | ||||
| 7 | North West Bay River, Tas. | G. maculatus | 3 | −43.006 | 147.244 | White#11 | ||||
| 8 | Styx River Bushy Park, Tas. | G. truttaceus | 1 | 1 | −42.711 | 146.904 | FISHy4:MT5–1 | |||
| 9 | Derwent River, Tas. | G. truttaceus | 3 | −42.740 | 147.220 | M30:T8+ | ||||
| N. cleaveri | 3 | M30:C3+ | ||||||||
| 10 | Allens Creek #1, Tas. | G. truttaceus | 40 | 40 | −43.067 | 147.867 | GALAX2:A | |||
| G. brevipinnis | 3 | White#5 | ||||||||
| 11 | Allans Creek #2, Tas. | G. truttaceus | 8 | 8 | 1 | −43.072 | 147.890 | FISHy4:MT5–5 A | ||
| 12 | Fortescue Lagoon Creek, Tas. | G. truttaceus | 42 | 42 | −43.133 | 147.950 | GALAX2:F | |||
| N. cleaveri | 2 | White#26 | ||||||||
| 13 | Prosser River, Tas. | G. truttaceus | 3 | 3 | 1 | −42.607 | 147.699 | White#14 | ||
| G. maculatus | 3 | 1 | White#13 | |||||||
| 14 | Swan River, Tas. | G. fontanus | 3 | 4 | −42.017 | 148.083 | White#10 | |||
| 15 | Cygnet River, Tas. | G. brevipinnis | 2 | −41.942 | 147.877 | UTas:Gb01+ | ||||
| 16 | Last River tributary, Tas. | G. maculatus | – | 2 | −41.125 | 148.206 | FISH98:MT47+ | |||
| 17 | Boobyalla River, Tas. | G. truttaceus | 1 | 1 | −41.032 | 147.822 | FISH98:MT53 | |||
| 18 | Lake Sorrell, Tas. [L] | G. auratus | 40 | 40 | −42.144 | 147.175 | GALAX2:S | |||
| 19 | Lake Crescent, Tas. [L] | G. auratus | 8 | 8 | 5 | −42.167 | 147.167 | White#16 | ||
| G. auratus | 3 | 3 | White#7 | |||||||
| 20 | Woods Lake, Tas. [L] | G. tanycephalus | 3 | 3 | 3 | −42.065 | 147.015 | GALAX2:W | ||
| 21 | Arthurs Lake, Tas. [L] | G. tanycephalus | 3 | 3 | −41.980 | 146.880 | GALAX2:Wm | |||
| G. tanycephalus | 3 | 3 | 3 | 6 | White#19 | |||||
| P. mesotes | 3 | 3 | White#3 | |||||||
| 22 | near Shannon Lagoon, Tas. [L] | G. truttaceus | 8 | 8 | 5 | −41.985 | 146.754 | FISHy4:MT5–2 | ||
| 23 | Great Lake, Tas. [L] | G. brevipinnis | 1 | 1 | −41.845 | 146.696 | White#21 | |||
| G. brevipinnis | 2 | UTas:GL | ||||||||
| P. dissimilis | 3 | 3 | White#22 | |||||||
| P. eleotroides | 3 | 2 | White#23 | |||||||
| 24 | Carters Lake, Tas. [L] | G. truttaceus | 6 | 6 | 3 | −41.862 | 146.535 | White#8 | ||
| G. brevipinnis | 3A | 1 | White#17 | |||||||
| P. julianus | 3 | 2 | White#18 | |||||||
| 25 | Isabella Lagoon, Tas. [L] | G. truttaceus | 40 | 40 | 3 | −41.879 | 146.489 | GALAX2:I | ||
| 26 | Clarence Lagoon, Tas. [L] | G. johnstoni | 3 | 2 | −42.086 | 146.318 | White#24 | |||
| 27 | Rocky River, Tas. | G. brevipinnis | 1 | −41.651 | 145.237 | UTas:Fish-1+ | ||||
| 28 | Thornton River, Tas. | G. brevipinnis | 1 | −41.317 | 144.805 | UTas:Fish-12 | ||||
| 29 | Lawson Rivulet, Tas. | G. brevipinnis | 1 | −41.112 | 145.190 | FISH98:MT17+ | ||||
| 30 | near Wynyard, Tas. | G. truttaceus | 5 | 5 | 1 | −40.972 | 145.699 | FISHy4:MT5–6B | ||
| N. cleaveri | 3 | FISHy4:MT5–8 | ||||||||
| 31 | near Montagu, Tas. | G. truttaceus | 8 | 8 | 1 | −40.783 | 144.882 | FISH98:MT-78 | ||
| 32 | Katoomba Creek, NSW [L] | G. olidus | 3 | −33.679 | 150.307 | White#25 | ||||
| 33 | McIvor River, Vic. [L] | G. truttaceus | 2A | 2 | 3 | −36.899 | 144.686 | FISHy4:MDt-1+ | ||
| 34 | Bald Hills Creek, Vic. | G. truttaceus | 8 | 8 | 1 | −38.751 | 145.978 | PU02–75GT | ||
| 35 | Barongarook Creek | G. truttaceus | 8 | 8 | 1 | −38.345 | 143.594 | PU02–90GT | ||
| 36 | Ellard Creek, SA | G. truttaceus | 1 | −38.052 | 140.959 | E350:CY75+ | ||||
| 37 | Pick Swamp, SA | G. truttaceus | 6 | 6 | 1 | −38.046 | 140.894 | E350:CY69 | ||
| 38 | Angove River, WA | G. truttaceus | 10 | 10 | 1 | −34.923 | 118.152 | FISHy4:Gt2+ | ||
| 39 | Goodga River, WA | G. truttaceus | 11 | 11 | 1 | −34.939 | 118.083 | FISHy4:Gt25+ | ||
| Total | 63 | 168 | 91 | 265 | 85 |
Allozyme datasets are listed by year. [L], landlocked site. cytb sample sizes exclude the 15 Genbank sequences that were not generated by the study of Morgan et al. (2016).
AA single F1 hybrid, confirmed by allozyme profiling, was also found at each of these two sites (Site 24, G. truttaceus × G. brevipinnis; Site 33, G. truttaceus × G. oliros).
Composite map showing the location of all sites surveyed across the various molecular studies. Site numbers mirror those used in Table 1.
General allozyme methods
Allozyme electrophoresis on muscle homogenates was undertaken as described elsewhere (Richardson et al. 1986; Hammer et al. 2007). Homogenates were successfully screened for various combinations of the enzymes employed in our other studies on galaxiids (Adams et al. 2014; Morgan et al. 2016). Histochemical stain recipes plus enzyme, locus and allozyme nomenclature all follow Hammer et al. (2007).
We employed a range of different approaches to analyse the allozyme data, depending on the dataset under consideration. For the initial overview study, we generated a neighbour-joining (NJ) tree among all individuals, constructed from a pairwise matrix of Roger’s genetic distances (Rogers’ R). This NJ tree was rooted using N. cleaveri, identified as the earliest-branching species among this group based on mtDNA and nDNA sequence data (Burridge et al. 2012). We also calculated the pairwise number of fixed differences and unbiased Nei’s distances (Nei Ds) among all species to assess their distinctiveness and for cross-referencing with the NJ tree among individuals. For the 2012 study we used principal co-ordinates analysis (PCoA) to assess the genetic affinities of individuals, independent of locality or taxon. Analysis of the ‘2022’ dataset involved generating an unrooted NJ network among sites on the basis of unbiased Nei Ds. All details for these methods have been published previously (Hammer et al. 2007; Adams et al. 2014).
mtDNA sequencing
As the majority of published mtDNA sequences for the Australian galaxiids involve the cytochrome b (cytb) gene, we constructed our mtDNA gene tree using a combination of newly generated cytb sequences plus Genbank-held exemplars for all taxa and most of the populations profiled in the allozyme studies. Importantly, we confirmed (Jonathan Waters, pers. comm.) that the G. pedderensis sequences published by Burridge et al. (2012) were obtained from a single individual, directly sourced from the population of G. pedderensis that was translocated into Lake Oberon prior to the species becoming extinct in the wild (Chilcott et al. 2013). Our final cytb dataset also included sequences for two individuals collected in 2014 by one of us (C. P. Burridge) from the type locality for G. niger.
Some of the new sequences were generated and edited using the methods and primers detailed in Morgan et al. (2016), whereas others were obtained using protocols that differed only in the choice of primers (those described in Nielsen et al. 1994), and edited using software Geneious Prime (ver. 2021.2, see https://www.geneious.com). All new sequences have been deposited in GenBank, with Accession numbers OQ738621–OQ738684 and OQ738809–OQ738811. The final gene tree was generated using RAxML (ver. 8.2.12, see https://github.com/stamatak/standard-RAxML; Stamatakis 2014) on the CIPRES cluster (see http://www.phylo.org/; Miller et al. 2010) by using the model GTRGAMMA and searching for the best-scoring maximum likelihood (ML) tree. Bootstrapping was set to finish on the basis of the autoMRE majority-rule criterion.
Results
1985 allozyme overview study
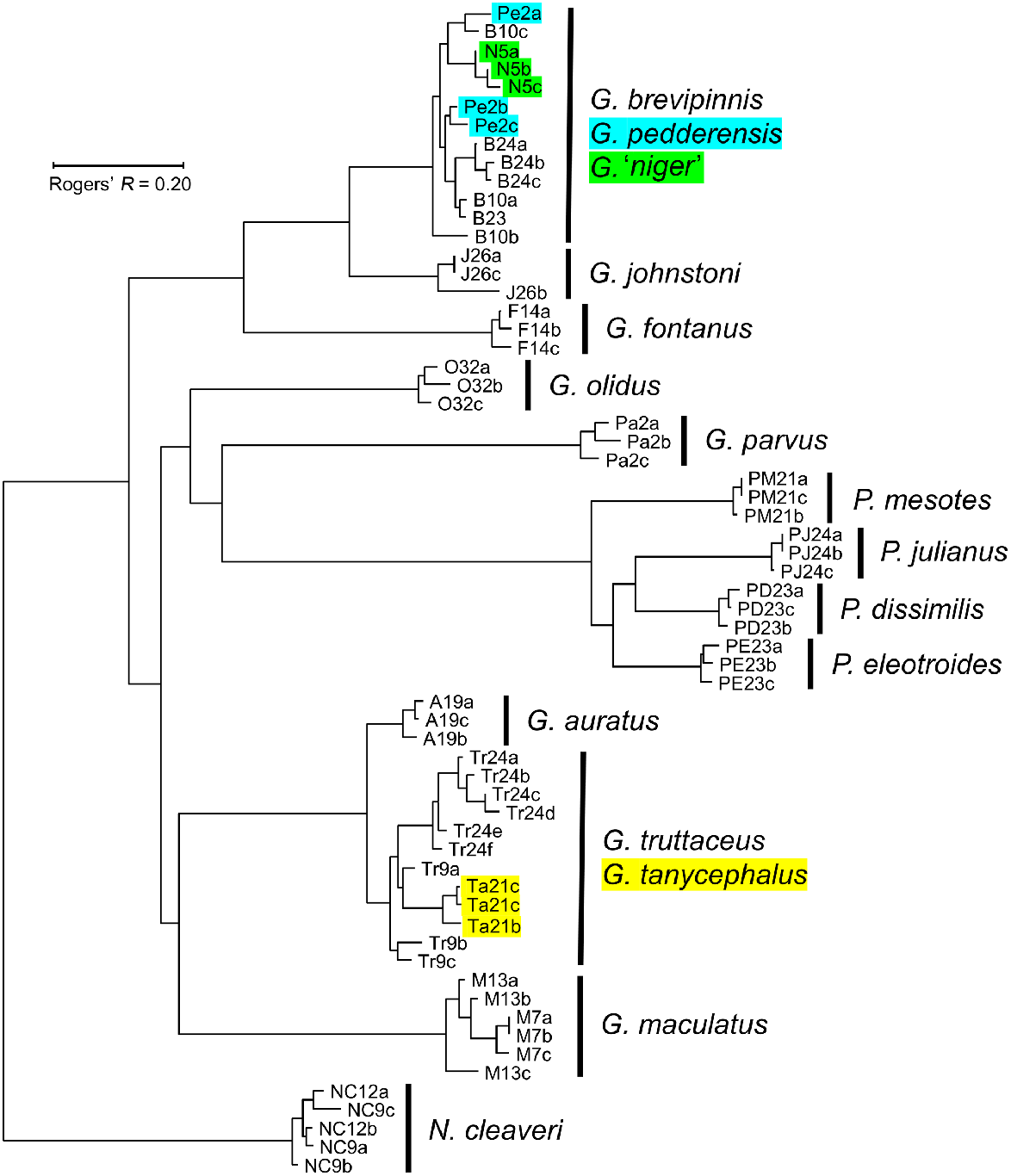
The final dataset for the initial overview study comprised the allozyme profiles of 63 individuals at 43 putative loci. Excluded from this dataset was a single fish, allozymically identified as a G. truttaceus × G. brevipinnis F1 hybrid (both species being present at that site; Table 2). Allozyme frequencies for each locus and species are summarised in Supplementary Table S1. Most of the 16 nominal species (15 Tasmanian species plus G. olidus) appeared as distinct clusters within the NJ tree (Fig. 3), the exceptions being one cluster comprising G. brevipinnis–G. pedderensis–G. niger and the other involving G. truttaceus–G. tanycephalus–G. auratus. This outcome is supported by the number of pairwise fixed differences among species (Table 3), with every species readily diagnosable by multiple fixed differences (range 6–34) except for the two clusters described above (range 0–3).
Neighbour-joining tree among the 63 individuals included in the 1985 allozyme overview study, rooted using N. cleaveri. Individuals are labelled by a unique species code (a one- or two-letter abbreviation) plus site; multiple individuals from the same site are labelled alphabetically. Highlighted individuals: supplied as G. pedderensis (blue); supplied as G. niger (green); G. tanycephalus (yellow).
| Species | Pe | B | N | J | F | O | Pa | A | T | Ta | M | NC | PM | PJ | PE | PD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G. pedderensis (Pe) | – | 0.01 | 0.03 | 0.18 | 0.43 | 0.67 | 0.98 | 0.63 | 0.68 | 0.73 | 0.78 | 0.94 | 1.36 | 1.70 | 1.24 | 1.44 |
| G. brevipinnis (B) | 0 | – | 0.04 | 0.17 | 0.43 | 0.68 | 0.98 | 0.63 | 0.68 | 0.70 | 0.77 | 0.92 | 1.33 | 1.67 | 1.21 | 1.41 |
| G. niger (N) | 0 | 1 | – | 0.18 | 0.46 | 0.65 | 1.08 | 0.63 | 0.68 | 0.73 | 0.73 | 0.89 | 1.34 | 1.67 | 1.22 | 1.42 |
| G. johnstoni (J) | 6 | 6 | 7 | – | 0.46 | 0.62 | 0.97 | 0.69 | 0.70 | 0.75 | 0.77 | 0.95 | 1.32 | 1.52 | 1.18 | 1.38 |
| G. fontanus (F) | 13 | 14 | 15 | 15 | – | 0.74 | 1.18 | 0.87 | 0.89 | 0.94 | 0.79 | 0.99 | 1.15 | 1.41 | 1.15 | 1.10 |
| G. olidus (O) | 21 | 20 | 20 | 20 | 22 | – | 0.65 | 0.54 | 0.52 | 0.56 | 0.68 | 0.80 | 1.02 | 1.03 | 0.93 | 0.94 |
| G. parvus (Pa) | 25 | 25 | 26 | 26 | 27 | 20 | – | 0.63 | 0.61 | 0.64 | 0.84 | 1.25 | 1.20 | 1.22 | 1.17 | 1.04 |
| G. auratus (A) | 19 | 19 | 19 | 21 | 24 | 16 | 18 | – | 0.08 | 0.09 | 0.55 | 0.89 | 1.15 | 1.29 | 1.04 | 1.06 |
| G. truttaceus (T) | 20 | 20 | 19 | 21 | 23 | 15 | 18 | 3 | – | 0.06 | 0.52 | 0.95 | 1.23 | 1.34 | 1.14 | 1.16 |
| G. tanycephalus (Ta) | 21 | 21 | 20 | 22 | 26 | 16 | 18 | 3 | 2 | – | 0.57 | 0.86 | 1.24 | 1.32 | 1.14 | 1.14 |
| G. maculatus (M) | 22 | 21 | 20 | 21 | 22 | 21 | 23 | 17 | 16 | 17 | – | 0.92 | 1.19 | 1.29 | 1.09 | 1.05 |
| N. cleaveri (NC) | 24 | 24 | 23 | 25 | 25 | 21 | 27 | 23 | 24 | 23 | 24 | – | 1.17 | 1.03 | 1.32 | 1.18 |
| P. mesotes (PM) | 31 | 30 | 31 | 31 | 29 | 26 | 28 | 29 | 30 | 29 | 28 | 28 | – | 0.30 | 0.23 | 0.22 |
| P. julianus (PJ) | 34 | 33 | 34 | 33 | 32 | 27 | 29 | 30 | 30 | 30 | 30 | 26 | 10 | – | 0.24 | 0.20 |
| P. eleotroides (PE) | 29 | 28 | 29 | 28 | 28 | 25 | 27 | 27 | 28 | 28 | 26 | 28 | 7 | 8 | – | 0.15 |
| P. dissimilis (PD) | 31 | 30 | 31 | 31 | 28 | 25 | 25 | 27 | 28 | 27 | 26 | 28 | 8 | 8 | 6 | – |
Lower triangle, number of fixed differences (allowing a cumulative 10% tolerance for any shared allozymes); upper triangle, unbiased Nei distances.
Under the usually reliable assumption that phylogenetic relatedness is roughly correlated with overall genetic similarity, we infer from Fig. 3 the following results: (1) Galaxias johnstoni and G. fontanus appear to be sister species to the G. brevipinnis ‘complex’, (2) the four species of Paragalaxias comprise a monophyletic genus, (3) G. parvus and G. maculatus are not closely related to any other included species, (4) Tasmanian Galaxias species may not be monophyletic, and (5) the genus Galaxias is likely to be paraphyletic.
2012 allozyme study of G. truttaceus species group
The final allozyme dataset for this study comprised 91 individuals profiled at 57 putative loci (i.e. the dataset of Morgan et al. (2016) plus landlocked sites of Tr22 (n = 8) and Tr33 (n = 2)). A single fish identified by its allozyme profile as a G. truttaceus × Galaxias oliros F1 hybrid (Site Tr33, Table 2) was excluded from this dataset. Allozyme frequencies by locus for each of the 16 sites are provided in Table S2. A PCoA of these 91 individuals displayed three primary clusters, corresponding to the three member species in this group. Conventional measures of among-species genetic divergence are similar for all pairwise combinations (Nei D range: 0.17–0.24); however, only G. auratus is fully diagnosable by fixed differences (Fig. 4). In contrast, G. tanycephalus and G. truttaceus are only marginally diagnosable by fixed differences, particularly when the latter is partitioned by habitat type (see next section).
Scatterplot of the relative scores in the first two dimensions for the principal coordinates analysis of 91 G. truttaceus group individuals, based on the 2012 allozyme study. Species are identified by a unique symbol corresponding to the cluster label. Symbols shown in green represent fish from landlocked or lake populations (blue circles for G. truttaceus being fish from ‘coastal’ sites that are presumed diadromous). A pairwise genetic-distance matrix among the major groups and subgroups is shown in the boxed inset; lower-left triangle shows number of fixed differences (numbers in parentheses allow a 10% tolerance for all shared alleles summed); upper right triangle shows unbiased Nei Ds.
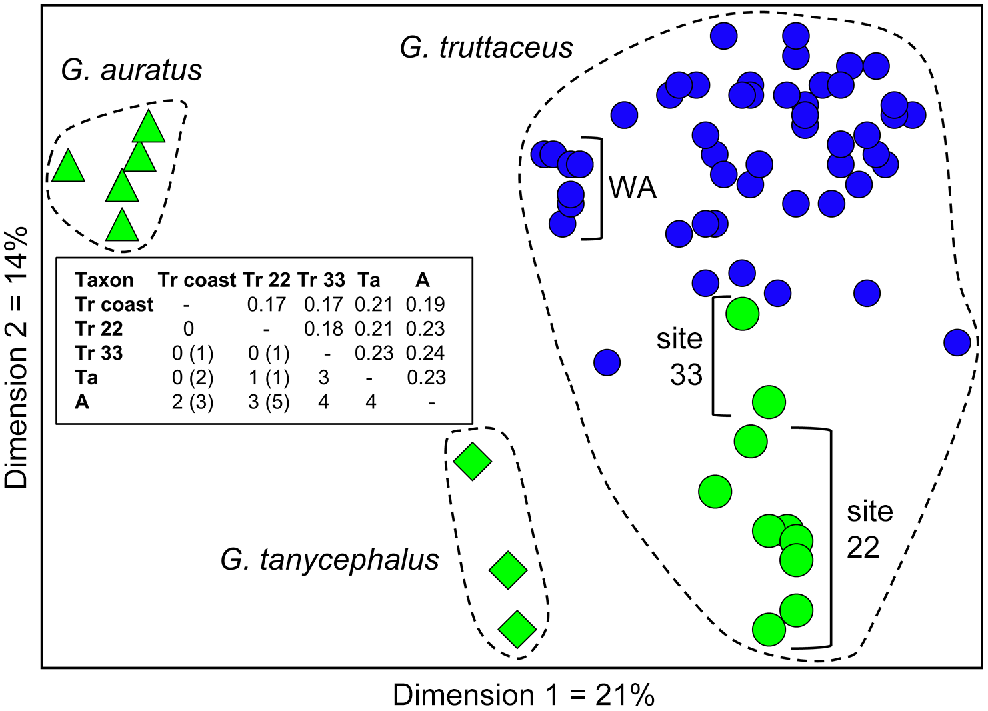
As expected for a widespread species, G. truttaceus exhibited far greater genetic diversity than was present in its two lacustrine congeners. However, less predictably, a substantial proportion of this allozyme diversity (i.e. as shown in PCoA Dimension 2, Fig. 4) does not correlate with geographic distance (e.g. over 3500 km between WA and Tasmanian sites) but instead correlates well with whether the site is landlocked or ‘coastal’ (e.g. with easy access to the ocean). Indeed, putting aside species boundaries, a significant proportion of the genetic diversity in this complex clearly resides in landlocked habitats.
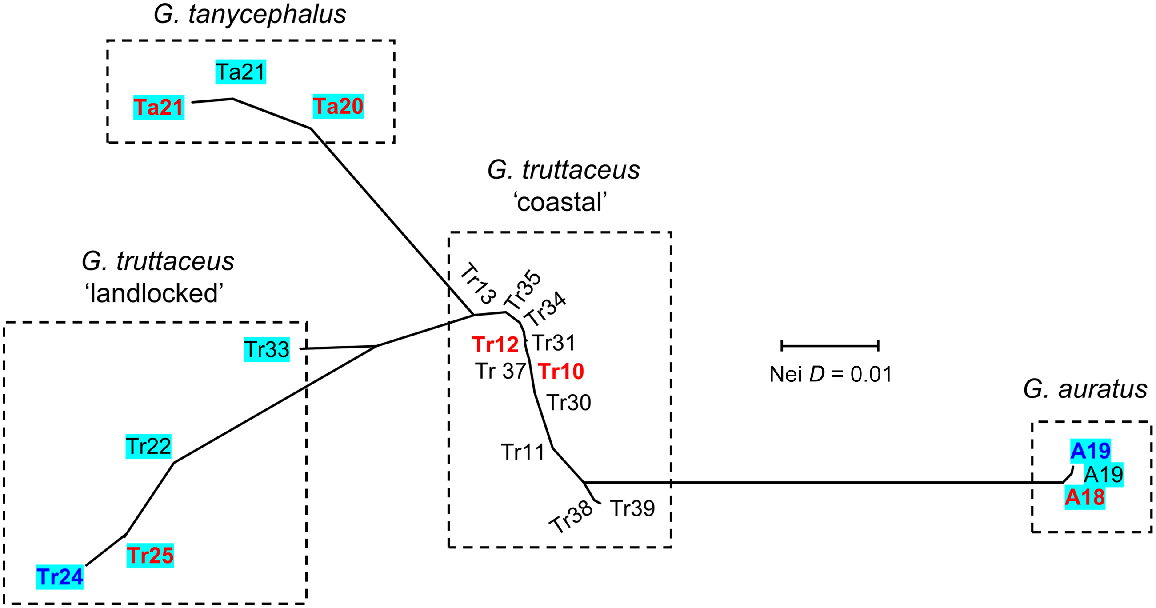
2022 combined allozyme analysis of G. truttaceus species group
The final 2022 allozyme dataset comprised 265 individuals at the 20 loci found to be polymorphic in the G. truttaceus species group (Ovenden and White 1990; Ovenden et al. 1993). Allele frequencies at all 21 sampling events across 19 sites are summarised in Table S3 (Sites Ta21 and A19 were independently sampled in each stand-alone allozyme screen). Four clusters corresponding to G. auratus, G. tanycephalus, coastal G. truttaceus and landlocked G. truttaceus are evident from the NJ network (Fig. 5) and PCoA (Fig. 4), with the mainland Tr 33 site again being the least divergent of the latter group. Importantly, this same pattern holds even after the inclusion of additional landlocked and coastal sites for G. truttaceus (Tr24 and Tr25 are lake populations; Tr10 and Tr12 are coastal sites), plus the addition of one extra site for both G. auratus (A18) and G. tanycephalus (Site Ta20; Site Ta21 also represented by a second sampling event).
Unrooted neighbour-joining network for the combined 2022 allozyme dataset (all populations of the G. truttaceus species group, profiled at 20 polymorphic loci). Populations represented in the 1985 study are in bold blue text; those in the 1988 study (Ovenden and White 1990; Ovenden et al. 1993) are in bold red text. Landlocked populations are highlighted in blue.
Matrilineal genealogy for the Tasmanian galaxiids
The final mtDNA dataset comprised 100 cytb sequences of length 1120 bp, representing all species and most populations. This included 68 novel sequences, 17 Genbank sequences directly linked to the 2012 allozyme study (Morgan et al. 2016), and 15 Genbank sequences from other sources. RAxML recovered a single tree and the rapid bootstrap search ended after 450 replicates (Fig. 6). This tree shares many features with the original allozyme overview study (Fig. 3). Both analyses support the monophyly of (1) G. johnstoni and G. fontanus with the G. brevipinnis ‘complex’, (2) the four species of Paragalaxias, and (3) the three species in the G. truttaceus species group. Likewise, both show that G. maculatus and G. parvus are distinctive early branching lineages, and both infer the genus Galaxias overall and Tasmanian Galaxias in particular as being unlikely to be monophyletic. A final point of similarity was the inability to distinguish G. niger from G. brevipinnis; indeed, the same cytb haplotype was present in both G. niger and the G. brevipinnis collected from Reservoir Lakes in 2014 (Site 5; Fig. 6).
Maximum-likelihood tree, based on 1120 bp of cytb and rooted using N. cleaveri. Nodes with bootstrap support of 90% or more are indicated by an asterisk. As for Fig. 3, individuals are labelled using their abbreviated species code (Ol = G. oliros) plus a site code (if known); multiple individuals at a site are labelled numerically (.1, .2, etc.). Genbank exemplars (red) are labelled with their accession number and, where traceable, their source site (Site 2# was the translocated population of G. pedderensis, originally sourced from Site 2); all other sequences were listed in Genbank only as from ‘Tasmania’. Highlighted sequences: G. truttaceus from Site 22 (Shannon Lagoon; yellow); F1 hybrids (grey); G. pedderensis as included in the 1985 allozyme study (blue); G. niger (green).
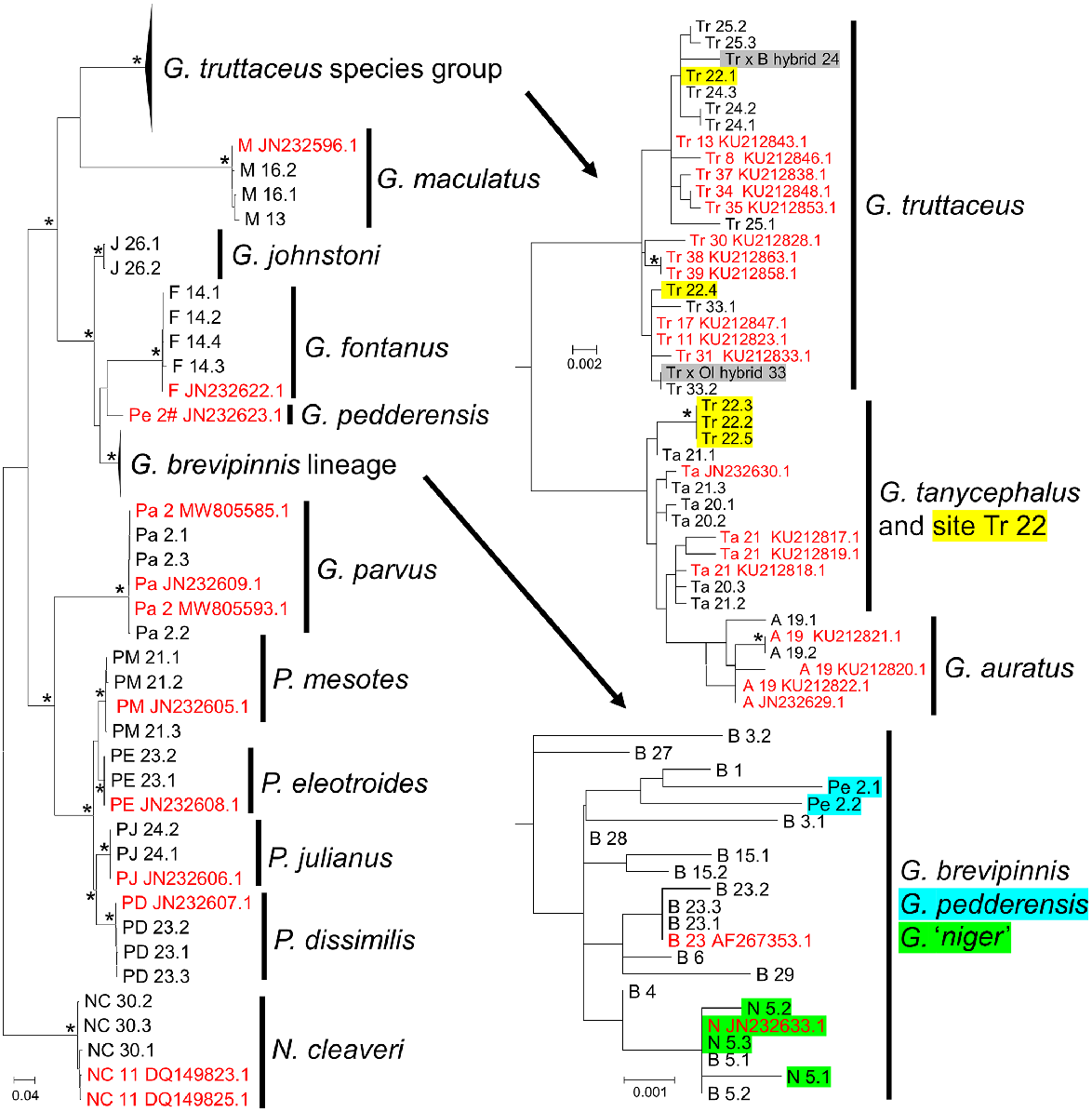
Notwithstanding the overall concordance of allozyme and mtDNA perspectives, there are two notable differences between them. First and foremost is the discrepancy between the cytb haplotypes displayed by the two G. pedderensis included in the 1985 allozyme study (Pe 2.1 and Pe 2.2, Fig. 6) when compared with the true-to-label cytb haplotype available on Genbank (accession number JN232623.1, from Burridge et al. 2012). Although both datasets confirm that G. pedderensis is closely related to G. brevipinnis, the mtDNA results suggest that the fish allozymed in 1985 may not have the same provenance as the separately sourced tissue sequenced by Burridge et al. (2012). Second, although both agree that the three members of the G. truttaceus species group are closely related, two modest discrepancies are evident; the nuclear markers suggest that G. tanycephalus and G. truttaceus are sister species (Fig. 3–5), whereas mtDNA places G. tanycephalus sister to G. auratus (Fig. 6). In addition, the mtDNA shows that the landlocked population near Shannon Lagoon (Site Tr22) is the only population of G. truttaceus that harbours cytb haplotypes from each of the two primary cytb subclades, which otherwise diagnose the G. truttaceus versus G. tanycephalus–G. auratus matrilineal dichotomy.
Discussion
Multi-locus genetic datasets (mostly involving allozyme profiling in the pre-genomics era) have already played an important role in species delineation and discovery for Australia’s freshwater fishes, and such datasets will inevitably become even more valuable into the future (Hammer et al. 2013b). Most researchers now accept that the gold standard for contemporary molecular systematics ought to centre around genomic data, with single nucleotide polymorphisms (SNPs) arguably the simplest and most cost-effective approach at present (Leaché and Oaks 2017; Georges et al. 2018). However, this acknowledgement does not diminish the ability of allozyme profiling to continue to provide key taxonomic and conservation insights for groups where genomic data are lacking (i.e. most of Australia’s freshwater fishes). Indeed, recent studies have now demonstrated that allozymes provide exceptional concordance with a companion SNP dataset for the delineation of all primary and admixed lineages in every group examined thus far (Gadopsis, Unmack et al. 2017; Hypseleotris, Unmack et al. 2019; Retropinna, Unmack et al. 2022; Craterocephalus, Mogurnda and Rhadinocentrus, M. Adams and P. J. Unmack, unpubl. data), and can even provide comparable insights into fine-scale population structure (Jusaitis and Adams 2005; Brooks et al. 2022).
Species boundaries in Tasmanian galaxiids
Despite occupying less than 1% of Australia’s total landmass, Tasmania is home to 25% of its threatened freshwater fishes (10 of the 40 species listed nationally as ‘extinct in the wild’, ‘critically endangered’, ‘endangered’, or ‘vulnerable’; Lintermans et al. 2020; Department of Climate Change, Energy, the Environment and Water 2022). All the threatened Tasmanian species are galaxiids and most lack even the most basic multi-locus genetic assessment of their taxonomic integrity. Furthermore, their threatened status limits routine contemporary sampling of tissues, thus placing additional value on our historic tissue collections. In this light, the genetic appraisals presented herein provide an important baseline assessment of species boundaries plus an independent perspective on evolutionary relationships in this highly threatened group (Adams et al. 2014; Raadik 2014; Page et al. 2018; Lintermans et al. 2020).
Both allozyme and mtDNA analyses were concordant in unequivocally supporting the taxonomic validity of the following Tasmanian galaxiids: G. fontanus, G. johnstoni, G. maculatus, G. parvus, N. cleaveri, Paragalaxias dissimilis, Paragalaxias eleotroides, Paragalaxias mesotes and Paragalaxias julianus (all Tasmanian endemics except for N. cleaveri and all of conservation concern apart from G. maculatus). These nine species were readily diagnosable from each other at multiple genes and, apart from a single F1 hybrid, displayed no evidence of hybridisation with co-occurring or sibling congeners. The remaining six species fell into one of two distinctive species groups, namely (1) a G. brevipinnis lineage (the diadromous G. brevipinnis plus the lacustrine endemics G. pedderensis and G. niger), or (2) the G. truttaceus complex (diadromous and landlocked G. truttaceus plus the lacustrine endemics G. auratus and G. tanycephalus).
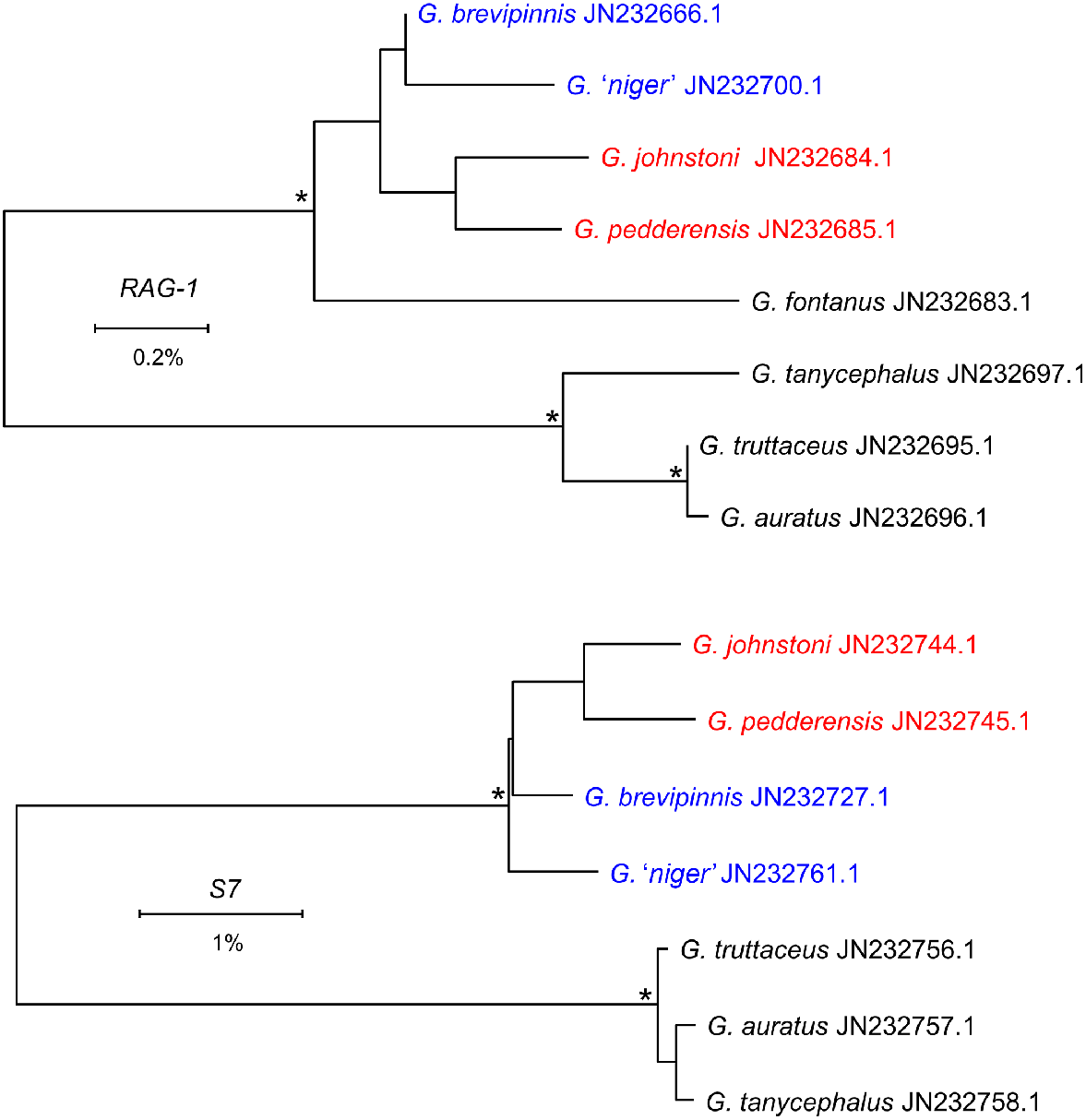
Our G. pedderensis and G. niger exemplars, collected in 1985 and assigned to these taxa primarily on the basis of being taken from the type locality for each species, were genetically indistinguishable from G. brevipinnis for both allozymes and cytb. Such an outcome has two possible explanations, namely (1) the fish collected in 1985 were actually G. brevipinnis and thus the real endemics were not sampled, or (2) both endemic ‘species’ are simply lacustrine populations of G. brevipinnis that have undergone discernable phenotypic divergence since being isolated from coastal populations. Unfortunately, no voucher specimens were collected at that time to distinguish between these alternatives. In the case of G. pedderensis, we suggest that the first explanation (i.e. genuine G. pedderensis were not collected in 1985) is the most tenable, for two reasons. First, the G. pedderensis sourced by Burridge et al. (2012) displayed a cytb haplotype that was sister to but distinct from all G. brevipinnis haplotypes (n = 16; Fig. 6) plus was genetically most similar to G. johnstoni rather than to G. brevipinnis for two nuclear genes (S7 and RAG-1; Fig. 7), and thus seems genuine. Second, historical records indicate that G. brevipinnis was known to have colonised Lake Pedder by the early 1980s, although intensive surveys failed to detect G. pedderensis in the lake itself after 1987 (Chilcott et al. 2013). Thus, it is plausible that the fish collected from Lake Pedder in 1985 were G. brevipinnis rather than G. pedderensis.
Neighbour-joining trees constructed using p-distances for G. brevipinnis and G. truttaceus complex species, on the basis of the published sequences for two nuclear genes of S7 and RAG-1 (Burridge et al. 2012). Sequences are labelled with their Genbank accession numbers. Nodes with bootstrap support of 90% or more are indicated by an asterisk.
In contrast to G. pedderensis, all available genetic data suggest that G. niger is likely to represent a darker, conspecific form of G. brevipinnis (see also Fulton 1990; McDowall and Fulton 1996; Hardie et al. 2006; Raadik 2014). Nevertheless, these data cannot exclude the possibility that, as for G. pedderensis, there was once an endemic galaxiid in Reservoir Lakes when first collected, but which subsequently became rare or was extirpated following colonisation by G. brevipinnis. This possibility could be confirmed or refuted in either of two ways. First, an explicit morphological comparison of the two species could re-assess the original assertion by Andrews (1985) that G. niger is readily diagnosable from G. brevipinnis on the basis of the latter’s large, paired fins and canine teeth (the two species were not otherwise compared). Second, it is now possible to generate genomic data from formalin-preserved voucher specimens, an approach that would help resolve this and many other taxonomic grey areas (Card et al. 2021), including any uncertainty surrounding G. pedderensis.
As well as confirming previously observed low levels of genetic divergence between G. truttaceus and its sister species G. auratus and G. tanycephalus (Ovenden et al. 1993; Morgan et al. 2016), our study further explored the relationships among these species by including allozyme and mtDNA assessments of additional landlocked G. truttaceus (Sites Tr22, Tr24, Tr25 in Tasmania, and Tr 33 from the Murray–Darling Basin; Fig. 2, 4–6). Together these analyses showed a consistent pattern whereby landlocked G. truttaceus display levels of nuclear genetic divergence from their diadromous coastal counterparts that mirror those between the three species themselves (Fig. 5). Importantly, whereas most landlocked G. truttaceus individuals exhibited the cytb subclade unique to this species, three of the five fish from Shannon Lagoon (Site Tr24) possessed a haplotype belonging to the subclade representing the two landlocked species (Fig. 6). Given how similar both subclades are to one another (average cytb sequence divergence = 2% and minimal divergence at S7 and RAG-1; Fig. 7), such an outcome either reflects lineage sorting among recently diverged taxa or post-isolation gene flow, both further indicators of the close affinities among these three species.
Evolutionary relationships
Our nuclear and matrilineal datasets complement and support the comprehensive, family-wide phylogeny of Burridge et al. (2012). Both studies found that the Tasmanian galaxiids are a heterogeneous assembly of evolutionarily distinct species (herein G. maculatus, G. parvus and N. cleaveri, but also including G. pusilla and L. sealii), plus three distinctive and monophyletic clades, namely, Paragalaxias, the G. brevipinnis complex (herein G. brevipinnis, G. ‘niger’, G. pedderensis, G. johnstoni and G. fontanus, plus many other galaxiids from New Zealand and New Caledonia), and the G. truttaceus complex (G. truttaceus, G. auratus and G. tanycephalus). Both studies also demonstrated that the genus Galaxias is not monophyletic, particularly with respect to Paragalaxias, which, along with G. parvus, appears to have closer evolutionary ties to members of the G. olidus complex in south-eastern Australia (Fig. 3; Burridge et al. 2012).
Isolation and landlocking as drivers of speciation
As Burridge et al. (2012) have already published a thorough exploration of evolutionary patterns, biogeographic affinities and life-history characteristics for the Galaxiidae, further discussion here will be limited to one key evolutionary theme applicable to this group, namely landlocking. Detailed studies of galaxiids, both in Australia and elsewhere, have demonstrated that the establishment of a non-migratory landlocked population from a diadromous, vagile ancestor has often driven rapid morphological, ecological and genetic divergence in the newly founded population which, if left untainted by subsequent gene flow, may ultimately result in speciation (Humphries 1990; Waters and Wallis 2001; Zattara and Premoli 2005; Waters et al. 2010). This phenomenon is most notable in the extended clade of species containing the diadromous G. brevipinnis and its many congeners (especially in New Zealand and, as further demonstrated herein, for Tasmania; Burridge et al. 2012). Moreover, it may also explain speciation events in two of Australia’s other galaxiid species complexes, one involving the diadromous G. maculatus and its non-migratory sister species G. rostratus and G. occidentalis, and the other being the G. truttaceus complex in Tasmania.
Our genetic data for the G. truttaceus complex showed a range of populations at differing stages along the path from landlocking to speciation. Mirroring an earlier morphological study (Humphries 1990), lacustrine populations of G. truttaceus displayed levels of genetic divergence from their diadromous counterparts that approached those among the three described species within the complex. Given that the budding of isolated populations from a vagile ancestor seems to be a common life-history characteristic of galaxiids, isolated populations of G. truttaceus and other species ought to be priority candidates for any genomic search for cryptic species or to identify high-value populations for conservation. A landlocked population of G. truttaceus in the Murray–Darling Basin (Site Tr33), variously regarded as either a bait bucket introduction or long-lost native population (Humphries 2009), was genetically intermediate between lacustrine and diadromous, which implies that it may either be native to the region or display a founder effect after translocation. This prediction requires mtDNA combined with SNPs or similar genomic data to confirm which scenario is in play.
Conservation perspectives
Freshwater fishes are well known for their tendency to hybridise with congeners (Verspoor and Hammart 1991; Hammer et al. 2013b), particularly after experiencing anthropogenic habitat modification (McFarlane and Pemberton 2019). We detected two F1 hybrids in this study, both involving sympatric species from quite different evolutionary lineages (G. brevipinnis and G. truttaceus, Site 24; G. truttaceus and G. oliros, Site 33) and neither identified a priori on the basis of external appearance. Unacknowledged introgression from widespread common species can seriously affect conservation efforts for rarer and narrowly distributed species (Guildea et al. 2015; Frankham et al. 2017), particularly if managers unwittingly choose only admixed populations for conservation management or as source populations for future translocations.
In recognition of these concerns, Adams et al. (2011) advocated that congeneric species should initially be included in any ‘best practice’ conservation genetic or genomic study of an imperilled freshwater fish if at least one of the following criteria was satisfied for the target species and its congener:
currently sympatric or parapatric, currently allopatric but with a reasonable likelihood of prior sympatry or parapatry, allopatric but co-occurring in a natural corridor (e.g. a river) capable of facilitating gene flow, or conspecific prior to a taxonomic revision [Adams et al. 2011, p. 768].
With conservation concerns over so many Tasmanian galaxiids, our study effectively satisfies this recommendation by demonstrating that all vulnerable species were, in the 1980s, readily diagnosable at multiple unlinked loci except for those involved in the G. brevipinnis–pedderensis–‘niger’ and G. truttaceus–tanycephalus–auratus complexes. Nevertheless, as anthropogenic-directed catchment changes, accidental and deliberate translocations, and climate change have seen G. brevipinnis colonise new catchments in Tasmania and elsewhere (e.g. the Murray–Darling Basin; Waters et al. 2002), conservation managers should always be mindful of the potential for hybridisation and cryptic introgression between congeneric species.
Future molecular research priorities
In the current age of genomics, it is likely that all future molecular genetic explorations of species boundaries, population structure, and wildlife conservation will aspire to large-scale SNP datasets (Georges et al. 2018). One such study is already being undertaken on taxonomic integrity and population structure in the Swan River galaxias G. fontanus (Bruce Deagle, CSIRO, pers. comm.) and the fine-scale assessments provided for the few remaining natural populations and their translocated counterparts will be invaluable in guiding future management efforts. We advocate comparable studies for all of Tasmania’s imperilled galaxiids, using the evolutionary and taxonomic affinities presented herein to guide which studies can be stand-alone and which require the inclusion of a few samples of the relevant congeners (e.g. G. brevipinnis ought to be included in any conservation genomic study of G. pedderensis). Such studies can be non-invasive (Le Vin et al. 2011) and need not involve large sample sizes to be highly informative, provided they include all known populations and, if possible, historic exemplars (Baverstock and Moritz 1996; Card et al. 2021).
A second molecular research priority ought to be the contemporary genomic sampling of all landlocked populations of apparently common species, to assess the possibility they represent unacknowledged ‘cryptic’ species (whether truly cryptic or simply not identifiable a priori morphologically). Even if shown to be conspecific, the levels of genetic diversity contained in landlocked populations are often disproportionally high compared with those in other, more connected populations. Such populations merit recognition as having high value conservation and evolutionary potential should the species suffer future declines in abundance.
Data availability
The allozyme data are available on Dryad (doi:10.5061/dryad.jdfn2z3gf). mtDNA newly collected during this study is available from Genbank, with accession numbers OQ738621–OQ738684 and OQ738809–OQ738811.
Conflicts of interest
Peter J. Unmack is an editor for Marine and Freshwater Research but did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Marine and Freshwater Research encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Acknowledgements
We are indebted to Robert White as the initial driving force and primary provider of frozen tissues for the 1985 and 1988 allozyme studies and to Jenny Ovenden for her assistance in organising the 1988 allozyme study. We also thank Ross Andrews, Brigitte Winton, and Terry Reardon for technical assistance in the allozyme laboratory.
References
Adams M, Wedderburn SD, Unmack PJ, Hammer MP, Johnson JB (2011) Use of congeneric assessment to reveal the linked genetic histories of two threatened fishes in the Murray–Darling Basin, Australia. Conservation Biology 25, 767-776.
| Crossref | Google Scholar |
Adams M, Page TJ, Hurwood DA, Hughes JM (2013) A molecular assessment of species boundaries and phylogenetic affinities in Mogurnda (Eleotridae): a case study of cryptic biodiversity in the Australian freshwater fishes. Marine and Freshwater Research 64, 920-931.
| Crossref | Google Scholar |
Adams M, Raadik TA, Burridge CP, Georges A (2014) Global biodiversity assessment and hyper-cryptic species complexes: more than one species of elephant in the room? Systematic Biology 63, 518-533.
| Crossref | Google Scholar |
Andrews AP (1985) A new species of Galaxias (Pisces: Galaxiidae) from southern Tasmania. Papers and Proceedings of The Royal Society of Tasmania 119, 55-60.
| Crossref | Google Scholar |
Arthington AH, Milton D, McKay RJ (1983) Effects of urban development and habitat alterations on the distribution and abundance of native and exotic freshwater fish in the Brisbane region, Queensland. Australian Journal of Ecology 8, 87-101.
| Crossref | Google Scholar |
Bray DJ (2018) Family GALAXIIDAE. In ‘Fishes of Australia’. (Museums Victoria) Available at https://fishesofaustralia.net.au/home/family/126 [Verified 13 July 2023]
Brooks E, Slender AL, Cu S, Breed MF, Stangoulis JCR (2022) A range-wide analysis of population structure and genomic variation within the critically endangered spiny daisy (Acanthocladium dockeri). Conservation Genetics 23, 1027-1037.
| Crossref | Google Scholar |
Burridge CP, McDowall RM, Craw D, Wilson MVH, Waters JM (2012) Marine dispersal as a pre-requisite for Gondwanan vicariance among elements of the galaxiid fish fauna. Journal of Biogeography 39, 306-321.
| Crossref | Google Scholar |
Card DC, Shapiro B, Giribet G, Moritz C, Edwards SV (2021) Museum genomics. Annual Review of Genetics 55, 633-659.
| Crossref | Google Scholar |
Chilcott S, Freeman R, Davies PE, Crook DA, Fulton W, Hamr P, Jarvis D, Sanger AC (2013) Extinct habitat, extant species: lessons learned from conservation recovery actions for the Pedder galaxias (Galaxias pedderensis) in south-west Tasmania, Australia. Marine and Freshwater Research 64, 864-873.
| Crossref | Google Scholar |
Coleman RA, Pettigrove V, Raadik TA, Hoffmann AA, Miller AD, Carew ME (2010) Microsatellite markers and mtDNA data indicate two distinct groups in dwarf galaxias, Galaxiella pusilla (Mack) (Pisces: Galaxiidae), a threatened freshwater fish from south-eastern Australia. Conservation Genetics 11, 1911-1928.
| Crossref | Google Scholar |
Coleman RA, Hoffman AA, Raadik TA (2015) A review of Galaxiella pusilla (Mack) (Teleostei: Galaxiidae) in south-eastern Australia with a description of a new species. Zootaxa 4021, 243-281.
| Crossref | Google Scholar |
Cook BD, Kennard MJ, Real K, Pusey BJ, Hughes JM (2011) Landscape genetic analysis of the tropical freshwater fish Mogurnda mogurnda (Eleotridae) in a monsoonal river basin: importance of hydrographic factors and population history. Freshwater Biology 56, 812-827.
| Crossref | Google Scholar |
Cook BD, Adams M, Unmack PJ, Burrows D, Pusey BJ, Perna C, Hughes JM (2017) Phylogeography of the mouth-brooding freshwater fish Glossamia aprion (Apogonidae) in northern and eastern Australia: historical biogeography and allopatric speciation. Biological Journal of the Linnean Society 121, 833-848.
| Crossref | Google Scholar |
Crowl TA, Townsend CR, Mcintosh AR (1992) The impact of introduced brown and rainbow trout on native fish: the case of Australasia. Reviews in Fish Biology and Fisheries 2, 217-241.
| Crossref | Google Scholar |
Department of Climate Change, Energy, the Environment and Water (2022) Species Profile and Threats Database: EPBC Act List of Threatened Fauna. Available at https://www.environment.gov.au/cgi-bin/sprat/public/publicthreatenedlist.pl?wanted=fauna [Verified 13 July 2023]
Dudgeon D, Arthington AH, Gessner MO, Kawabata Z-I, Knowler DJ, Lévêque C, Naiman RJ, Prieur-Richard A-H, Soto D, Stiassny MLJ, Sullivan CA (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biological Reviews 81, 163-182.
| Crossref | Google Scholar |
Duncan JR, Lockwood JL (2001) Extinction in a field of bullets: a search for causes in the decline of the world’s freshwater fishes. Biological Conservation 102, 97-105.
| Crossref | Google Scholar |
Faulks LK, Kerezsy A, Unmack PJ, Johnson JB, Hughes JM (2017) Going, going, gone? Loss of genetic diversity in two critically endangered Australian freshwater fishes, Scaturiginichthys vermeilipinnis and Chlamydogobius squamigenus, from Great Artesian Basin springs at Edgbaston, Queensland, Australia. Aquatic Conservation: Marine and Freshwater Ecosystems 27, 39-50.
| Crossref | Google Scholar |
Georges A, Adams M (1996) Electrophoretic delineation of species boundaries within the short-necked freshwater turtles of Australia (Testudines: Chelidae). Zoological Journal of the Linnean Society 118, 241-260.
| Crossref | Google Scholar |
Georges A, Gruber B, Pauly GB, White D, Adams M, Young MJ, Kilian A, Zhang X, Shaffer HB, Unmack PJ (2018) Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Molecular Ecology 27, 5195-5213.
| Crossref | Google Scholar |
Guildea C, Hitchen Y, Duffy R, Dias PJ, Ledger JM, Snow M, Kennington WJ (2015) Introgression threatens the survival of the critically endangered freshwater crayfish Cherax tenuimanus (Decapoda: Parastacidae) in the wild. PLoS ONE 10, e0121075.
| Crossref | Google Scholar |
Hammer MP, Adams M, Unmack PJ, Walker KF (2007) A rethink on Retropinna: conservation implications of new taxa and significant genetic sub-structure in Australian smelts (Pisces: Retropinnidae). Marine and Freshwater Research 58, 327-341.
| Crossref | Google Scholar |
Hammer MP, Bice CM, Hall A, Frears A, Watt A, Whiterod NS, Beheregaray LB, Harris JO, Zampatti BP (2013a) Freshwater fish conservation in the face of critical water shortages in the southern Murray–Darling Basin, Australia. Marine and Freshwater Research 64, 807-821.
| Crossref | Google Scholar |
Hammer MP, Unmack PJ, Adams M, Raadik TA, Johnson JB (2014) A multigene molecular assessment of cryptic biodiversity in the iconic freshwater blackfishes (Teleostei: Percichthyidae: Gadopsis) of south-eastern Australia. Biological Journal of the Linnean Society 111, 521-540.
| Crossref | Google Scholar |
Hammer MP, Allen GR, Martin KC, Adams M, Ebner BC, Raadik TA, Unmack PJ (2018) Revision of the Australian wet tropics endemic rainbowfish genus Cairnsichthys (Atheriniformes: Melanotaeniidae), with description of a new species. Zootaxa 4413, 271-294.
| Crossref | Google Scholar |
Hammer MP, Allen GR, Martin KC, Adams M, Unmack PJ (2019a) Two new species of dwarf rainbowfishes (Atheriniformes: Melanotaeniidae) from northern Australia and southern New Guinea. Zootaxa 4701, 201-234.
| Crossref | Google Scholar |
Hammer MP, Adams M, Thacker CE, Johnson JB, Unmack PJ (2019b) Comparison of genetic structure in co-occurring freshwater eleotrids (Actinopterygii: Philypnodon) reveals cryptic species, likely translocation and regional conservation hotspots. Molecular Phylogenetics and Evolution 139, 106556.
| Crossref | Google Scholar |
Hammer MP, Taillebois L, King AJ, Crook DA, Wedd D, Adams M, Unmack PJ, Hoese DF, Bertozzi T (2021a) Unravelling the taxonomy and identification of a problematic group of benthic fishes from tropical rivers (Gobiidae: Glossogobius). Journal of Fish Biology 99, 87-100.
| Crossref | Google Scholar |
Hammer MP, Adams M, Unmack PJ, Hassell KL, Bertozzi T (2021b) Surprising Pseudogobius: molecular systematics of benthic gobies reveals new insights into estuarine biodiversity (Teleostei: Gobiiformes). Molecular Phylogenetics and Evolution 160, 107140.
| Crossref | Google Scholar |
Hardie SA, Jackson JE, Barmuta LA, White RWG (2006) Status of galaxiid fishes in Tasmania, Australia: conservation listings, threats and management issues. Aquatic Conservation: Marine and Freshwater Ecosystems 16, 235-250.
| Crossref | Google Scholar |
Harvey MS, Rix MG, Framenau VW, Hamilton ZR, Johnson MS, Teale RJ, Humphreys G, Humphreys WF (2011) Protecting the innocent: studying short-range endemic taxa enhances conservation outcomes. Invertebrate Systematics 25, 1-10.
| Crossref | Google Scholar |
Hoese DF, Hammer MP (2021) A review of the Glossogobius giuris complex in Australia, with wider discussion on nomenclature and possible synonymies. Zootaxa 4974, 79-115.
| Crossref | Google Scholar |
Humphries P (1990) Morphological variation in diadromous and landlocked populations of the spotted galaxias, Galaxias truttaceus, in Tasmania, south-eastern Australia. Environmental Biology of Fishes 27, 97-105.
| Crossref | Google Scholar |
Humphries P (2009) Wilhelm Blandowski’s contribution to ichthyology of the Murray–Darling Basin, Australia. Proceedings of the Royal Society of Victoria 121, 90-108.
| Crossref | Google Scholar |
Jerry DR (2008) Phylogeography of the freshwater catfish Tandanus tandanus (Plotosidae): a model species to understand evolution of the eastern Australian freshwater fish fauna. Marine and Freshwater Research 59, 351-360.
| Crossref | Google Scholar |
Jusaitis M, Adams M (2005) Conservation implications of clonality and limited sexual reproduction in the endangered shrub Acanthocladium dockeri (Asteraceae). Australian Journal of Botany 53, 535-544.
| Crossref | Google Scholar |
Larson HK, Hammer MP (2021) A revision of the gobiid fish genus Pseudogobius (Teleostei, Gobiidae, Tridentigerinae), with description of seven new species from Australia and South-east Asia. Zootaxa 4961, 1-85.
| Crossref | Google Scholar |
Le Vin AL, Adam A, Tedder A, Arnold KE, Mable BK (2011) Validation of swabs as a non-destructive and relatively non-invasive DNA sampling method in fish. Molecular Ecology Resources 11, 107-109.
| Crossref | Google Scholar |
Leaché AD, Oaks JR (2017) The utility of single nucleotide polymorphism (SNP) data in phylogenetics. Annual Review of Ecology, Evolution, and Systematics 48, 69-84.
| Crossref | Google Scholar |
Leis JM, Gomon MF, Larson HK (2007) Australian fish taxonomists – an endangered species. AMSA Bulletin 176, 32-33.
| Google Scholar |
Lintermans M (2004) Human-assisted dispersal of alien freshwater fish in Australia. New Zealand Journal of Marine and Freshwater Research 38, 481-501.
| Crossref | Google Scholar |
Lintermans M, Geyle HM, Beatty S, Brown C, Ebner BC, Freeman R, Hammer MP, Humphreys WF, Kennard MJ, Kern P, Martin K, Morgan DL, Raadik TA, Unmack PJ, Wager R, Woinarski JCZ, Garnett ST (2020) Big trouble for little fish: identifying Australian freshwater fishes in imminent risk of extinction. Pacific Conservation Biology 26, 365-377.
| Crossref | Google Scholar |
Lundberg JG, Kottelat M, Smith GR, Stiassny MLJ, Gill AC (2000) So many fishes, so little time: an overview of recent ichthyological discovery in continental waters. Annals of the Missouri Botanical Garden 87, 26-62.
| Crossref | Google Scholar |
McDowall RM, Frankenberg RS (1981) The galaxiid fishes of Australia (Pisces: Galaxiidae). Records of the Australian Museum 33, 443-605.
| Crossref | Google Scholar |
McFarlane SE, Pemberton JM (2019) Detecting the true extent of introgression during anthropogenic hybridization. Trends in Ecology & Evolution 34, 315-326.
| Crossref | Google Scholar |
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In ‘Proceedings of the Gateway Computing Environments Workshop (GCE)’, 14 November 2010, New Orleans, LA, USA. INSPEC Accession Number 11705685. (IEEE) doi:10.1109/GCE.2010.5676129
Morgan DL, Unmack PJ, Beatty SJ, Ebner BC, Allen MG, Keleher JJ, Donaldson JA, Murphy J (2014) An overview of the ‘freshwater fishes’ of Western Australia. Journal of the Royal Society of Western Australia 97, 263-278.
| Google Scholar |
Morgan DL, Beatty SJ, Close PG, Allen MG, Unmack PJ, Hammer MP, Adams M (2016) Resolving the taxonomy, range and ecology of biogeographically isolated and critically endangered populations of an Australian freshwater galaxiid, Galaxias truttaceus. Pacific Conservation Biology 22, 350-359.
| Crossref | Google Scholar |
Mossop KD, Adams M, Unmack PJ, Smith Date KL, Wong BBM, Chapple DG (2015) Dispersal in the desert: ephemeral water drives connectivity and phylogeography of an arid-adapted fish. Journal of Biogeography 42, 2374-2388.
| Crossref | Google Scholar |
Nielsen JL, Tupper D, Thomas WK (1994) Mitochondrial DNA polymorphism in unique runs of Chinook Salmon (Oncorhynchus tshawytscha) from the Sacramento–San Joaquin River Basin. Conservation Biology 8, 882-884.
| Crossref | Google Scholar |
Ovenden JR, White RW (1990) Mitochondrial and allozyme genetics of incipient speciation in a landlocked population of Galaxias truttaceus (Pisces: Galaxiidae). Genetics 124, 701-716.
| Crossref | Google Scholar |
Ovenden JR, White RWG, Adams M (1993) Mitochondrial and allozyme genetics of two Tasmanian galaxiids (Galaxias auratus and G. tanycephalus, Pisces: Galaxiidae) with restricted lacustrine distributions. Heredity 70, 223-230.
| Crossref | Google Scholar |
Page TJ, Stevens MI, Adams M, Foster R, Velasco-Castrillón A, Humphreys WF (2018) Multiple molecular markers reinforce the systematic framework of unique Australian cave fishes (Milyeringa: Gobioidei). Australian Journal of Zoology 66, 115-127.
| Crossref | Google Scholar |
Raadik TA (2014) Fifteen from one: a revision of the Galaxias olidus Günther, 1866 complex (Teleostei, Galaxiidae) in south-eastern Australia recognises three previously described taxa and describes 12 new species. Zootaxa 3898, 1-198.
| Crossref | Google Scholar |
Raadik TA (2019a) Upland Galaxias: Galaxias sp. nov. ‘Hunter’. In ‘The IUCN Red List of Threatened Species 2019’. e.T128972888A128972891. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/128972888/128972891 [Verified 13 July 2023]
Raadik T (2019b) ‘Moroka’ Galaxias: Galaxias sp. nov. ‘Moroka’. In ‘The IUCN Red List of Threatened Species 2019’. e.T128972928A128972932. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/128972928/128972932 [Verified 13 July 2023]
Raadik T (2019c) Galaxias sp. nov. ‘Morwell’. In ‘The IUCN Red List of Threatened Species 2019’. e.T128972916A128972919. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/128972916/128972919 [Verified 13 July 2023]
Raadik T (2019d) ‘Yalmy’ Galaxias: Galaxias sp. nov. ‘Yalmy’. In ‘The IUCN Red List of Threatened Species 2019’. e.T128972900A128972908. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/128972900/128972908 [Verified 13 July 2023]
Reid AJ, Carlson AK, Creed IF, Eliason EJ, Gell PA, Johnson PTJ, Kidd KA, MacCormack TJ, Olden JD, Ormerod SJ, Smol JP, Taylor WW, Tockner K, Vermaire JC, Dudgeon D, Cooke SJ (2019) Emerging threats and persistent conservation challenges for freshwater biodiversity. Biological Reviews 94, 849-873.
| Crossref | Google Scholar |
Schmidt DJ, Crook DA, Macdonald JI, Huey JA, Zampatti BP, Chilcott S, Raadik TA, Hughes JM (2014) Migration history and stock structure of two putatively diadromous teleost fishes, as determined by genetic and otolith chemistry analyses. Freshwater Science 33, 193-206.
| Crossref | Google Scholar |
Shelley JJ, Delaval A, Le Feuvre MC (2017) A revision of the grunter genus Syncomistes (Teleostei, Terapontidae, Syncomistes) with descriptions of seven new species from the Kimberley region, northwestern Australia. Zootaxa 4367, 1-103.
| Crossref | Google Scholar |
Shelley JJ, Swearer SE, Adams M, Dempster T, Le Feuvre MC, Hammer MP, Unmack PJ (2018) Cryptic biodiversity in the freshwater fishes of the Kimberley endemism hotspot, northwestern Australia. Molecular Phylogenetics and Evolution 127, 843-858.
| Crossref | Google Scholar |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313.
| Crossref | Google Scholar |
Thacker CE, Geiger DL, Unmack PJ (2022) Species delineation and systematics of a hemiclonal hybrid complex in Australian freshwaters (Gobiiformes: Gobioidei: Eleotridae: Hypseleotris). Royal Society Open Science 9, 220201.
| Crossref | Google Scholar |
Unmack PJ (2016) Update on saving running river rainbowfish. Fishes of Sahul 30, 1025-1032.
| Google Scholar |
Unmack PJ, Hammer MP, Adams M, Dowling TE (2011) A phylogenetic analysis of pygmy perches (Teleostei: Percichthyidae) with an assessment of the major historical influences on aquatic biogeography in southern Australia. Systematic Biology 60, 797-812.
| Crossref | Google Scholar |
Unmack PJ, Bagley JC, Adams M, Hammer MP, Johnson JB (2012) Molecular phylogeny and phylogeography of the Australian freshwater fish genus Galaxiella, with an emphasis on dwarf galaxias (G. pusilla). PLoS ONE 7, e38433.
| Crossref | Google Scholar |
Unmack PJ, Hammer MP, Adams M, Johnson JB, Dowling TE (2013) The role of continental shelf width in determining freshwater phylogeographic patterns in south-eastern Australian pygmy perches (Teleostei: Percichthyidae). Molecular Ecology 22, 1683-1699.
| Crossref | Google Scholar |
Unmack PJ, Sandoval-Castillo J, Hammer MP, Adams M, Raadik TA, Beheregaray LB (2017) Genome-wide SNPs resolve a key conflict between sequence and allozyme data to confirm another threatened candidate species of river blackfishes (Teleostei: Percichthyidae: Gadopsis). Molecular Phylogenetics and Evolution 109, 415-420.
| Crossref | Google Scholar |
Unmack PJ, Adams M, Bylemans J, Hardy CM, Hammer MP, Georges A (2019) Perspectives on the clonal persistence of presumed ‘ghost’ genomes in unisexual or allopolyploid taxa arising via hybridization. Scientific Reports 9, 4730.
| Crossref | Google Scholar |
Unmack PJ, Adams M, Hammer MP, Johnson JB, Gruber B, Gilles A, Young M, Georges A (2022) Plotting for change: an analytical framework to aid decisions on which lineages are candidate species in phylogenomic species discovery. Biological Journal of the Linnean Society 135, 117-137.
| Crossref | Google Scholar |
Verspoor E, Hammart J (1991) Introgressive hybridization in fishes: the biochemical evidence. Journal of Fish Biology 39(Suppl. A), 309-334.
| Crossref | Google Scholar |
Waters JM, Wallis GP (2001) Cladogenesis and loss of the marine life-history phase in freshwater galaxiid fishes (Osmeriformes: Galaxiidae). Evolution 55, 587-597.
| Crossref | Google Scholar |
Waters JM, White RWG (1997) Molecular phylogeny and biogeography of the Tasmanian and New Zealand mudfishes (Salmoniformes: Galaxiidae). Australian Journal of Zoology 45, 39-48.
| Crossref | Google Scholar |
Waters JM, Shirley M, Closs GP (2002) Hydroelectric development and translocation of Galaxias brevipinnis: a cloud at the end of the tunnel? Canadian Journal of Fisheries and Aquatic Sciences 59, 49-56.
| Crossref | Google Scholar |
Waters JM, Rowe DL, Burridge CP, Wallis GP (2010) Gene trees versus species trees: reassessing life-history evolution in a freshwater fish radiation. Systematic Biology 59, 504-517.
| Crossref | Google Scholar |
Welsh SA, Jerry DR, Burrows DW (2014) A new species of freshwater eel-tailed catfish of the genus Tandanus (Teleostei: Plotosidae) from the wet tropics region of eastern Australia. Copeia 2014, 136-142.
| Crossref | Google Scholar |
Welsh SA, Jerry DR, Burrows DW, Rourke ML (2017) A new species of freshwater eel-tailed catfish of the genus Tandanus (Teleostei: Plotosidae) from coastal rivers of mid-northern New South Wales, Australia. Copeia 105, 229-236.
| Crossref | Google Scholar |
Whiterod NS, Hammer MP, Barnes TC, Tucker M, Adams M, Raadik TA (2020) Clear as mud: the ecology and conservation of a secretive wetland fish (Neochanna cleaveri: Galaxiidae) in a heavily altered landscape. Wetlands Ecology and Management 28, 779-795.
| Crossref | Google Scholar |
World Wildlife Fund (2020) The World’s forgotten fishes. (WWF International) Available at https://www.wwf.ch/sites/default/files/doc-2021-02/World%27s%20Forgotten%20Fishes%20%28REPORT%20FINAL%29.pdf
Zattara EE, Premoli AC (2005) Genetic structuring in Andean landlocked populations of Galaxias maculatus: effects of biogeographic history. Journal of Biogeography 32, 5-14.
| Crossref | Google Scholar |


