Nuclear trafficking dynamics of Bromodomain-containing protein 7 (BRD7), a switch/sucrose non-fermentable (SWI/SNF) chromatin remodelling complex subunit, in porcine oocytes and cleavage-stage embryos
Jennifer S. Crodian A * , Bethany M. Weldon A * , Yu-Chun Tseng A , Birgit Cabot A and Ryan Cabot A B
A B
A Department of Animal Sciences, Purdue University, 270 South Russell Street, West Lafayette, IN, 47907, USA.
B Corresponding author. Email: rcabot@purdue.edu
Reproduction, Fertility and Development 31(9) 1497-1506 https://doi.org/10.1071/RD19030
Submitted: 22 January 2019 Accepted: 30 March 2019 Published: 13 May 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
In the work presented here, we investigated how bromodomain-containing protein 7 (BRD7), a subunit associated with switch/sucrose non-fermentable (SWI/SNF) chromatin remodelling complexes, is trafficked between cellular compartments during embryo development. SWI/SNF complexes are multi-subunit complexes that contain a core catalytic subunit (SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily A, member 4, or member 2; SMARCA4 or SMARCA2) and a collection of additional subunits that guide the complexes to their appropriate loci; BRD7 is one of these additional subunits. We hypothesised that BRD7 is exported from the nuclei of porcine oocytes and embryos in a Chromosome Region Maintenance 1 (CRM1)-dependent manner and imported into the nuclei using the karyopherin α/β1 heterodimer. Porcine oocytes and embryos were treated with inhibitors of CRM1-mediated nuclear export and karyopherin α/β1-mediated nuclear import to test this hypothesis. An RNA interference assay and a dominant negative overexpression assay were also performed to determine if karyopherin α7 serves a specific role in BRD7 trafficking. Our findings indicate that BRD7 shuttles between nuclear and cytoplasmic compartments during cleavage development. The shuttling of BRD7 indicates that it serves a unique role in remodelling chromatin during this developmental window.
Additional keywords: epigenetic, importin, karyopherin, trafficking, SWI/SNF.
Introduction
Dynamic changes in chromatin structure, which can impact developmental competence, occur during the first week of mammalian embryo development. These early changes include remodelling of epigenetic marks, which include heritable covalent and non-covalent modifications to DNA that impact gene expression. Changes in chromatin structure that impact transcription must be tightly regulated to ensure successful embryo development.
Epigenetic marks accumulated during germ cell development are remodelled during the cleavage stage to prepare for further development (Sasaki and Matsui 2008). Both covalent and non-covalent chromatin modifications take place during early embryo development. An example of a covalent epigenetic modification is the methylation of either DNA or histone proteins. DNA can be methylated at the cytosine-5 position. DNA methylation is dramatically remodelled on a global level during cleavage development and is required for successful embryo development (Li et al. 1992).
Non-covalent chromatin modifications involve repositioning and restructuring nucleosomes (Bartholomew 2014). Chromatin remodelling machinery utilises the energy released from ATP hydrolysis to reposition nucleosomes to facilitate activation or repression of transcription. The switch/sucrose non-fermentable (SWI/SNF) chromatin remodelling complexes are one group of ATP-dependent chromatin remodelling complexes; they are utilised extensively during embryonic development in mammalian embryos (Bultman et al. 2006).
The large, multi-subunit SWI/SNF complexes contain a catalytic subunit, either SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily A, member 4 (SMARCA4; also known as Brahma-related gene 1, BRG1) or SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily A, member 2 (SMARCA2; also known as Brahma, BRM); these catalytic subunits have ATPase activity (Clapier and Cairns 2009). A functional SWI/SNF chromatin remodelling complex contains a collection of BRG1-associated factors (BAFs) in addition to the catalytic subunit. The specific combination of BAFs that associate with the catalytic subunit creates unique SWI/SNF complexes that act at discrete loci within the genome (Clapier and Cairns 2009).
SWI/SNF chromatin remodelling complexes participate in a myriad of cellular processes. It has been shown that SWI/SNF-mediated chromatin remodelling is necessary for mammalian embryo development. When SMARCA4 is knocked out in murine embryos, they fail to develop past the blastocyst stage (Bultman et al. 2000). When SMARCA2 is knocked out in embryos, the embryos develop to full term but display a mild overgrowth phenotype as adults (Reyes et al. 1998). SMARCA4 also appears to be required for zygotic genome activation and trophectoderm development (Bultman et al. 2006; Wang et al. 2010). In addition to these two catalytic subunits, SWI/SNF complexes contain a core group of subunits that include BAF155, BAF170 and SMARCB1 (SNF5). These BAFs are also required for development (Euskirchen et al. 2012).
Each BAF subunit has a unique role within a given SWI/SNF chromatin remodelling complex. The subunits that make up the functional core of the SWI/SNF complex are essential for full remodelling activity; downregulation of these subunits results in defects in transcription (Phelan et al. 1999). The AT-rich interacting domain (ARID) family of BAF subunits are utilised for DNA binding (Wilsker et al. 2005) and are needed for appropriate cell cycle control (Nagl et al. 2005). Bromodomain-containing domain 7 (BRD7) is a polybromo-associated factor (PBAF) that is found in both undifferentiated embryonic stem cells and differentiated cells (Kaeser et al. 2008). In embryonic stem cells, BRD7 and ARID1A work to control the expression of SWI/SNF target genes (Kaeser et al. 2008). BRD7 is required for successful embryo development; studies involving a BRD7 knockout mouse model revealed that BRD7-null embryos die by mid-gestation (Kim et al. 2016).
Previous work from our group has shown that BRD7 undergoes a change in intracellular distribution during progression in development from immature, germinal vesicle (GV)-stage oocyte to blastocyst-stage embryo (Cabot et al. 2017). Approximately 50% of GV-stage oocytes presented a predominately cytoplasmic localisation of BRD7, with the other 50% possessing a nuclear enrichment of BRD7. Pronuclear-stage embryos displayed similar findings with half of the pronuclear-stage embryos examined presenting BRD7 restricted to the cytoplasm, whereas the other 50% of oocytes presented BRD7 in both nuclear and cytoplasmic compartments. BRD7 was found to be predominantly cytoplasmic in embryos at later cleavage stages (4-cell and blastocyst stages of development). The variable pattern of BRD7 intracellular localisation suggests that BRD7 may shuttle dynamically between nuclear and cytoplasmic compartments, allowing BRD7-containing SWI/SNF complexes to exist transiently.
Movement of intracellular proteins between the nucleus and cytoplasm is mediated by a host of transport factors. Chromosome Region Maintenance 1 (CRM1) is a nuclear export factor that functions to carry proteins that possess a nuclear export signal (NES) out of the nucleus. The NESs recognised by CRM1 are part of the primary amino acid sequence in proteins and are typically short (~10 amino acids) and rich in leucine residues (Fukuda et al. 1997). CRM1-mediated nuclear export occurs in GV-stage porcine oocytes and throughout cleavage development, but CRM1-mediated export is not required for development until the 4-cell stage of development (Cabot et al. 2002).
One of the most documented transport pathways is nuclear import mediated by the karyopherin α/β1 heterodimer (also referred to as importin α/β; Görlich et al. 1996). Karyopherin α functions as an adaptor and binds to proteins that contain a nuclear localisation signal (NLS). Classical NLSs typically consist of short stretches of basic amino acids, typically lysine residues. These NLSs exist in either monopartite or bipartite arrangements. The former is exemplified by the NLSs found in the SV40 large T antigen (PKKKRKV), whereas the latter is characterised by two small clusters of basic amino acids separated by 10 amino acids. The NLS from the protein nucleoplasmin (PAATKKAGQAKKK) is an example of a bipartite NLS (Dingwall and Laskey 1991). Once karyopherin α binds to the NLS region of the cargo, it forms a trimeric complex with karyopherin β1. The karyopherin α/β1–cargo trimeric complex then moves through the nuclear pore complex (NPC) by interactions with the NPC proteins. Once the trimeric complex enters the nucleus, karyopherin β1 binds to the guanosine triphosphate (GTP)-bound form of the GTPase Ran; the binding of Ran-GTP causes the complex to disassociate (Ribbeck et al. 1998). The karyopherin α and β1 proteins are exported back to the cytoplasm where they will bind another cargo containing an NLS signal.
Because intracellular proteins are trafficked to cellular compartments by various transport receptors, we wanted to determine if CRM1-mediated nuclear export and karyopherin α/β1-mediated import were involved in transporting BRD7 within porcine oocytes and cleavage-stage embryos. We hypothesised that BRD7 is exported from the nuclei of porcine oocytes and embryos in a CRM1-dependent manner and imported using the karyopherin α/β1 heterodimer. To test this hypothesis, GV-stage porcine oocytes and 4-cell-stage porcine embryos were incubated with leptomycin B (LMB), an inhibitor of CRM1-mediated nuclear export and with ivermectin (IVER), an inhibitor of karyopherin α/β1-mediated import. In addition, we selectively blocked karyopherin α/β1-mediated import mediated by a specific karyopherin α subtype, KPNA7, which has been shown to be exclusively expressed in mammalian oocytes and early cleavage-stage embryos.
Materials and methods
All chemicals were obtained from Sigma-Aldrich unless stated otherwise.
Porcine oocyte collection
Ovaries from prepubertal gilts were collected from a local meat processing facility, rinsed in a saline solution containing 75 μg mL−1 penicillin and 50 μg mL−1 streptomycin at 37°C and transported in an insulated container to the laboratory for processing. Antral ovarian follicles were manually aspirated with a sterile 18-gauge needle and 10-mL syringe; follicular fluid was pooled and held at 39°C. Cumulus–oocyte complexes (COCs) were allowed to settle by gravity, follicular fluid was discarded and COCs were resuspended in a 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES)-buffered medium containing 0.01% polyvinyl alcohol (PVA; Abeydeera et al. 1998). Cumulus–oocyte complexes with multiple layers of intact cumulus cells were selected for experiments. Cumulus cells were removed from selected COCs immediately after selection for immunocytochemical staining assays involving GV-stage oocytes.
In vitro maturation
For experiments involving RNA microinjection (green fluorescent protein, GFP, or dominant negative karyopherin α7, KPNA7) and KPNA7 small interfering RNA (siRNA) injection into porcine embryos, oocytes were first matured in groups of 50–100 COCs in 500 µL of Medium 199 containing 0.14% PVA, 10 ng mL−1 epidermal growth factor, 0.57 mM cysteine, 0.5 IU mL−1 FSH, 0.5 IU mL−1 LH, 20 ng mL−1 leukaemia inhibitory factor (LIF; MilliporeSigma), 20 ng mL−1 insulin-like growth factor 1 (IGF1; Prospec Protein Specialists) and 40 ng mL−1 fibroblast growth factor 2 (FGF2; PeproTech) as previously described (Yuan et al. 2017). The COCs were matured for 42–44 h in a humidified incubator (100% humidity) at 39°C with 5% CO2 in air. Cumulus cells were removed from matured COCs by vortexing in 0.1% hyaluronidase in a HEPES-buffered medium containing 0.01% PVA for 4 min. For experiments involving embryos incubated with leptomycin B and ivermectin, groups of 50–100 COCs were placed in 500 µL of Medium 199 containing 0.14% PVA, 10 ng mL−1 epidermal growth factor, 0.57 mM cysteine, 0.5 IU mL−1 FSH and 0.5 IU mL−1 LH as previously described (Abeydeera et al. 1998). The COCs were matured for 42–44 h in a humidified incubator (100% humidity) at 39°C with 5% CO2 in air. Cumulus cells were removed from matured COCs by vortexing in 0.1% hyaluronidase in a HEPES-buffered medium containing 0.01% PVA for 4 min.
In vitro fertilisation and embryo culture
Denuded oocytes were transferred to a modified Tris-buffered medium (mTBM) and fertilised as previously described (Abeydeera and Day 1997). Fresh semen was collected from a mature boar with proven fertility housed at Purdue’s Animal Science Research and Educational Centre, extended with a commercial semen extender (EnduraGuard Plus; Mofa Global) and stored at 17.5°C until use for in vitro fertilisation. The spermatozoa were washed three times in Dulbecco’s phosphate-buffered saline (PBS) and then resuspended in mTBM. The sperm suspension was added to droplets containing 15–30 mature oocytes such that the final concentration of spermatozoa during gamete co-incubation was 5 × 105 spermatozoa mL−1. The gametes were allowed to co-incubate for 5 h and were then transferred to porcine zygotic medium 3 (PZM3), an embryo culture medium (Yoshioka et al. 2002) supplemented with 3 mg mL−1 bovine serum albumin (BSA).
Leptomycin B culture experiments
Leptomycin B (LMB) was added to in vitro maturation medium (7 nM LMB final concentration in ethanol; LMB treatment) and denuded GV-stage oocytes were incubated for 12 h. As a control, ethanol was added to in vitro maturation medium (1 μL ethanol per 500 μL maturation medium; 0.2% ethanol, ETOH treatment) and denuded GV-stage oocytes were incubated for 12 h. Following the 12-h incubation, all cells were fixed in 3.7% paraformaldehyde for 1 h, subjected to an immunocytochemical staining protocol and evaluated using confocal microscopy (Nikon A1R-MP; Nikon).
For experiments involving porcine embryos, oocytes were matured and fertilised in vitro; embryos were placed in embryo culture medium for 48 h. After 48 h, 4-cell-stage embryos were moved to embryo culture medium contained either 7 nm LMB (LMB treatment) or 0.2% ethanol (ETOH treatment) for 12 h. Following treatment, 4-cell-stage embryos were fixed in 3.7% paraformaldehyde, subjected to an immunocytochemical staining protocol and evaluated using confocal microscopy. To determine the quality of the embryos produced in each biological replicate, a group of LMB and ETOH embryos was also cultured to Day 6 after gamete co-incubation. Replicates in which control (ETOH-treated) embryos failed to yield blastocyst-stage embryos by Day 6 after fertilisation were excluded from our analysis.
Ivermectin culture experiments
Porcine GV-stage oocytes and 4-cell-stage embryos were cultured with 500 μM ivermectin (Alfa Aesar) to determine if the karyopherin α/β1 import pathway controlled the trafficking of BRD7 at these developmental stages. GV-stage oocytes were added to maturation medium that contained either 500 µM ivermectin in dimethyl sulphoxide (DMSO; IVER treatment) or a control group that contained 4.0% DMSO (DMSO treatment). Following 12 h of incubation, all cells were fixed in 3.7% paraformaldehyde for 1 h, subjected to an immunocytochemical staining protocol and evaluated using confocal microscopy. For experiments involving porcine embryos, oocytes were matured and fertilised in vitro; immediately after gamete co-incubation the embryos were moved to embryo culture medium for 48 h. After 48 h, 4-cell-stage embryos were moved to embryo culture medium containing either 500 µM ivermectin (IVER) or 4.0% dimethyl sulfoxide (DMSO). After 12 h of incubation, 4-cell-stage embryos were fixed in 3.7% paraformaldehyde, subjected to an immunocytochemical staining protocol and evaluated using confocal microscopy. To determine the quality of the embryos produced in each biological replicate, a group of IVER and DMSO embryos was also cultured to Day 6 after gamete co-incubation. Replicates in which control (DMSO) embryos failed to yield blastocyst-stage embryos by Day 6 after fertilisation were excluded from our analysis.
Ivermectin dose titration
Porcine oocytes were incubated in varying concentrations (25–500 μM) of ivermectin, an inhibitor of karyopherin α/β1-mediated import, to determine the effective concentration of ivermectin necessary to block nuclear import using a reporter protein injection assay. Briefly, Alexa 488-labelled glutathione S-transferase (GST)-NLS was injected into GV-stage porcine oocytes that were incubated for 4 h in ivermectin (or DMSO, control). Injected oocytes were cultured in oocyte maturation medium for 24 h after microinjection in the presence of the respective concentrations of ivermectin. Following culture, oocytes were fixed in 3.7% paraformaldehyde for 1 h and subsequently washed three times in PBS containing 0.1% Tween-20 (PBST); 15 min for each wash. Cells were permeabilised by incubation in PBS containing 1% Triton X-100 for 1 h; oocytes were washed three times in PBST and then stained with Hoechst 33342 (5 µg mL−1) for 20 min. Cells were then mounted on slides with 20 μL Vectashield (Vector Laboratories, Inc.) and examined by a confocal microscope (Nikon A1R-MP).
GST-NLS production
A recombinant fusion protein consisting of glutathione S-transferase (GST) and the nuclear localisation signal (NLS) from the SV40 T-antigen (GST-NLS) was produced in BL21 cells as described previously (Cabot and Prather 2003). Briefly, transfected bacteria were grown in liquid culture medium at 37°C to an optical density between 0.5 and 0.8 at 600 nm; protein expression was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12 h at 18°C. Liquid cultures were cooled to 4°C following induction; bacteria were pelleted by centrifugation at 10 000g at 4°C for 15 min. Bacterial pellets were resuspended in 20 mL PBS containing 0.3% Triton X-100 and protease inhibitors (Complete Protease Inhibitor Cocktail; Roche) and lysed by sonication. Bacterial lysates were centrifuged at 12 000g for 15 min at 4°C; supernatants were then incubated with a glutathione–agarose column at 4°C for 2 h. The column was then washed four times with PBS containing 1% Triton X-100, proteins were eluted with 10 mM reduced glutathione and eluted proteins were labelled with Alexa488, quantified with the Bradford assay and stored at −80°C until use.
Dominant negative KPNA7
A dominant negative KPNA7 (DN-KPNA7) construct was generated by deleting the 5′ end of the porcine KPNA7 open reading frame (Wang et al. 2012; National Center for Biotechnology Information (NCBI) reference sequence NM_001163411.1), which encodes the import β binding (IBB) domain. A forward oligonucleotide primer (5′-CACCAGGAAAATGCTGTCCCAGGAgGAGG) and reverse oligonucleotide primer (5′- GCCAGCGTCTTCTTCCTCACTGAAGT) were used to amplify the remaining open reading frame of KPNA7 downstream of the IBB domain by polymerase chain reaction (PCR). The PCR product was cloned into the Gateway-enabled entry vector, pENTR/s.d./D-TOPO (Invitrogen), sequenced to verify its identity and recombined into the Gateway pcDNA-pDEST53 destination vector (Invitrogen) to yield an N-terminal GFP fusion protein. This vector was linearised by restriction enzyme digestion with StuI and used as the template for in vitro mRNA synthesis using the mMessage mMachine kit (Thermo-Fisher) following the manufacturer’s directions. Aliquots of purified in vitro-transcribed mRNA (750 μg mL−1) were stored at −80°C until use.
RNA interference-mediated knockdown of KPNA7
Interfering RNAs were used to knockdown the KPNA7 transcript as previously described (Wang et al. 2012). Briefly, custom-made Stealth siRNAs targeting porcine KPNA7 (5′-CAUGAAUGCUUAACGCCCUUAACAA and 5′-UUGUUAAGGGCGUUAAGCAUUCAUG) and control siRNAs (5′-CAUCGUAAAUUCCGCAUUCAAGCAA and 5′-UUGCUUGAAUGCGGAAUUUACGAUG) were synthesised by a commercial vendor (Invitrogen). Duplexed RNAs were diluted in RNase-free water to a concentration of 20 µM; aliquots were frozen at −20°C. Aliquots of interfering RNAs were diluted to 1 µM in RNase-free water immediately before microinjection.
Microinjection
Spermatozoa were removed from presumptive zygotes by vortexing embryos in 0.1% hyaluronidase in HEPES-buffered medium for 4 min after gamete co-incubation. For mRNA injections, presumptive zygotes were then divided into three treatments: DN-KPNA7 mRNA injection, GFP mRNA injection and non-injected controls. Injection pipettes were loaded with 5 μL of mRNA; a Femtojet microinjector (Eppendorf) was used to perform microinjection. Embryos that lysed immediately after microinjection were discarded and excluded from analysis. After injection, surviving embryos were placed in PZM3 embryo culture medium and cultured for 46 h. Cleaved embryos in all treatment groups were fixed in 3.7% paraformaldehyde and processed to detect BRD7 immunocytochemically.
For siRNA experiments, presumptive zygotes were assigned to one of three treatments: KPNA7 siRNA injected, nonsense siRNA injected and non-injected controls. Injection pipettes were loaded with 5 μL of 1 μM siRNA; a Femtojet microinjector was used to perform microinjection. Embryos that lysed immediately after microinjection were discarded and excluded from analysis. After injection, surviving embryos were placed in PZM3 embryo culture medium and cultured for 46 h. Cleaved embryos in all treatment groups were fixed in 3.7% paraformaldehyde and processed to detect BRD7 immunocytochemically.
Immunocytochemical analysis
Oocytes and embryos were processed according to an established immunocytochemical staining protocol (Cabot et al. 2017). Briefly, oocytes and embryos were fixed in 3.7% paraformaldehyde for 1 h at 4°C. After fixation the embryos and oocytes were washed three times in PBS containing 0.1% Tween-20 for 30 min at 4°C. Oocytes and embryos were stored in 500 µL PBST and stored for up to 3 days at 4°C until further processing. All incubations were carried out at 4°C. Fixed oocytes and embryos were permeabilised with 1% Triton X-100 in PBS for 1 h. Embryos and oocytes were then placed in blocking solution consisting of 0.1 M glycine, 1% goat serum, 0.01% Triton X-100, 1% powdered nonfat dry milk, 0.5% BSA and 0.02% sodium azide in PBS (Prather and Schatten 1992). Oocytes and embryos were probed with a commercially available antibody directed against BRD7 (Abcam) diluted 1 : 500 in PBS containing 0.1% Tween-20 for 16 h then washed three times in PBS containing 0.1% Tween-20 (30 min for each wash). Embryos and oocytes were stained with secondary antibody (fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG), which was diluted 1 : 500 in PBS containing 0.1% Tween-20 for 16 h, then washed three times in PBS containing 0.1% Tween-20 (30 min for each wash). For the dominant negative KPNA7 ectopic expression studies the secondary antibody used was tetramethylrhodamine (TRITC)-conjugated anti-rabbit IgG. Embryos and oocytes were stained with 5 µg mL−1 Hoechst 33342 in PBS for 15 min, mounted on slides using Vectashield mounting medium (Vector Laboratories, Inc.) and examined using confocal microscopy (Nikon A1R-MP; Nikon).
Quantification of intracellular distribution of BRD7 signal in oocytes and embryos
Images were taken on a confocal microscope equipped with a camera, with a max fluorescence below 4000 optical units. Photos were taken of an optical section of oocytes and embryos at 461 nm and 495 nm in order to capture Hoechst 33342 and FITC staining respectively. Analytical imaging software (Nikon Elements; Nikon Corporation) was used to analyse the images taken at the confocal microscope for the sum intensity of fluorescence of nuclear localisation in comparison to cytoplasmic localisation by using regions of interest (ROI) around each respective area. The nuclear ROI was determined based on Hoechst staining that showed the edges of the nucleus. The cytoplasmic ROI was determined based on the edge of the oocyte or blastomere. The nuclear-to-cytoplasmic ratio was calculated by dividing the sum intensity by the ROI area for the nuclear and cytoplasmic regions. The intensity was measured in terms of optical density using the Nikon Elements software and recorded as analogue to digital units (ADU).
Statistical analyses
The ratio of nuclear-to-cytoplasmic intensity was calculated on each individual oocyte or embryo in the ivermectin and leptomycin B culture experiments. A higher ratio indicated an increased nuclear localisation. The measurements of intracellular localisation of BRD7 were analysed by two-way ANOVA using R-Studio (R-Studio). Significance was determined by a two-sample t-test. A P value less than 0.05 was considered to be significant. A Chi-square test was used to detect differences pertaining to the intracellular distribution of BRD7 in both the KPNA7 RNA interference and dominant negative KPNA7 overexpression assays.
Results
Effect of LMB on BRD7 localisation in porcine oocytes
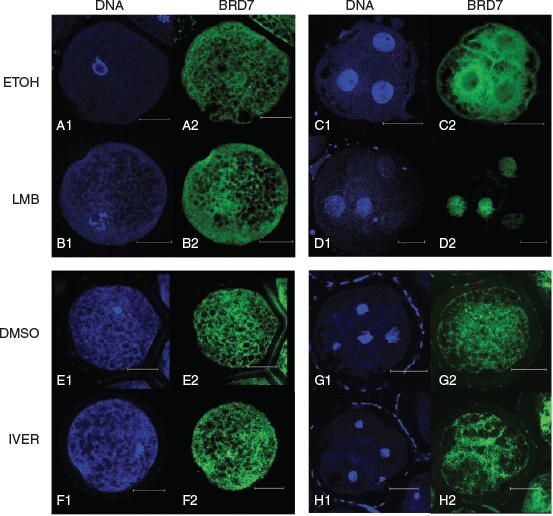
Two putative nuclear export signals (NESs) were found within porcine BRD7 (UniProt accession number I3 L640) using an online NES prediction platform sponsored by The Netherlands Cancer Institute (http://research.nki.nl/fornerodlab/NES-Finder.htm, accessed 23 April 2019). The first presumptive NES begins at amino acid 193 (N’-IEELKDNFKL) and the second NES begins at amino acid 237 (N’-IQSLKQSIDF). No significant differences between localisation patterns of BRD7 were detected in GV-stage oocytes in the respective LMB and ETOH treatment groups. BRD7 was found to have an even distribution between nuclear and cytoplasmic compartments in all oocytes examined (n = 9/9, n = 13/13 for LMB and ETOH treatments respectively). The nuclear intensity of the ETOH treatment group was 1.17 ± 0.37 ADU µm−1 and the average nuclear intensity of the LMB treatment group was 1.09 ± 0.26 ADU µm−1. Representative images are shown in Fig. 1.
|
Effect of LMB on BRD7 localisation in 4-cell-stage porcine embryos
The distribution of BRD7 was significantly different between LMB and ETOH treatment groups. Embryos in the ETOH treatment displayed two distribution patterns: either even distribution of BRD7 between the nucleus and cytoplasm (n = 8/15) or a predominantly cytoplasmic localisation (n = 7/15). Embryos in the LMB treatment group possessed a dramatic nuclear localisation of BRD7 (n = 7/7). The average nuclear intensity in the ETOH treatment group was 1.17 ± 0.29 ADU µm−1 and the average nuclear intensity in the LMB treatment group was 10.72 ± 2.82 ADU µm−1 (LMB vs ETOH, P < 0.05). Representative images are shown in Fig. 1.
Effect of ivermectin on BRD7 localisation in porcine oocytes and embryos
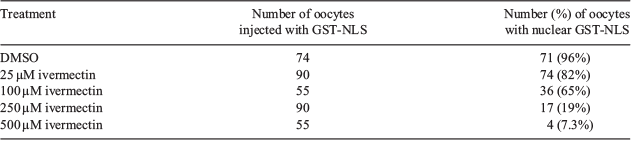
Multiple putative nuclear localisation signals (NLSs) were identified within BRD7 (UniProt accession number I3 L640) using an online NLS prediction platform (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi, accessed 23 April 2019; Table 1). A dose titration experiment was performed to identify the effective dose of ivermectin necessary to block karyopherin α/β1-mediated nuclear import. Our results show that a concentration of 500 μM ivermectin was able to block nuclear accumulation of a fluorescently labelled reporter protein in 93% of GV-stage oocytes (Table 2).

|
|
No significant differences were found between IVER- and DMSO-treated oocytes. BRD7 was found to have an even distribution between nuclear and cytoplasmic compartments in the majority of oocytes examined (n = 21/33, n = 20/25 for DMSO and IVER treatments respectively). The remaining oocytes showed a predominantly cytoplasmic localisation (n = 12/33, n = 5/25 for DMSO and IVER treatments respectively). The nuclear intensity of the DMSO treatment group was 0.95 ± 0.55 ADU µm−1 and the average nuclear intensity of the IVER treatment group was 0.88 ± 0.39 ADU µm−1. No significant differences between localisation patterns of BRD7 were detected in 4-cell-stage embryos in the respective DMSO or IVER treatment groups. BRD7 was found to have a ubiquitous staining throughout the nucleus and cytoplasm in blastomeres of all embryos examined (n = 7/7, n = 7/7 for DMSO and IVER treatments respectively). The nuclear intensity of the DMSO treatment group was 3.25 ± 1.07 ADU µm−1 and the average nuclear intensity of the IVER treatment group was 3.20 ± 1.60 ADU µm−1. Representative images are shown in Fig. 1.
RNA interference-mediated knockdown of KPNA7 restricts BRD7 nuclear localisation
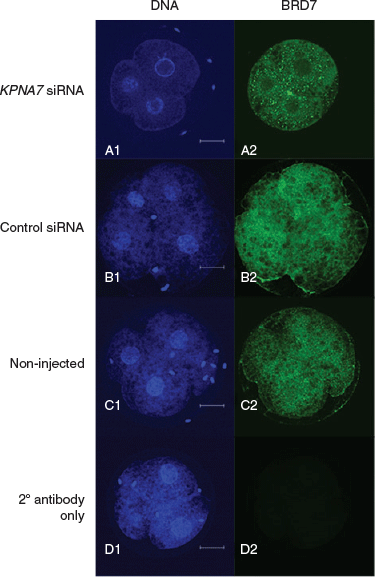
We found a significant difference in the BRD7 intracellular localisation patterns in porcine embryos 46 h after treatment with KPNA7 interfering RNAs. While the majority of embryos in the control siRNA treatment (n = 16/22) and non-injected embryo treatment (n = 11/15) revealed an even distribution of BRD7 between nuclear and cytoplasmic compartments, the majority of embryos in the KPNA7 siRNA treatment (n = 24/28) showed a reduced amount of BRD7 in the nucleus as compared with the cytoplasm (KPNA7 siRNA vs control siRNA and non-injected, P < 0.05). Representative images are shown in Fig. 2.
|
Expression of a dominant negative KPNA7 restricts BRD7 nuclear localisation
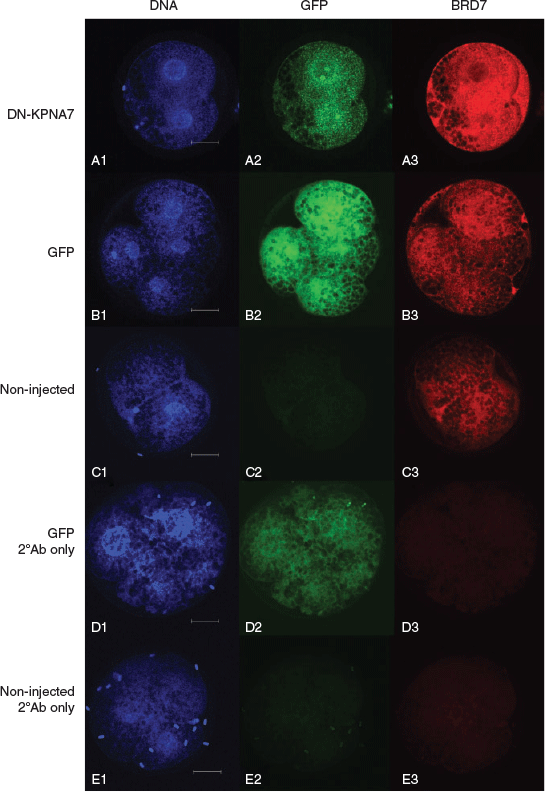
We found a significant difference in the BRD7 intracellular localisation patterns in porcine embryos 46 h after injection with mRNA encoding DN-KPNA7. All embryos in the GFP and non-injected treatment groups revealed detectable BRD7 either with equal distribution between the nuclear and cytoplasmic compartments (n = 3/8 GFP, n = 16/18 non-injected) or with greater intensity within the nucleus (n = 5/8 GFP, n = 2/18 non-injected). BRD7 staining was enriched in the cytoplasm in embryos expressing DN-KPNA7 (n = 6/17), evenly distributed between the nucleus and cytoplasm (n = 9/17) or enriched in the nucleus (n = 2/17; DN-KPNA7 vs GFP and non-injected, P < 0.05). Representative images are shown in Fig. 3.
|
Discussion
SWI/SNF chromatin remodelling complexes play an essential role during early embryogenesis, impacting both zygotic genome activation and trophectoderm differentiation (Bultman et al. 2006). The collection of BAF subunits that associate with one another determines the activity of a given SWI/SNF chromatin remodelling complex. Determining the mechanisms that regulate the intracellular localisation of discrete BAF subunits will help us to understand which SWI/SNF complexes can exist in the nucleus at specific time points during early embryogenesis.
Because the intracellular localisation of BRD7 was found to change over the course of development from immature oocyte to cleavage-stage embryo, we wanted to determine which nuclear trafficking pathways played a role in regulating the intracellular localisation of BRD7. We identified classical NLSs recognised by the karyopherin α/β1 heterodimer (Table 1) and NESs recognised by CRM1 within porcine BRD7; Zhou et al. (2011) had also reported that BRD7 contained an LMB-sensitive NES. As these two pathways have been shown to be operational in porcine oocytes and embryos (Cabot et al. 2002; Cabot and Prather 2003) we hypothesised that they were involved in trafficking BRD7.
It was interesting to find that LMB treatment led to nuclear accumulation of BRD7 in 4-cell-stage embryos but not GV-stage oocytes, as CRM1-mediated nuclear export has been shown to operate at both of these developmental stages (Cabot et al. 2002). While CRM1-mediated export can occur in porcine oocytes, this activity was ascertained by using a microinjection assay involving fluorescently labelled, recombinant protein (Cabot et al. 2002). The kinetics of this transport system have not been established in porcine cells, therefore it is possible that the activity of CRM1-mediated export differs between the GV-stage oocyte and the 4-cell-stage embryo. It is also possible for multiple intracellular proteins to associate with one another and mask nuclear localisation signals. In addition, post-translational modifications that impact protein folding may occlude localisation signals at discrete stages of development. Other members of the karyopherin β family of transport receptors also mediate intracellular trafficking. Karyopherin β2, for example, binds to the M9 domain (found in the primary amino acid sequence of intracellular cargos) and transports proteins in a bidirectional manner across the nuclear envelope (Bonifaci et al. 1997). Although we did not detect an M9 domain within BRD7, it is plausible that BRD7 utilises additional transport receptors to translocate between the nuclear and cytoplasmic compartments.
Aside from the possibilities outlined above, we speculate that the lack of nuclear accumulation of BRD7 in response to LMB treatment in porcine oocytes was due to attenuated BRD7 nuclear import at this stage of development. If BRD7 is not trafficked into the nucleus, then inhibiting its export would show no change in intracellular distribution. We first attempted to address this question with an ivermectin-based assay to block nuclear import. Ivermectin has been shown to be an effective inhibitor of karyopherin α/β1-mediated nuclear import (Wagstaff et al. 2012). Although Wagstaff et al. (2012) determined that a 25 μM concentration of ivermectin was sufficient to block karyopherin α/β1-mediated nuclear import in HeLa cells, the dose titration performed in this report (Table 2) revealed that an ivermectin concentration of 500 μM of ivermectin was necessary to inhibit karyopherin α/β1-mediated import.
The fact that no change in BRD7 intracellular localisation was observed following ivermectin treatment in either GV-stage oocytes or 4-cell-stage embryos was somewhat surprising. It is possible that ivermectin is simply not 100% effective in blocking karyopherin α/β1-mediated nuclear import in porcine oocytes and embryos at the concentration used in our study. Alternatively, although ivermectin can block karyopherin α/β1-mediated nuclear import, it is possible that ivermectin preferentially disrupts trafficking of only a subset of karyopherin α subtypes. Karyopherin α subtypes are known to be differentially expressed in various cell types and discrete developmental stages (Wang et al. 2012). We therefore chose to target a specific karyopherin α-subtype by two complementary methods to further test the hypothesis that karyopherin α/β1-mediated nuclear import regulates the intracellular localisation of BRD7. We focussed on karyopherin α7 (KPNA7), as KPNA7 appears to be an oocyte- and early embryo-specific karyopherin α subtype that is required for embryogenesis (Tejomurtula et al. 2009; Wang et al. 2012).
The dominant negative KPNA7 mutant that we generated lacks the IBB domain, which is required for the interaction between karyopherin α and karyopherin β1, and retains the armadillo (ARM) repeats, which are responsible for binding NLS-bearing cargo (Lott and Cingolani 2011). Overexpression of the dominant negative KPNA7 should be able to bind its respective cargo in the cytoplasm, but be unable to transport the cargo to the nucleus. While our results clearly show that BRD7 is redistributed in 4-cell-stage embryos as a result of the presence of dominant negative KPNA7, it is possible that the redistribution may be an indirect effect of ectopic protein overexpression. We used a previously reported RNA interference approach to reduce KPNA7 levels in porcine embryos (Wang et al. 2012). As shown in Figs 2 and 3, both approaches to modulate KPNA7 levels resulted in a redistribution of BRD7. These experiments suggest a role for KPNA7 in BRD7 import, but these experiments do not exclude the possibility that other karyopherin α subtypes also participate in BRD7 import.
We have previously shown that BRD7 is largely restricted to the cytoplasm at the 4-cell stage of development (Cabot et al. 2017). The new data presented here suggest that the steady-state localisation of BRD7 at the 4-cell stage is cytoplasmic because the rate of CRM1-mediated nuclear export exceeds the rate of karyopherin α/β1-mediated nuclear import. This apparent shuttling of BRD7 between the nucleus and cytoplasm at this stage of development was surprising and may reflect a unique role for BRD7 during the 4-cell stage of development.
Zygotic genome activation occurs during the 4-cell stage in the porcine embryo (Anderson et al. 1999). The shuttling of BRD7 at this stage may reflect a role that BRD7-containing SWI/SNF complexes serve in modifying chromatin structure in preparation for the global increase in transcription observed at the 4-cell stage. Previously published data have shown that CRM1-mediated nuclear export occurs before the 4-cell stage (Cabot et al. 2002). It is possible that BRD7-containing SWI/SNF complexes also serve a role in regulating the low level of transcription before the 4-cell stage. We hypothesise that BRD7 participates in chromatin remodelling events during the 4-cell stage of development that are distinct from those that occur in the GV-stage oocyte.
Porcine embryos used in the nuclear export assay we reported here were cultured in the presence of LMB for 12 h. This incubation time does not allow us to ascertain how rapidly (or frequently) a given SWI/SNF subunit transits between the nucleus and cytoplasm. There are some differences in localisation patterns between what we have reported here and what was previously published by (Cabot et al. 2017). For instance, we previous reported that BRD7 shows a cytoplasmic localisation in the majority of embryos at the 4-cell stage of development, whereas 47% of our control embryos had a cytoplasmic localisation of BRD7 and 53% revealed an even distribution between the nucleus and cytoplasm. This subtle difference is likely due to the different age of the 4-cell-stage embryos in the two studies. Four-cell-stage embryos were fixed 48 h after gamete mixing in the former study (Cabot et al. 2017); 4-cell-stage embryos in the present study were collected 48 h after gamete mixing and then incubated for 12 h before fixation. This added incubation puts embryos much closer to the onset of mitosis. It is plausible that the shuttling of BRD7 may serve a role in directing specific SWI/SNF complexes to genomic loci at discrete time points during the cell cycle or in remodelling chromatin to mediate aspects of zygotic genome activation.
SWI/SNF chromatin remodelling complexes are known to serve critical roles in the events of zygotic genome activation and trophectoderm formation. The findings in this work add to our understanding of how discrete SWI/SNF complex subunits are partitioned between the nucleus and cytoplasm in cleavage-stage embryos. Understanding the mechanism by which these protein subunits are partitioned within cells of the cleavage-stage embryo will help us determine the types of SWI/SNF complexes that exist at discrete time points during development. Future investigations that examine how BRD7 gains entry to the nucleus and how transcript profiles change when the intracellular localisation or the levels of BRD7 are altered will help us to determine the networks that are regulated by SWI/SNF chromatin remodelling. This new knowledge will improve our understanding of mammalian embryogenesis and provide a means to develop new and novel approaches to improve current conditions for handling embryos in vitro.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors acknowledge the use of the facilities of the Bindley Bioscience Center, a core facility of the NIH-funded Indiana Clinical and Translational Sciences Institute. We would also like to thank Indiana Packers Corporation for supplying the porcine ovaries used in this study, as well as all of the support from our laboratory members, Hayly M. Goebel, Marta Kwiatkowska and Charlie C. Zhang. We also want to thank our statistical consultant, Qi Lu, and the Purdue Statistical Consulting Service for assistance in analysing the data presented in this manuscript. This project was supported, in part, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (award number R01HD084309). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
Abeydeera, L. R., and Day, B. N. (1997). In vitro penetration of pig oocytes in a modified Tris-buffered medium: effect of BSA, caffeine and calcium. Theriogenology 48, 537–544.| In vitro penetration of pig oocytes in a modified Tris-buffered medium: effect of BSA, caffeine and calcium.Crossref | GoogleScholarGoogle Scholar | 16728149PubMed |
Abeydeera, L. R., Wang, W. H., Prather, R. S., and Day, B. N. (1998). Maturation in vitro of pig oocytes in protein-free culture media: fertilization and subsequent embryo development in vitro. Biol. Reprod. 58, 1316–1320.
| Maturation in vitro of pig oocytes in protein-free culture media: fertilization and subsequent embryo development in vitro.Crossref | GoogleScholarGoogle Scholar | 9603270PubMed |
Anderson, J. E., Matteri, R. L., Abeydeera, L. R., Day, B. N., and Prather, R. S. (1999). Cyclin B1 transcript quantitation over the maternal to zygotic transition in both in vivo and in vitro-derived 4-cell stage porcine embryos. Biol. Reprod. 61, 1460–1467.
| Cyclin B1 transcript quantitation over the maternal to zygotic transition in both in vivo and in vitro-derived 4-cell stage porcine embryos.Crossref | GoogleScholarGoogle Scholar | 10569990PubMed |
Bartholomew, B. (2014). Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 83, 671–696.
| Regulating the chromatin landscape: structural and mechanistic perspectives.Crossref | GoogleScholarGoogle Scholar | 24606138PubMed |
Bonifaci, N., Moroianu, J., Radu, A., and Blobel, G. (1997). Karyopherin 2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94, 5055–5060.
| Karyopherin 2 mediates nuclear import of a mRNA binding protein.Crossref | GoogleScholarGoogle Scholar | 9144189PubMed |
Bultman, S., Gebuhr, T. C., Yee, D., Mantia, C. L., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., and Magnuson, T. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295.
| A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes.Crossref | GoogleScholarGoogle Scholar | 11163203PubMed |
Bultman, S. J., Gebuhr, T. C., Pan, H., Svoboda, P., Schultz, R. M., and Magnuson, T. (2006). Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 20, 1744–1754.
| Maternal BRG1 regulates zygotic genome activation in the mouse.Crossref | GoogleScholarGoogle Scholar | 16818606PubMed |
Cabot, R. A., and Prather, R. S. (2003). Cleavage stage porcine embryos may have differing developmental requirements for karyopherins α2 and α3. Mol. Reprod. Dev. 64, 292–301.
| Cleavage stage porcine embryos may have differing developmental requirements for karyopherins α2 and α3.Crossref | GoogleScholarGoogle Scholar | 12548662PubMed |
Cabot, R. A., Hannink, M., and Prather, R. S. (2002). CRM1-mediated nuclear export is present during porcine embryogenesis, but is not required for early cleavage. Biol. Reprod. 67, 814–819.
| CRM1-mediated nuclear export is present during porcine embryogenesis, but is not required for early cleavage.Crossref | GoogleScholarGoogle Scholar | 12193389PubMed |
Cabot, B., Tseng, Y.-C., Crodian, J. S., and Cabot, R. (2017). Differential expression of key subunits of SWI/SNF chromatin remodeling complexes in porcine embryos derived in vitro or in vivo. Mol. Reprod. Dev. 84, 1238–1249.
| Differential expression of key subunits of SWI/SNF chromatin remodeling complexes in porcine embryos derived in vitro or in vivo.Crossref | GoogleScholarGoogle Scholar | 29024220PubMed |
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304.
| The biology of chromatin remodeling complexes.Crossref | GoogleScholarGoogle Scholar | 19355820PubMed |
Dingwall, C., and Laskey, R. E. (1991). Nuclear targeting sequences-a consensus? Trends Biochem. Sci. 16, 478–481.
| Nuclear targeting sequences-a consensus?Crossref | GoogleScholarGoogle Scholar | 1664152PubMed |
Euskirchen, G., Auerbach, R. K., and Snyder, M. (2012). SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287, 30897–30905.
| SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions.Crossref | GoogleScholarGoogle Scholar | 22952240PubMed |
Fukuda, M., Asano, S., Nakamura, T., Adachi, M., and Nature, Y.-M. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311.
| CRM1 is responsible for intracellular transport mediated by the nuclear export signal.Crossref | GoogleScholarGoogle Scholar | 9384386PubMed |
Görlich, D., Henklein, P., Laskey, R. A., and Hartmann, E. (1996). A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15, 1810–1817.
| A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus.Crossref | GoogleScholarGoogle Scholar | 8617226PubMed |
Kaeser, M. D., Aslanian, A., Dong, M.-Q., Yates, J., and Emerson, B. M. (2008). BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254–32263.
| BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells.Crossref | GoogleScholarGoogle Scholar | 18809673PubMed |
Kim, Y., Andrés Salazar Hernández, M., Herrema, H., Delibasi, T., and Park, S. W. (2016). The role of BRD7 in embryo development and glucose metabolism. J. Cell. Mol. Med. 20, 1561–1570.
| The role of BRD7 in embryo development and glucose metabolism.Crossref | GoogleScholarGoogle Scholar | 27444544PubMed |
Li, E., Bestor, T., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926.
| Targeted mutation of the DNA methyltransferase gene results in embryonic lethality.Crossref | GoogleScholarGoogle Scholar | 1606615PubMed |
Lott, K., and Cingolani, G. (2011). The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592.
| The importin β binding domain as a master regulator of nucleocytoplasmic transport.Crossref | GoogleScholarGoogle Scholar | 21029753PubMed |
Nagl, N. G., Patsialou, A., Haines, D. S., Dallas, P. B., Beck, G. R., and Moran, E. (2005). The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 65, 9236–9244.
| The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest.Crossref | GoogleScholarGoogle Scholar | 16230384PubMed |
Phelan, M. L., Sif, S., Narlikar, G. J., and Kingston, R. E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253.
| Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits.Crossref | GoogleScholarGoogle Scholar | 10078207PubMed |
Prather, R. S., and Schatten, G. (1992). Construction of the nuclear matrix at the transition from maternal to zygotic control of development in the mouse: an immunocytochemical study. Mol. Reprod. Dev. 32, 203–208.
| Construction of the nuclear matrix at the transition from maternal to zygotic control of development in the mouse: an immunocytochemical study.Crossref | GoogleScholarGoogle Scholar | 1497870PubMed |
Reyes, J. C., Barra, J., Muchardt, C., Camus, A., Babinet, C., and Yaniv, M. (1998). Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17, 6979–6991.
| Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α).Crossref | GoogleScholarGoogle Scholar | 9843504PubMed |
Ribbeck, K., Lipowsky, G., Kent, H. M., Stewar, M., and Gorlich, D. (1998). NTF2 mediates nuclear import of Ran. EMBO J. 17, 6587–6598.
| NTF2 mediates nuclear import of Ran.Crossref | GoogleScholarGoogle Scholar | 9822603PubMed |
Sasaki, H., and Matsui, Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140.
| Epigenetic events in mammalian germ-cell development: reprogramming and beyond.Crossref | GoogleScholarGoogle Scholar | 18197165PubMed |
Tejomurtula, J., Lee, K. B., Tripurani, S. K., Smith, G. W., and Yao, J. (2009). Role of importin alpha8, a new member of the importin alpha family of nuclear transport proteins, in early embryonic development in cattle. Biol. Reprod. 81, 333–342.
| Role of importin alpha8, a new member of the importin alpha family of nuclear transport proteins, in early embryonic development in cattle.Crossref | GoogleScholarGoogle Scholar | 19420384PubMed |
Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., and Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443, 851–856.
| Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus.Crossref | GoogleScholarGoogle Scholar | 22417684PubMed |
Wang, K., Sengupta, S., Magnani, L., Wilson, C. A., Henry, R. W., and Knott, J. G. (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One 5, e10622.
| Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts.Crossref | GoogleScholarGoogle Scholar | 21209939PubMed |
Wang, X., Park, K. E., Koser, S., Liu, S., Magnani, L., and Cabot, R. A. (2012). KPNA7, an oocyte-and embryo-specific karyopherin subtype, is required for porcine embryo development. Reprod. Fertil. Dev. 24, 382–391.
| KPNA7, an oocyte-and embryo-specific karyopherin subtype, is required for porcine embryo development.Crossref | GoogleScholarGoogle Scholar | 22281085PubMed |
Wilsker, D., Probst, L., Wain, H. M., Maltais, L., Tucker, P. W., and Moran, E. (2005). Nomenclature of the ARID family of DNA-binding proteins. Genomics 86, 242–251.
| Nomenclature of the ARID family of DNA-binding proteins.Crossref | GoogleScholarGoogle Scholar | 15922553PubMed |
Yoshioka, K., Suzuki, C., Tanaka, A., Anas, I., and Iwamura, S. (2002). Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol. Reprod. 66, 112–119.
| Birth of piglets derived from porcine zygotes cultured in a chemically defined medium.Crossref | GoogleScholarGoogle Scholar | 11751272PubMed |
Yuan, Y., Spate, L. D., Redel, B. K., Tian, Y., Zhou, J., Prather, R. S., and Roberts, R. M. (2017). Quadrupling efficiency in production of genetically modified pigs through improved oocyte maturation. Proc. Natl. Acad. Sci. USA 114, E5796–E5804.
| Quadrupling efficiency in production of genetically modified pigs through improved oocyte maturation.Crossref | GoogleScholarGoogle Scholar | 28673989PubMed |
Zhou, M., Guo, C., Li, X., He, J., Xu, X., Wang, H., Tang, K., Cao, L., and Li, G. (2011). Definition and function identification of nucleus export signal of BRD7. Zhong Nan Da Xue Xue Bao Yi Xue Ban 36, 634–639.
| 21873788PubMed |
* These authors contributed equally to this paper.


