Towards a new online species-information system for legumes
Anne Bruneau A M , Leonardo M. Borges
A M , Leonardo M. Borges  B , Robert Allkin C , Ashley N. Egan
B , Robert Allkin C , Ashley N. Egan  D L , Manuel de la Estrella
D L , Manuel de la Estrella  E , Firouzeh Javadi F , Bente Klitgaard
E , Firouzeh Javadi F , Bente Klitgaard  G , Joseph T. Miller
G , Joseph T. Miller  H , Daniel J. Murphy
H , Daniel J. Murphy  I , Carole Sinou
I , Carole Sinou  A , Mohammad Vatanparast
A , Mohammad Vatanparast  J and Rong Zhang K
J and Rong Zhang K
A Institut de Recherche en Biologie Végétale and Département de Sciences Biologiques, Université de Montréal, 4101 Sherbrooke Est, Montréal, QC, H1X 2B2, Canada.
B Universidade Federal de São Carlos, Departamento de Botânica, Rodovia Washington Luís, quilômetro 235, São Carlos, SP, 13565-905, Brazil.
C Biodiversity Informatics and Spatial Analysis Department, Royal Botanic Gardens, Kew, Richmond, Surrey, TW9 3AE, UK.
D Department of Biosciences, Aarhus University, Ny Munkegade 116, DK-8000 Aarhus, Denmark.
E Departamento de Botánica, Ecología y Fisiología Vegetal, Campus de Rabanales, Universidad de Córdoba, E-14071, Córdoba, Spain.
F Institute of Decision Science for a Sustainable Society, Kyushu University, 744 Motooka, Nishiku, Fukuoka, 819-0395, Japan.
G Identification and Naming Department, Biodiversity Informatics and Spatial Analysis Department, Royal Botanic Gardens, Kew, Richmond, Surrey, TW9 3AE, UK.
H Global Biodiversity Information Facility, 15 Universitetparken, DK-2100 Copenhagen, Denmark.
I Royal Botanic Gardens Victoria, Birdwood Avenue, Melbourne, Vic. 3004, Australia.
J Department of Geosciences and Natural Resource Management, Rolighedsvej 23, DK-1958 Frederiksberg C, University of Copenhagen, Denmark.
K Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, No.132 Lanhei Road, Kunming, 650201, PR China.
L Present address: Department of Biology, Utah Valley University, 800 W University Parkway, Orem, UT 84058, USA.
M Corresponding author. Email: anne.bruneau@umontreal.ca
Australian Systematic Botany 32(6) 495-518 https://doi.org/10.1071/SB19025
Submitted: 2 April 2019 Accepted: 25 July 2019 Published: 1 October 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
The need for scientists to exchange, share and organise data has resulted in a proliferation of biodiversity research-data portals over recent decades. These cyber-infrastructures have had a major impact on taxonomy and helped the discipline by allowing faster access to bibliographic information, biological and nomenclatural data, and specimen information. Several specialised portals aggregate particular data types for a large number of species, including legumes. Here, we argue that, despite access to such data-aggregation portals, a taxon-focused portal, curated by a community of researchers specialising on a particular taxonomic group and who have the interest, commitment, existing collaborative links, and knowledge necessary to ensure data quality, would be a useful resource in itself and make important contributions to more general data providers. Such an online species-information system focused on Leguminosae (Fabaceae) would serve useful functions in parallel to and different from international data-aggregation portals. We explore best practices for developing a legume-focused portal that would support data sharing, provide a better understanding of what data are available, missing, or erroneous, and, ultimately, facilitate cross-analyses and direct development of novel research. We present a history of legume-focused portals, survey existing data portals to evaluate what is available and which features are of most interest, and discuss how a legume-focused portal might be developed to respond to the needs of the legume-systematics research community and beyond. We propose taking full advantage of existing data sources, informatics tools and protocols to develop a scalable and interactive portal that will be used, contributed to, and fully supported by the legume-systematics community in the easiest manner possible.
Additional keywords: data exchange, data standards, genetic data, nomenclature, occurrence data, phylogenetic data, specialist data curation, taxonomic backbone, trait data.
Introduction
The need for scientists to exchange and share data and improvements in technology has resulted in a marked increase in the number of research-data portals over the past few decades (Fecher et al. 2015; Cicero et al. 2017). These portals have varying objectives and functions, but are generally considered informatics ‘systems that provide remote access to data repositories for discovery and distribution of reference data, the upload of new data for analysis or integration, and data sharing for collaborative analysis’ (Chard et al. 2018, p. 1). This proliferation of data portals is also apparent in systematics and taxonomy, where researchers have adopted informatics to organise and make data publicly available (e.g. O’Leary and Kaufman 2011; Benson et al. 2012; A. Goswami, see http://phenome10k.org/, accessed 31 May 2019). In contrast to some research communities where there can be reticence about sharing data (e.g. Tenopir et al. 2011; Poisot et al. 2019), collections-based research groups are accustomed to open data and are embracing new ways of sharing data and collaborating, which is changing the face of how biodiversity science is conducted (Cicero et al. 2017; Heaton 2018). Cyber-infrastructures have had a major impact on taxonomy and helped revitalise the discipline by allowing quick access to bibliographic information, nomenclatural and other biological data, and specimen information (Wheeler et al. 2004). This is clearly illustrated by the continued development and maintenance of portals dedicated to taxonomic literature (e.g. Biodiversity Heritage Library), nomenclature and synonymy (e.g. Catalogue of Life, Tropicos, International Plant Names Index, The Plant List, World Checklist of Selected Plant Families), georeferenced specimen data (e.g. Global Biodiversity Information Facility), taxon lists and hierarchies, phylogenies, images, and other associated biodiversity data (links to resources mentioned in the text are in Appendix 1).
From the late 1990s onward, several international taxonomic research communities developed taxon-centric portals that grouped information about a particular taxon into readily accessible web pages and databases (e.g. ants, spiders, fish, vertebrates, and various plant families). For example, AntWeb and the Global Ants Database (Parr et al. 2017) integrate different datasets in a single platform, curated by ant experts. There has been another trend, particularly in the past decade, to group species data into international projects that focus on a particular data type. On these platforms, specific queries are submitted to extract information pertaining to a particular taxon or list of taxa. The question then becomes, whether taxon-centric portals remain pertinent (and viable) for data sharing, storage, aggregation and analysis, and, if so, how such taxon-specific portals can efficiently interface with more general and comprehensive information systems. Here, we argue that a community of researchers focused on a particular taxon has the interest, commitment, collaborative links and, above all, the knowledge to sustain and maintain such a body of data. Only they can ensure the quality of data for downstream analyses and novel research and contribute effectively to more general data providers. Taxon-centric portals, thus, continue to serve a useful function in parallel to, and different from, but closely linked to large data-aggregation portals.
An online species-information system for Leguminosae
Legume systematists pioneered using informatics to share data, and, particularly, taxonomic information, with the development in 1985 of the International Legume Data Information System (ILDIS; described later). However, due to lack of continuity of resources and support, as well as evolving software protocols, increased data complexity, and the need for distributed data curation, ILDIS has not been curated by the legume community for more than 20 years. The ILDIS taxonomic data are now deployed through the Catalogue of Life and other ILDIS data are visible through Plants of the World Online. This example points to the synergy between taxon-specific community projects and higher-level international initiatives, feeding quality-controlled data to them and benefitting from the data aggregation and standardisation provided by them, but also raises the question of how to sustain taxon-centric initiatives, which is something we address in this paper.
The legume-systematics community has recently expressed interest in developing a new portal dedicated to deployment of knowledge, information and data pertinent to researchers and others interested in legumes. In 2010, the Legume Phylogeny Working Group was founded, and has since published three papers under this name, including a new subfamily classification for the family (Legume Phylogeny Working Group 2017). This research community has been collaborating and sharing data for several decades, since the first International Legume Conference in 1978 held at the Royal Botanic Gardens, Kew (RBG Kew), even though the exchange of information occurs informally rather than through a dedicated portal.
Here, we explore best practices for development of a legume portal to enable data sharing and a better understanding of what data are available, missing, or erroneous, and ultimately facilitate cross-analyses and collaboration within the legume-systematics community and with other stakeholders. Our objective is to take full advantage of existing data sources, informatics tools and protocols to develop an easily manageable, scalable and interactive portal that will be built, maintained and contributed to primarily by the legume-systematics community. We discuss what resources currently exist that are pertinent to our needs, explore established taxon-based portals to determine which features are of interest and most useful to ensure long-term sustainability and utility, and discuss lessons learned from past projects. We propose a vision and a road map for the development of a Leguminosae (Fabaceae) species-information portal.
Target audience and data cycle
A taxon-centric portal is a powerful tool that allows access to different types of integrated data for a particular taxon. To maximise the relevance of such a portal and its sustainability (lifespan), it is important to properly define potential users, stakeholders and what kind of data are useful to them. It is clear to us that systematists and evolutionary biologists are the main focal group of a data portal for legumes. However, Leguminosae is an economically and ecologically important plant family, of general interest to crop breeders, farmers, pharmacists, horticulturists, timber merchants, ecologists, conservationists and the general public, as well as many other scientific communities. All these groups need access to reliable, scientifically validated and well-structured information about legumes, a need that can be met only by participation of the legume-systematics community. We suggest that the interests of legume systematists and of more general users often overlap and that this synergy can drive and facilitate the development of an online information system for Leguminosae.
Legume systematists produce information about taxonomy, geographic distributions, trait diversity (molecular, morphological and others), ecology, phylogeny and evolution of legume species and higher taxa. At the same time, this same information is also used by legume systematists. When using information, systematists often curate and update data produced by other scientists, a step necessary to support accurate downstream analyses. This process results in data-quality improvements, providing a service to specialised data providers and ensuring their systems become more effective and meaningful (Costello et al. 2013). Thus, during routine work, systematists can improve the quality of information on legumes available to themselves and others.
Generalist users of a data portal are interested in accessing and using information for onward analysis or to answer many straightforward questions. The type of information varies, but it is important that it be of high quality and reliable. Users aiming to identify species, for example, require simple tools that work well, whether identification keys or photo guides. Biogeographers need accurately georeferenced specimens. Conservationists need to know the full distributions of threatened or endangered species. Government officials need to identify and control invasive species reliably and verifiably (e.g. Binggeli 1996; van Kleunen et al. 2015). All users need names to be applied accurately and unambiguously. However, the general user faces practical problems in finding information across multiple disconnected systems and in resolving discrepancies or differences of opinion between systems. This disconnection and overdispersion of information is a problem shared with systematists.
The shared common needs of the legume-systematics community, other scientists, and more general users highlight the value of a taxon-based portal that aggregates high-quality, accurately curated information centred on names and verifiable vouchers (Fig. 1). Such a system is important for legume systematists themselves, but also critical as an authoritative reference for people accessing information about legumes indirectly through other information systems (e.g. Global Biodiversity Information Facility (GBIF), Encyclopedia of Life (EOL), Wikipedia and others).

|
Past initiatives for managing legume taxonomic data
The size, diversity, ubiquity, and economic and biological importance of the Leguminosae has prompted abundant research by systematists and other biologists. The Advances in Legume Systematics series, first published in 1981, fostered global collaboration among legume systematists. The prominence and importance of the family and early emergence of a collaborative research network meant that legumes were the focus for early initiatives to develop and deploy information technologies for capturing, sharing and disseminating taxonomic and other data and information. Indeed, legume researchers often led the way in developing methods to manage and share taxonomic data.
Early days
Led by Frank Bisby and Richard White in Southampton, UK, in the late 1980s, the Vicieae Database Project built a monographic information resource for the ~600 species of tribe Vicieae (Adey et al. 1984). The project was established to explicitly prototype taxonomic-information management and test software and technologies available at that time, to demonstrate what was possible and where development was necessary. Software limitations required creation of separate, albeit interlinked, database modules. These included a ‘taxonomic nomenclatural backbone’, ‘geographical distribution’, ‘morphological descriptions’, ‘chemical constituents’ and ‘bibliography’, which were linked with newly developed software tools. All data entry and management were undertaken by a small centralised team at a single institution and, in the pre-internet era, project outputs comprised printed catalogues, identification keys generated using DELTA software (Dallwitz 1993), and research papers describing and discussing methodologies (Allkin 1984; White 1984).
Technical success of the Vicieae Project led to two parallel developments. The first involved the first attempts to use data modelling for management of taxonomic monographic data (Allkin and White 1988; White et al. 1993) and to build software capable of managing species checklists and descriptive data that could interact with specimen-collection management and DELTA identification software. These initiatives led to establishment of some of the first taxonomic data standards adopted by the then newly formed Taxonomic Databases Working Group (TDWG; e.g. Allkin and White 1993; Wieczorek et al. 2009). The second important development was the International Legume Database and Information Service (ILDIS).
International Legume Database and Information Service (ILDIS)
The more ambitious ILDIS initiative aimed to build a comprehensive checklist of the entire legume family. ILDIS was to be built in phases (Zarucchi et al. 1993). Phase 1 involved construction of a taxonomic checklist including full synonymy, low-resolution data on geographic distributions (botanical ‘countries’) and a modest set of life-history traits. The bulk of the original ILDIS species checklist was built through a series of non-overlapping regional checklists compiled from the literature (e.g. Lock 1989). Phase 2 sought to add depth to that taxonomic framework by including morphological, chemical (Bisby et al. 1994) and other data. However, lack of funding mostly prevented progress with Phase 2, other than definition of the data standards to be used, as well as, for example, the inclusion of root-nodulation data (e.g. Faria et al. 1989; Sprent 2001). Independently, accounts of seeds and fruit morphology of legumes were assembled (Gunn 1984, 1991; Kirkbride et al. 2003a, 2003b); however, these were never incorporated into ILDIS.
Implementation of ILDIS required database software to manage the core checklist and other more sophisticated and varied linked data types in Phase 2. It required well-defined data standards and terminological controls and software capable of enforcing these, as well as data-exchange mechanisms (export, import and integration) to enable datasets from collaborating centres to be merged and aggregated. These, in turn, led to development of further data standards and exchange protocols (Allkin et al. 1992; White and Allkin 1992) that became of wider utility in biological-data management (e.g. Hollis and Brummitt 1992; Berendsohn et al. 2011). A strength of ILDIS was that each observation relating to a record or plant name was linked to the source(s) from which the data were obtained, facilitating acknowledgement, validation and analysis. Each database and associated publication was compiled using Alice software (Allkin and Winfield 1993; Bisby 1993) and ILDIS data standards, and the datasets were subsequently merged into the central ILDIS database.
With efforts from staff at RBG Kew, Missouri Botanical Garden and Reading University, the ILDIS species checklist (Phase 1) was completed, but ILDIS was not subsequently maintained, owing, in part, to funding constraints. The ILDIS coordinators then focused on the ambitious wider challenge of creating a checklist of all plants and, ultimately, all species. Species 2000 and its product, the Catalogue of Life (Bisby et al. 2006), grew out of the ILDIS initiative.
Legumes of the World Online (LOWO)
In 2005, Lewis and colleagues published Legumes of the World (Lewis et al. 2005). This landmark encyclopedia, which included contributions from 24 legume systematists, provided a brief illustrated account of each genus and its position within the family classification.
Because Legumes of the World was published as a hardcopy book, updates were not easily possible. Nevertheless, the structure of the book and the consistent format of generic accounts lent itself to digitisation and the contents were made available as ‘Legumes of the World Online’ (LOWO). This digital version allowed free online access and flexible browsing of genera using either the taxonomic structure of the family or phylogenetic diagrams, and removed constraints on the numbers of images. However, the most significant advantage was that the RBG Kew legume researchers and those at other institutions (e.g. Royal Botanic Gardens Edinburgh) were able to curate, update and extend the content of LOWO. New genera and images were added, species counts refined, species names for uses and photographs revised and extended, corrections made to data in many tribes and accounts of some genera were revised and new genera added. In 2017, most of the data in LOWO were copied to Kew’s angiosperm-wide Plants of the World Online portal, which offers no means of editing these data. However, the generic backbone in LOWO and the functionality implemented provide a basis to initiate the development of a new legume data portal.
Current Leguminosae portals
Although other legume-focused portals are ongoing and pertinent to systematics and taxonomic research, no portal currently exists for deployment and aggregation of legume-species information. For example, the International Legume Database of Nodulation (ILDON; Appendix 1) builds on one of the original ILDIS Phase 2 modules for root-nodulation data and on the legume species checklist in the World Checklist of Selected Plant Families (see below). A second example is the Legume Information System (LIS), focused on legume crops, which integrates genetic, genomic and trait data across legume species, enabling cross-species genomic and transcript comparisons and facilitating crop improvement (Gonzales et al. 2005; Dash et al. 2016). Other legume portals, focused on particular clades (e.g. Acacia Mill., Leucaena Benth.), genomic resources, chemical data, and on cultivated or economically important legume taxa, are listed in Appendix 1.
Web resources presently available for plant systematics and evolution
Over the past three decades, numerous online resources have been developed that facilitate the work of taxonomists and systematists. These include taxonomic and nomenclatural resources and databases, geospatial portals based on data from collections and observations, databases focused on traits, morphology, chemistry, DNA sequences, full or partial genomes, repositories for datasets and phylogenetic trees, and phylogenetic assemblers (Appendix 1). In practice, systematists often will need to consult multiple resources to access the desired information.
Taxonomic resources
Taxonomic and nomenclatural resources are of the following two main types: curated original datasets and aggregators that merge data from multiple sources. Both types tend to be under the auspices of large natural-history museums or international consortia of research institutions, and respond to particular and sometimes specific needs (lists of plant names, synonyms, reference literature, basic descriptive information). Some of these resources include taxonomic and nomenclatural data that have not been updated recently, and some use older technologies that offer limited functionality. Because of this diversity and inconsistency, researchers and curators currently need to consult several different taxonomic and nomenclatural resources to arrive at nomenclatural decisions and gather information. Here, we provide a short description of the main resources that are presently available. The first four are curated databases, whereas the The Plant List, Catalogue of Life and World Flora Online are taxonomic aggregators.
The International Plant Names Index (IPNI) is a complete and actively maintained catalogue, and default reference, for all scientific names of vascular plants. It provides information on authorship of names (including standard abbreviations) and their date and place of publication, but not on the status, i.e. currently accepted names and synonyms. Increasingly, journals (e.g. Phytokeys and Kew Bulletin) are automating addition of new names to IPNI. IPNI has links to the Biodiversity Heritage Library (BHL) and a new β version of IPNI links the names more explicitly to protologues in online articles. IPNI is actively curated by RBG Kew, Harvard University and the Australian National Herbarium (see Appendix 1 for statistics for this and other resources). It is the modern descendant of Index Kewensis that was active from 1893 to 2000, of the Gray Card Index, which was computerised in 1992, and of the Australian Plant Name Index, which was published in 1991.
The World Checklist of Selected Plant Families (WCSP) is compiled at RBG Kew. For each accepted name, it lists full synonymy and geographic distribution derived from published literature. It also presents information on alternative taxonomic classifications where these are known. New plant names are derived directly from IPNI. Importantly, WCSP is actively curated (~250 000 edits annually); once a family list has been compiled, it enters a peer review process by taxonomic experts. The Leguminosae species checklist included in WCSP is complete for 98% of all legume species (May 2019) and is now in the review phase with various specialists contacted. The legume checklist is, therefore, not yet published on the WCSP website, but is visible through Plants of the World Online.
Plants of the World Online (POWO) is a taxon-based portal that uses taxonomic data from WCSP and IPNI and publishes additional information provided by RBG Kew and its partners, such as descriptions (from floras or monographs), static maps and links to other databases. POWO also includes common names and information on uses, habit and other descriptive information derived from ILDIS and from Legumes of the World Online.
Tropicos, from the Missouri Botanical Garden, is a rich source of botanical information particularly for the New World, derived from the herbarium collections of Missouri Botanical Garden, other international collections and published floras. Each entry includes the distribution, specimens, images (when available), and synonymy from certain publications. Tropicos presents conflicting taxonomies for some groups of plants. Initially created for internal use at Missouri Botanical Garden, Tropicos was made publicly available online c. 25 years ago.
The Plant List (TPL) was published in 2012 as a ‘working list of plant species’ in response to Target 1 of the Global Strategy for Plant Conservation (GSPC). The Plant List provides a checklist with synonyms of all vascular plants and bryophytes following APGIII (Angiosperm Phylogeny Group 2009). The data in TPL were aggregated from WCSP, Tropicos and monographic datasets for Compositae, Rosaceae and Leguminosae (ILDIS). Aggregating data from these different sources employed automated mechanisms to detect conflicting taxonomic views among and within overlapping datasets. These conflicts were resolved using a logical rule set, which attempted to replicate standard botanical decision making on the basis of the information available; however, automating this process inevitably led to some errors. Time constraints to meet GSPC targets precluded expert taxonomic review manually. The Plant List provides for search, browse and data-download functions. Taxonomic decisions are labelled with one, two or three stars, depending on the relative confidence or reliability of that taxonomic judgement. The Plant List has become the most popular plant checklist (1.7 million unique users per year), despite the uncertainties implicit in its underlying data. It is, for example, linked to R packages, such as ‘Taxonstand’ (Cayuela et al. 2012), which allows automated standardisation of taxonomic names in bioinformatics pipelines using TPL nomenclature. Despite its popularity, TPL suffers from data errors and gaps inherited from its contributors and introduced when merging multiple plant lists automatically. Most importantly, TPL has not been updated since 2013.
Catalogue of Life (COL) is another aggregator site that brings together (mostly non-overlapping) checklist datasets from diverse sources for all living organisms. The COL supports several biodiversity and conservation information services such as the Global Biodiversity Information Facility (GBIF), the Encylopedia of Life (EOL) and the IUCN Red List of threatened species. COL provides web services for querying their database. As for TPL, the legume data in COL derive from ILDIS and are not being actively curated.
The recently launched World Flora Online (WFO) is a compendium of the world’s plant species developed by an international consortium that aggregates data from published floristic accounts. World Flora Online currently employs TPL as its taxonomic backbone and plans to address later the conflicting taxonomies presented in the flora accounts included.
In addition to the curated databases listed above, many countries or geographic regions, as well as communities of taxonomic specialists, have developed and maintain curated online species lists with synonyms, vernacular names, maps, images and local uses, among others (e.g. Australian Plant Census, VASCAN, Anthos, African Plant Database).
Biodiversity portals, specimen information and occurrence data
Biological collections are the central source of large volumes of biodiversity information and their importance for biological research and conservation is widely acknowledged (e.g. Meineke et al. 2018). Accessing historical data in herbaria and or other natural-history collections traditionally meant visiting the physical collection or borrowing specimens. With the development of databasing, imagery and the internet, remote access to specimens has become a viable and efficient way of consulting collections for most purposes. Institutional portals arose in the early 2000s, or earlier, giving access to some collections. Soon after, the power of aggregating data, so as to efficiently allow users to discover and analyse these data from a single portal, led to the development of data aggregators and standards for publication.
GBIF is the most comprehensive biodiversity data aggregator, harvesting data from an extensive network of national and regional aggregators. This type of aggregation would not be efficient or possible without standardised data formats and protocols. In 2009, the Biodiversity Information Standard, formerly the TDWG, formalised the Darwin Core vocabulary as a standard to publish biodiversity data (Wieczorek et al. 2009). With the recent development of citizen science, much larger datasets can be generated from observations, and several initiatives have contributed to the publication of over a billion (109) observation data points now available on GBIF (e.g. eBirds, Swedish Species Observation System, iNaturalist). In the early 1990s, Mexico (CONABIO), Costa Rica (INBio) and Australia (ERIN) led the way in developing biodiversity portals and platforms that aggregated data at a national level, acting as central biodiversity information resources for these countries. In 2001, the governance structure of GBIF was established and many other countries established national portals from which data could be channelled to GBIF.
Of particular importance, the Atlas of Living Australia (ALA) portal, has emerged as a model in the biodiversity informatics community. Computationally complex, but with the source code fully available, ALA provides numerous modules, entry search points, analytical tools and dense information about Australia’s biodiversity and taxa. The biodiversity informatics community has been adopting and adapting the ALA model for developing national biodiversity portals (Living Atlases Community). Clearly, it is advantageous that the ALA has available a curated list of ‘accepted’ taxonomic names existing side-by-side with its biodiversity portal (i.e. the Australian Plant Census), but there are possibilities within ALA to incorporate alternative taxonomies and, thus, for other communities and countries to use the powerful ALA bioinformatic model.
Genetic, morphological and trait data
Several different online tools have been developed to share molecular, morphological and functional-trait data. Molecular data are commonly shared through one of the interconnected partners of the International Nucleotide Sequence Database Collaboration (INSDC), NCBI’s GenBank (Benson et al. 2012), the European Nucleotide Archive, and the DNA Databank of Japan (Appendix 1). There are more limited resources linking biodiversity-data portals to molecular data. One such initiative is the Barcode of Life (BoLD; Ratnasingham and Hebert, 2007), which links specimen records with DNA barcodes for various groups of organisms, on a user defined ‘project’ basis.
The web includes other data resources on plant traits, such as chromosome numbers (IPCN), mass spectral data (e.g. MassBank; Horai et al. 2010) and metabolites (KNApSAcK; Afendi et al. 2012). However, morphological data are still mostly restricted to descriptions in taxonomic works, or tabulated datasets in the form of tables or supplementary files accompanying publications. This is true for both categorical and continuous data, but the situation is worse for the latter. Authors of morphometric studies usually present tables only with mean and variance values for each variable. Categorical-data matrices, commonly used in phylogenetic studies, are usually available from journal websites or shared as nexus files in TreeBASE (ver. 2, see https://www.treebase.org/treebase-web/home.html, accessed 31 May 2019). Two other options for publishing, searching and downloading morphological data are the TRY Plant Trait Database (https://www.try-db.org, accessed 31 May 2019; Kattge et al. 2011) and MorphoBank (ver. 3.0, M. A. O’Leary and S. Kaufman, see http://www.morphobank.org, accessed 31 May 2019; O’Leary and Kaufman 2011). MorphoBank is dedicated to morphology and allows inclusion of continuous and categorical data, and supports uploading 2-D and 3-D images, including CT scans. TRY aggregates information on plant functional traits, including morphology, but consists only of tables of observations, which are made available only by request.
Biodiversity sampling and abiotic and biotic data are integrated in several ecologically oriented portals (e.g. NCEAS; OBIS; ILTER; Ocean Observatories; see Michener 2015), all of which aim for collaborative science and data sharing to further our understanding of biodiversity and ecology through time and space. For example, the National Ecological Observatory Network (NEON) provides comprehensive occurrence data and samples from field sites in the USA, collected using standard protocols. NEON includes data from terrestrial, airborne and aquatic environments, such as soil-microbe occurrences, LiDAR imagery, or chemical properties of groundwater.
Although there are several resources that deploy molecular and morphological data, fragmentation of information and lack of connection among databases is a problem. Researchers must also contend with challenges related to the uneven representation of different types of data and taxa, and with issues of taxonomic accuracy. Many of these data-focused resources lack a sound taxonomic or nomenclatural framework. No studies are available on the taxonomic reliability of morphological databases, but we expect that anatomical and morphological data, commonly produced by taxonomists, are more reliable, whereas the situation for chemical and wider functional-trait datasets may be similar to that of molecular databases (Bridge et al. 2003; Vilgalys 2003). Finally, specimen information is rarely linked to taxonomic databases or to collections, which hinders identification updates. As with molecular databases, most repositories for trait data include the option to provide specimen voucher information and it would be particularly powerful to connect these data to a well-curated nomenclature database through vouchers associated with unique identifiers in a taxon-centric portal.
Images and photos
In recent years, herbaria have made massive efforts to digitise specimen data and publish specimen images online (Soltis 2017). These images are published on collection websites, in aggregator portals such as national biodiversity portals, or through GBIF. A well-known example is Global Plants, a collaboration between JSTOR and some 300 herbaria that publishes ~2.5 million plant-specimen images with a strong focus on type specimens, but it is accessible only to paying members.
While museums and herbaria were embracing online data-sharing opportunities, several citizen-science portals were developed. These portals are a rich source of photographs and include millions of observations (e.g. iNaturalist; eBird, eButterfly, Pl@ntNet, to name just a few), which are identified by users themselves, by interaction between users and taxonomists (Bowser et al. 2014; van Horn et al. 2017), or by image-recognition softwares (Unger et al. 2016).
Computer-based image recognition of biological entities is rapidly improving (Nelson and Ellis 2018) and, in the near future, new machine-learning and artificial-intelligence approaches should facilitate identification of digitised herbarium specimens (Schuettpelz et al. 2017, Wäldchen and Mäder 2018) and of plant photographs (Gardiner and Bachman 2016; Kress et al. 2018; Younis et al. 2018). In the context of citizen science, Leafsnap (Kumar et al. 2012) uses visual recognition of leaves to help identify tree species, and morphological features are used to identify plant photographs in Pl@ntnet. Such citizen-science apps have the potential to contribute to the monitoring of biological dynamics, such as the effects of climate change and biological invasions (Kress et al. 2018); however, at present, they tend to misidentify less common species and species belonging to groups with less obvious morphological differences (e.g. grasses) or with a high intraspecific variability.
Taxonomists and field biologists often have accurately identified specimen-vouchered images of endemic or rare species. However, as with most botanical illustrations, which are confined to scientific publications, these high-quality images are rarely made available to online repositories or connected to other databases. A useful role of a legume-centric portal would be to connect names, photographs, illustrations and digitised vouchers in a scientific context, and make available, for the first time, a vast reservoir of legume imagery, which is currently locked away on individual hard drives. In turn, this could serve as a base for the development of better-performing image-recognition software and, consequently, lead to better tools for citizen-science identification.
Phylogenetic information
Deposition of datasets and inferred phylogenies is good practice for biodiversity research (Penev et al. 2017) and many journals now require this for publication, at least for molecular data (DNA-alignment matrices). Current methods for linking phylogenetic trees to underlying data include stand-alone databases such as TreeBASE, DRYAD and Figshare, as well as individual GitHub accounts. The next stage in using phylogenetic data is the integration of subtrees across phylogenetic databases. Open Tree of Life (OTOL; Hinchliff et al. 2015) is a good example that integrates analysis pipelines with taxonomy, to produce a supertree hypothesis of evolutionary relationships across life (Rees and Cranston 2017). Scripts and pipeline tools have been developed to enable interactive use and integration of OTOL into other databases and portals (e.g. Michonneau et al. 2016).
Integration of specimen, phylogenetic and spatial data can be particularly informative (Soltis et al. 2018). This can be performed in Phylolink, the successor to PhyloJIVE (Jolley-Rogers et al. 2014; Miller et al. 2019), which used Acacia as the pilot taxon. For example, it is possible to view the distributions of sister taxa in Phylolink, a feature that adds an evolutionary perspective to the commonly available mapping of specimen records. Also, species pages can be accessed from any terminal or node of a tree in Phylolink. Currently, Phylolink is built into the Atlas of Living Australia, where its visualisations can be merged with environmental layers (e.g. soil, climate) and serve as a base for comprehensive analyses in ecology and evolution.
Despite developments for integrating data and phylogenies, viewing and navigating large phylogenies remains a challenge. For instance, the current tree viewer in Phylolink does not work well for trees larger than a few hundred species. Large phylogenies are more easily viewed in OneZoom (Rosindell and Harmon 2012), which works fractally to zoom in and out of clades and travel along branches of a phylogeny, in addition to allowing incorporation of images and traits to the tree. The OneZoom web viewer currently provides phylogenies based on OTOL, including the Leguminosae, but the legume topology is not up-to-date. A caveat of the fractal approach of OneZoom is the lack of overview for the placement of a particular taxon because the level of resolution of the tree varies automatically. Other applications are being developed, such as Phlora, an iOS app (M. J. Sanderson, University of Arizona), which provides new ways of visualising and exploring phylogenetic trees with images associated to individual taxa, including legumes.
Taxon-based portals: examples from other taxa and lessons learned
In the early 1990s, several international initiatives, such as the Catalogue of Life partnership of Species2000 and the Integrated Taxonomic Information System (ITIS), advocated the need to rapidly index the world’s known species, to make biodiversity databases universally accessible, and to engage in a concerted effort to discover and describe the conservatively estimated 80% of species still unknown to science. At the same time, electronic cataloguing of specimens was seen as perhaps the most important innovation in natural-history collection management in the 20th century (Bisby 2000; Wilson 2000, 2003; Lawler 2001; Gewin 2002; Godfray 2002; Butler 2006). These developments resulted in the establishment of the Encyclopedia of Life project (EOL) and popularised the idea of electronic ‘species pages’, which synthesise and display known information about a taxon. Numerous taxon-centric portals were developed, each answering to specific user needs and built with different technologies (e.g. Scratchpads (see http://scratchpads.eu/, accessed 31 May 2019), Taxonomy Research and Information Network (TRIN), Symbiota (see http://symbiota.org/docs/, accessed 31 May 2019), TaxonWorks (see http://taxonworks.org/, accessed 31 May 2019), are examples of platforms built to share natural-history knowledge).
The landscape of biodiversity-data sharing has changed tremendously since then, and providing access to data and designing a taxon-centric portal today differs dramatically from models established 10 years ago. Nevertheless, critically viewing the strengths and weaknesses of existing portals (examples in Appendix 1) is important before designing a new legume portal that allows flexible development updating, that uses and takes advantage of the tools and services currently available, and is capable of evolving as required to remain relevant. Much can be learned from past developments.
General aspects
Design and aesthetics may not be the most important focus from a research perspective, but are critical to enable ease for finding and displaying information. The popularity of TPL, despite its erroneous and out-of-date data, for example, continues in part because of its simple interface that makes navigation intuitive and effective. The eMonocots portal (now included in Plants of the World Online) provided another example of an appealing design, with high-quality images and a database that was simple to access. Citizen-science portals such as eBird, eButterfly, Leafsnap, or the more generalist iNaturalist, all of which are easy to navigate and search for specific information, are other examples of appealing portals.
Several existing taxon-centric portals are static, lacking dynamic links to other data types or direct update of information (e.g. Gesneriaceae, eMonocots, POWO). Although these provide useful encyclopedias for information that is stable, they are of limited utility for research. Some more recently developed taxon portals provide access to varied types of information, including, for example, literature, occurrences (specimens or observations), molecular data, traits, phylogenetic information and images (e.g. Solanaceae Source, Global Ants Database, Atlas of Living Australia’s iconic species; Table 1, Fig. 2, Appendix 1). For example, several types of data in the ALA portal are obtained automatically from other databases and, thus, change as original sources are updated (Fig. 2). In the Global Ants Database, data submissions go through a quality check before being included in the database.
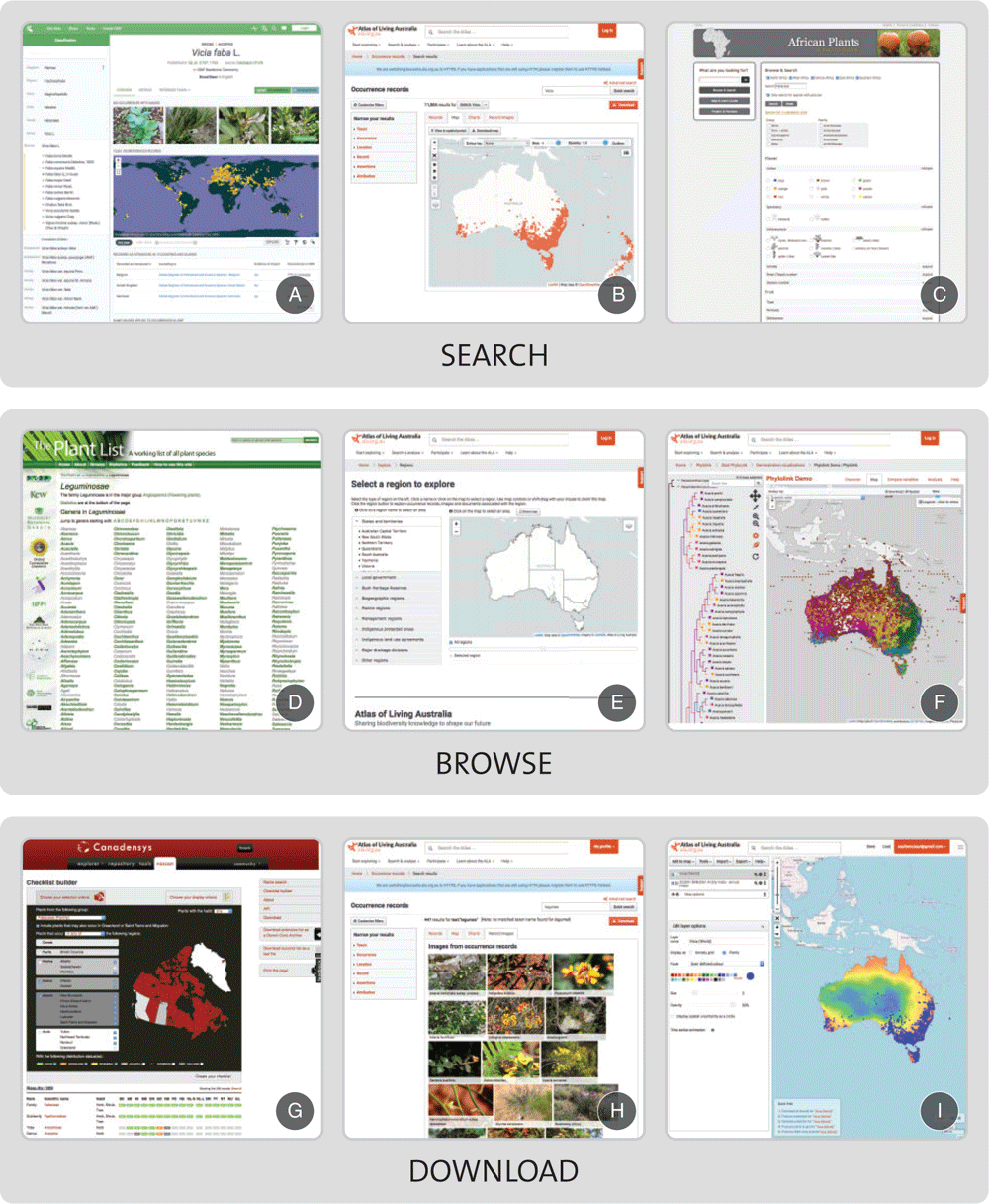
|
Building and sustaining data resources over time will require different approaches depending on the data. Direct data correction or entry is desirable in some instances, whereas updating the system through periodic data imports would be better for other data. Datasets that are entirely ‘static’ will quickly become out of date and redundant, but, for some purposes, a resource that is versioned, i.e. remaining static for a given period, may have advantages over a resource that is continually updated. Suffice to say, updating information and investing in data quality are desired features of a legume portal.
Data standards are required
ILDIS succeeded in bringing together data from multiple systematists and other legume scientists by communicating clearly and rigorously implementing terminological controls and data standards. The use of data standards is also integral to the success of GBIF and other large international projects such as the ALA community. Ensuring that contributors use and interpret terms consistently makes data retrieval and analysis possible. A limitation of LOWO (where the content derives from the text of a traditionally published book) is that a search for ‘medicinal’, for example, misses any genera used as ‘herbals’, and, more fundamentally, that the taxon descriptions were not developed with informatics standards in mind. Standardisation is needed, but it has to be applied having in mind not only organisation, but also discovery by data interconnection (Hobern et al. 2012).
Standards established by the Biodiversity Information Standards (TDWG) have focused on data-exchange protocols and formats such as Darwin Core. However, it is critical to control terminology for other categories in a way that is fully adopted by data providers and users. Although application programming interfaces (APIs; e.g. Miller et al. 2015) permitting data exchange are increasingly implemented in relevant information systems, the technology cannot resolve fundamental differences in how data values are used and interpreted within different systems. Thus, standards need to be discussed and agreed at an early stage of a new legume-portal initiative, and, if required, community-specific guidelines developed. We also should take advantage of ongoing development of ontologies (e.g. environment and habitat (Buttigieg et al. 2016); biomedical, plant phenotype and phenology, among others) when defining such standards. Finally, a clear dialogue is needed with large international data initiatives such as GBIF, which are moving towards establishing a high-level organising framework that deals in broad categories common to all taxa, collections and data, and the relationships between these entities, and into which taxon-specific communities can then incorporate their structured data.
A common taxonomic backbone
The essential starting point for any form of standardisation is an agreed list of accepted scientific names and synonyms. Databases using alternative accepted names for a species can still share data, provided a fully accepted and synonymised list is available. The World Checklist of Selected Plant Families appears to be fulfilling this role of developing a curated and peer-reviewed taxonomy that provides scientifically validated plant-species names. Presumably, a dialogue would be possible between a legume portal and WCSP in such a way as to ensure the most accurate taxonomy in both networks. This curated database should then become the source for other initiatives requiring a sound taxonomic list.
Central v. distributed data systems
The success of Tropicos, WCSP, GBIF and many other current digital initiatives demonstrate the merits of a coordination centre and clear institutional responsibility for building, and, more importantly, sustaining the digital infrastructure even though contributors may be based in many different institutions. The disadvantages of this model is the reliance on a single institution to provide core funding and infrastructure. A centralised endeavour also reduces the sense of joint ownership. This strongly centralised model differs from the ALA model, which, although developed by a single team of biodiversity informaticians in Australia, is now expanding to become a more distributed system, maintained by an active and dedicated community of researchers and programmers, the Living Atlases Community, deploying the system in their own network, and each contributing to its development to the extent possible with the resources available to them. Although GBIF is governed by a secretariat in Copenhagen, it also depends on an active international community who contributes data and tools, and participates in its governance.
Another major challenge for biodiversity cyber-infrastructures is long-term sustainability. Whereas the focus of all institutions changes over time, the rate of change may be greater within university departments that face pressures to develop new and innovative research initiatives and are reliant on (short-term) research funding. In the past, large well-established institutions, such as natural-history museums, botanical gardens and government agencies and departments, were viewed as more stable than were university departments. These institutions could count on core funding to curate their physical and digital assets, provided that these continued to be seen to fulfil the objectives of the organisation. Thus, over the decades, IPNI evolved from Index Kewensis and has been maintained and curated throughout that time almost entirely by RBG Kew to meet a wider community need, as originally envisaged by Charles Darwin whose donation helped fund the early development of Index Kewensis. However, the funding challenge is now a reality faced by most institutions, and this must be considered in the long-term sustainability of biodiversity and taxonomic cyber-infrastructures, which will require funds for practical tasks related to construction, maintenance and curation of datasets.
Accurate up-to-date information about legume species, well organised and in an easily retrievable form, would attract a wide range of users and, thus, create opportunities for wider financial support for a new legume portal. Fishbase, one of the most successful, globally most visited, taxon-based non-profit organisations, is an excellent example demonstrating how a wide user community has assisted Fishbase to secure funding for development and maintenance over more than 20 years. In addition, surveys of visitation rates and entry points for Fishbase, as well as for other identification resources (e.g. Neotropikey, see http://www.kew.org/science/tropamerica/neotropikey/key/neotropikey_quickstart.htm, accessed 31 May 2019), provide indications of user preferences and can be taken into account in the development of a sustainable legume portal.
Linking systems
The advent of universal data standards is facilitating transfer of data among information systems, but this remains challenging. Although this process is increasingly commonplace, it suffers when updates in one system are not reflected in all systems. This can often depend on refreshing data uploads by using an agreed frequency and protocol (or data dump). Application programming interfaces help link information systems that share common data so that edits and improvements in one system are automatically reflected in the other. One example is the display of GBIF specimen-distribution data on the webpage of EOL to portray the distribution of a species. As more data are available from APIs, it may be easier to link data and generate modular, user-specific data integrations and visualisations.
In 2018, the global biodiversity informatics community (Hobern et al. 2012, 2019) tasked GBIF to lead an alliance that would facilitate the seamless integration required for taxon-specific portals, including the development of interoperable modules from various national and international initiatives knitted together to meet portal-user needs. The Global Biodiversity Information Facility aims ‘to propose a collaborative approach for the global community for planning and agreeing on an optimal set of new or improved policies, data standards, processes, governance arrangements, software tools, informatics infrastructure investments and research programmes, with sufficient clarity to deliver an interoperable global infrastructure’ (Hobern et al. 2019, p. 6).
The social dimension
There are technical challenges inherent in building a new information system, particularly one linked to many existing platforms, and further challenges related to harnessing and curating content in a sustainable manner that meets our goals and research needs. However, what cannot be overstated are the social challenges in organising the ideas, contributions, data and efforts of the wider community and, in particular, in sustaining their interest and shared commitment over time. This begins by agreeing on a sufficiently clear and tightly defined purpose and target audience. Subsequently, some form of coordination or management group is inevitably required to oversee and support implementation. Sustaining such coordination has, arguably, been the most challenging issue facing earlier efforts and the single biggest factor in their lack of continuity. Any new project will require a considered proposal covering the multiple social aspects, including giving full credit and recognition for the work contributed.
Desired features of a legume portal
Having discussed currently available web resources, below we focus on desired features for a new legume portal, bearing in mind that the database structure would be centred on names and specimens (Fig. 1) and used both by legume systematists and a wider public, albeit not necessarily through the same interface.
General aspects
Conceptually, a legume-centric portal could be developed ‘simply’ by providing a taxonomic and nomenclatural core linked to external sources of information, managed and made visible through different interfaces. However, many uses by non-systematists rely on the ability to query, analyse or manipulate data from different sources or disciplines, and the legume-systematics community has expressed a need for a source of aggregated, integrated and curated information to facilitate research. A common overview of what data are available for which taxa would also help drive research by identifying and helping in filling knowledge gaps. Presenting data from multiple sources by a single platform might be achieved by presenting a summary of data for each taxon or by using more sophisticated tools for visualisation and analysis (Table 1, Fig. 2). A more useful unified resource could be achieved through the use of ontologies and adoption of semantic tools, providing powerful discovery instruments for our datasets (Deans et al. 2015).
One of the most important features of a legume portal is that the data are easy to maintain and contribute to. The system employed to curate these data should be robust and scalable, and should adopt data functions and tools developed by other initiatives (Fig. 1). Given the abundance of legume data already available (Table 1, Fig. 1), some of the data in the legume portal could be aggregated from existing databases; however, de novo databases will also be required. It is also important to ensure that the data collated in older publications or databases remain relevant, and, thus, to provide for future transfer and evolution of content.
An important communication tool in the legume-systematics community since 1974 is the legume-systematics newsletter, The Bean Bag (see https://www.kew.org/science/our-science/publications-and-reports/publications/the-bean-bag, accessed 31 May 2019). The legume portal could include a link to all the Bean Bag issues, as well as be a venue for announcing important meetings and conferences, or sharing ideas for research or seeking help from the community.
Taxonomic data
Central to any legume data portal is the need for an up-to-date list of accepted legume names and synonyms, being continuously curated by specialists, that can be viewed and used to aggregate data at various taxonomic levels (subfamily, clade, tribe, genera, species, infraspecific categories). The legume community is well advanced in this area and can call on the WCSP checklist to provide an initial checklist of accepted names and synonyms. It is equally clear that such a backbone will be fit for purpose only if it is actively curated by input from the legume community, but this holds true for most, if not all, biological databases.
Occurrence data
Geographic occurrence data have many uses, for biogeographical analyses, mapping taxa including invasive species, modelling species distributions under climate-change scenarios, and for assessing global rarity and IUCN threat categories. Cleaning and georeferencing specimen data are time consuming, even if tools for some degree of automatic cleaning are now available; currently, this process is often repeated by different researchers for the same collection or even for the same specimen. This is mainly because data aggregators such as GBIF do not currently provide a facility for feeding cleaned GBIF georeferenced data back for future use by other researchers. Ideally, GBIF and other aggregators would provide a workbench for taxonomic communities to curate associated sectors of the data. Regardless of how this will be achieved, hosting cleaned georeferenced legume data will undoubtedly facilitate legume research, while attracting additional users seeking reliable locality data for studies of global environmental change, phenology, climatology and various ecological modelling studies (e.g. Delisle et al. 2003; Soltis 2017; Soltis et al. 2018; Lang et al. 2019). The importance of this type of use has, for example, been documented for the online Australasian Virtual Herbarium (Cantrill 2018). Ultimately, the quality of species-distribution data is reliant on availability of published specimen data and because a legume portal would not publish georeferenced specimen data to GBIF, individual legume researchers and herbaria would have to continue to publish their specimen data in standardised Darwin Core format through their national GBIF provider nodes. That said, the legume-systematics community could engage with GBIF to discuss possibilities for returning validated taxon lists and enhanced georeferenced data.
Genetic, morphological and trait data
Legume systematists also regularly use sequence data obtained from molecular databases (e.g. GenBank). Effective pipelines for large-scale retrieval of GenBank data of particular taxa or clades are also available (e.g. PhyLoTA, Sanderson et al. 2008; SUPERSMART, Antonelli et al. 2017, Bennett et al. 2018; PyPHLAWD, Smith and Walker 2019) and could be integrated into the legume portal. The legume-systematics community could contribute to GenBank by providing an accurate and up-to-date taxonomic backbone (species list plus classification) for use by people submitting sequences to GenBank and by reconciling existing GenBank accessions with this new checklist. Also, of strong interest is the need to aggregate data on legume morphology, functional traits (including nodulation), phenology, ecology, habitat and chemistry. A large part of these data types is already gathered by systematists, and some data are in databases such as TRY and MorphoBank, but they are usually not standardised or centralised. To be optimally useful for the legume-research community and other users, trait and morphological data would be aggregated in a legume portal.
Phylogenetic information
Trying to understand biodiversity without considering evolutionary relationships is like viewing fine mosaic artistry as a pile of its individual tiles, i.e. the bigger picture is lost to view. An ideal legume portal would include a phylogenetic browser that would integrate individual phylogenies and their datasets or enable an overview of legume evolutionary relationships by tree grafting from numerous studies. An OTOL API allows a user to input a list of taxa and receive back a subset of a synthetic tree that contains only the taxa of interest. A legume portal could take advantage of this technology by always accessing the latest edition of the OTOL synthetic tree at the Leguminosae node. An advantage is that this legume community would then take ‘ownership’ of the curation of the Leguminosae on OTOL. Areas of missing data or poor resolution can be identified by the legume community and published trees identified, uploaded and curated in Open Tree to fill the gaps, or new studies initiated to generate the missing data. In the future, it will also be possible to place trait data on nodes and terminals of the tree (e.g. see https://www.phyloref.org/, accessed 31 May 2019).
Species pages
In addition to these more dynamic types of data and specifically identified as important for evolutionary and systematics research in legumes, a summary of current knowledge of individual species or clades is an asset both for research and more general users. Theoretically, ‘taxon descriptions’ could be artificially constructed by synthesising and summarising detailed and highly structured data records, which could be continuously refreshed on the basis of the underlying data. However, few structured data exist. Thus, there is a need for manually crafted text descriptions, which are easily understood and disseminated, but costly to maintain in synchrony with underlying data. To facilitate this task, the data available in Legumes of the World Online could be resurrected for basic information on legume genera and we could use flora accounts of legume species and monographs, many of which are already digitised and available. The ideal legume portal would link to species information, providing up-to-date and updateable morphological descriptions, geographic distributions, images and bibliographic references. This information could be presented at different hierarchical or phylogenetic clade levels, portraying the information at the level searched for. For example, the geographic distribution (country or regional) of a genus could be an aggregate of the distributions of the species in the genus. The same could be done for morphology, particularly with the development of semantic descriptions. Ultimately, the legume portal could become the source of species information for use in platforms of general use, such as Wikipedia, GBIF, and COL.
Identification keys
Interactive digital keys for legumes would be useful for a broad range of users, particularly ‘citizen scientists’ and government employees. In surveying more widely the communities of plant taxonomists and portal builders (including FishBase and Neotropikey, as noted above), it becomes evident that (1) technical characters, especially if not illustrated, are not suitable for most non-specialist users, (2) traditional dichotomous keys rely on single decision sequences, which become ineffective if the specimen lacks key characters, e.g. no flowers, (3) building full matrices for DELTA (see http://www.delta-intkey.com/www/overview.htm, accessed 31 May 2019) or Lucid (see http://www.lucidcentral.org/, accessed 31 May 2019) systems are costly and time consuming to build well and maintain, but offer the possibility of multiple use and analysis of those data, (4) image-based gallery filtering systems, such as provided in eMonocots, iNaturalist, and JSTOR Global, have some benefits but can be inefficient and imprecise despite popularity with non-scientist communities and, (5) despite tremendous progress in the artificial intelligence, it is not yet possible to exclusively use artificial intelligence for reliable and accurate species identification. One possible approach is to use a limited number of easily recognisable diagnostic characteristics to guide users through the portal. To work, species must be provided with short, coded descriptions including diagnostic characteristics (e.g. habit, leaf position and type, flower colour, geographical range, and uses), which could also be useful for and provided by the legume-systematics community. Exhaustive glossaries of legume terms exist already and could be translated into accepted ontological terms (e.g. Planteome, see http://planteome.org/, accessed 31 May 2019). Providing a complete glossary with images and linking it to the short descriptions would facilitate identification for an even wider range of user communities, and this can be achieved using tools designed for interactive description and identification (e.g. Xper3, see http://www.xper3.fr/, accessed 31 May 2019).
Appropriate technologies and tools for a dynamic legume portal
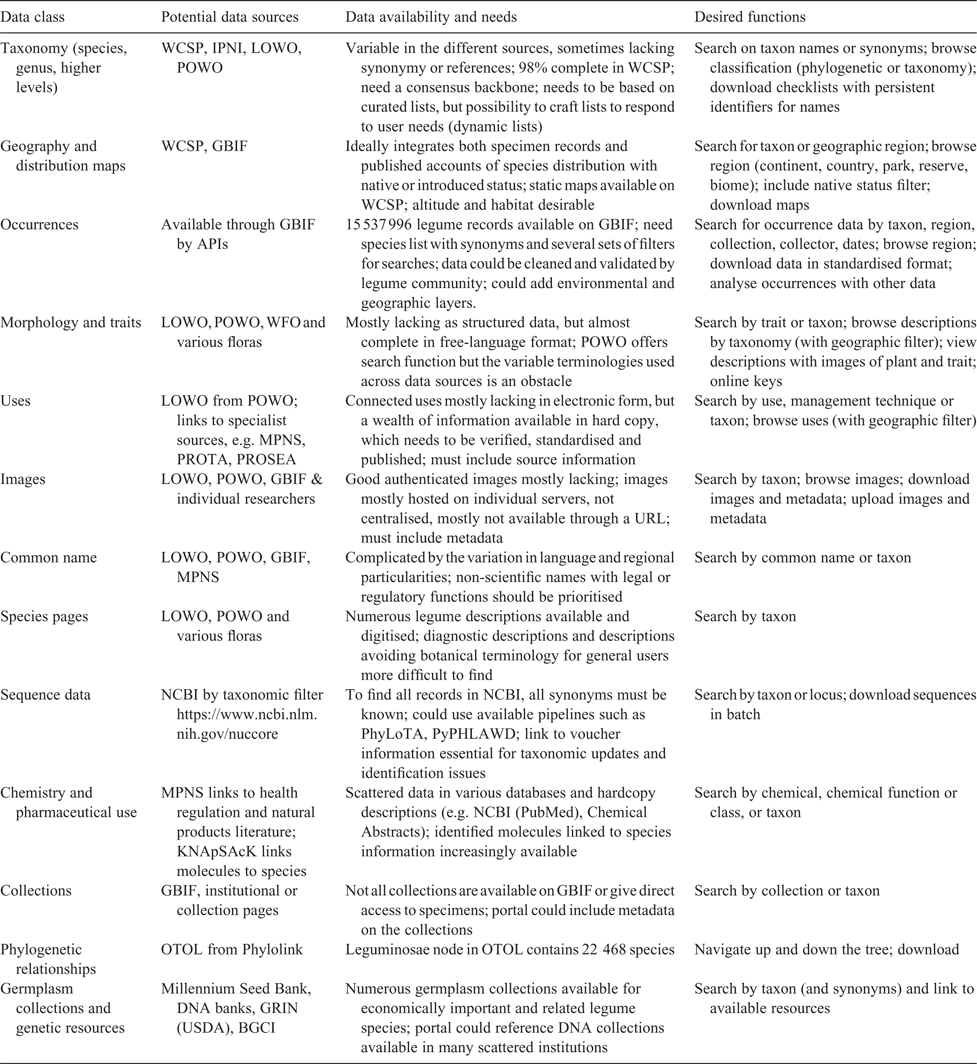
We have highlighted how taxon-based portals still have an important role in biological-data sharing and how a legume portal could be useful for the systematics community, and beyond. However, we still need to devise a plan to build a system that would be relevant, and that is scalable and sustainable. In this context, it is clear that information should be aggregated around specimens and the taxonomic classification system (Fig. 1), using accepted names and their synonyms, and, ideally, with internationally accepted unique identifiers for names and specimens. The modular nature of the portal would allow people to bypass taxonomy, but focusing on names facilitates data curation, which is the most desired feature of such an information system. To develop a legume portal, it is, first, necessary to define a basic model on top of which future developments can be made. Construction of the model relies on the following four main steps: (1) definition of the type of information to aggregate; (2) selection of online data sources; (3) development of scripts to centralise the information; and (4) production of an online graphical user interface (GUI) to retrieve and visualise information. In Table 1, we summarise potential data sources and functions available for data harvesting or for direct points to existing resources (see also Fig. 1, 2).
A broad selection and large number of organismic data are available; however, it will be necessary to select a smaller number of data types to begin with. Considering the nature of the portal, data on species names, occurrence data, DNA sequences, and morphological data are probably the most relevant in an initial phase (Table 1). These data can be automatically retrieved from existing databases, as has been done for Arabidopsis Heynh. proteomics data (Joshi et al. 2011), using data-harvesting tools. This method facilitates data aggregation; however, it is important to define priorities based on user needs.
It is important to remain open minded about how and where the tools for curating and linking these data are built. Provided a well-defined data structure is established and documented, then multiple tools might allow different sectors of the scientific community to contribute with their own expertise and data. These tools can operate in parallel and be replaced with improved tools over time. We should also avoid the assumption that a single dissemination interface will meet the needs of all audiences. A multiplicity of views for one set of data is feasible and only requires a modest investment. The significant costs are in collation, integration and curation of the data; ‘publishing’ that information is cheap, provided the data are reliable and well structured.
Data harvested from different sources will need to be stored in a central database, so as to provide easier and faster access to the information. This highlights the importance of having a central data store that is structured to meet the diverse needs and is well documented, and to develop relationships with existing data suppliers to permit automatic data extraction. Use and implementation of universally unique identifiers across different platforms could make the legume portal a workbench to more easily integrate data from different sources. Another recurring issue is long-term maintenance of the portal, which is necessary to keep up with informatics updates in the source databases (Stein 2003) as well as data content. As tools developed for a legume portal could be generic for any taxon and, thus, of wider interest, broad architectural planning could ease sustainability. Informatics tools that answer to the needs of the community are also likely to be available through open access and can be adopted, as can APIs that facilitate the use of services or gathering of data from public databases. Finally, collaboration between legume systematists and data scientists could facilitate writing of scripts and support of the cyber-infrastructure needed to create a database useful for answering varied scientific questions, as well as for other potential users.
Continued usage depends, in large part, on the ability of users to query, find and extract information and data easily and freely (Hobern et al. 2019). Because research objectives vary considerably and evolve quickly, data-export functions must be available for researchers to use and analyse the data as required. Thus, even though creation of a web interface is the last step of this development model, it is crucial for increasing the lifespan of a data portal. Thus, user experience, both of the systematics community and the broader public, has to be thoroughly explored during interface design. It is important that the information searched for, browsed through or downloaded is as up-to-date as possible (Table 1, Fig. 2). Although this can be a technological challenge, we can imagine a system that displays more stable data (e.g. morphological descriptions, images, general distribution) in a static but easily usable and updatable manner, while giving access to real-time or near real-time data from external datasets that change more rapidly. External data can be renewed on a regular basis by using APIs when available and when the data requests are not too voluminous, or by locally storing data when a user queries specific information (e.g. georeferenced specimen data from Vicia on GBIF).
Other functionalities can be included in subsequent versions of the portal. Legume researchers have expressed the desire to report back errors and to contribute new or cleaned data to data providers (Fig. 1). This feedback loop is important for community endorsement, so as to encourage involvement, and it can be seen as a contribution of the legume-systematics community to important data providers, such as GBIF, IPNI, GenBank, WCSP, The World Flora Online and POWO. Implementation of data cleaning, data transformation (when necessary) and export would, thus, be central to the portal development. The two last steps of this chain have to be automatic, whereas data cleaning can also be undertaken by users with available tools and pipelines (e.g. OpenRefine and R software packages). Human curation highlights the need to have unique identifiers both for data and people. Unique identifiers provide a means of tracking changes, crediting work, and contribute to a coherent database (Nelson et al. 2018). Additional steps during refinement of the portal would include increased interaction between datasets by APIs and implementation of controlled vocabularies and ontologies to facilitate data harvesting.
Creating and documenting a data model for a legume portal that contains a mixture of data curated locally and data harvested from elsewhere could enhance sustainability. This mixed model would allow third parties to provide data-curation or data-harvesting tools; allow different views of the data to be built for specific audiences; permit adoption of the same data standards, structures and software tools for other plant families (thus, reducing costs); and facilitate evolution of the software employed for curating and deploying the data, thereby reducing the risks of failure when technological progress requires upgrades to a large single system. Ideally, the portal would evolve together with technological advances, but it is important to keep in mind that curation and ease of use are central to connect taxonomists, citizens and data scientists. With that goal, we may be able to develop a sustainable legume portal.
Organisation, people, resources and sustainability
After outlining a development model, we can plan how to achieve our goal. Considering the task at hand, namely, to build an information portal for the third-largest family of flowering plants, we propose to use a working-group model followed by a design-sprint approach.
A working group will need to be established, with approximately a dozen people from varied institutions around the world, representing researchers with different expertise in systematics and including biodiversity informaticians and data programmers. Workshops will be organised to address issues noted above and to establish (1) target audiences and needs, (2) what data and tools we have and what we are missing, (3) benefits to users and to community building, (4) essential resources, business model and financial challenges, (5) a vision for long-term sustainability and (6) a governance model.
Once these important issues have been clarified, we can use design sprint to arrive at a prototype. In design sprint, a group of creators get together to develop a desirable product (Banfield et al. 2015; Knapp et al. 2016). The goal is to design the product, as well as to build and test a prototype, with a small set of users, particularly looking for flaws that would lead to failure of the project (Knapp et al. 2016). The creators finally move to actual production, or to fixing design problems, through an iteration of the design sprint. Once a prototype has been developed, it will need to be tested by a broad range of users with diverse expertise, so we can be sure the portal meets the needs of the legume-systematics community, as well as those of more general users. Regardless of the approach, the legume-portal development team will need to be fully aware of international initiatives in biodiversity informatics, so as to adapt existing and pertinent tools, pipelines and approaches, and also to contribute to ongoing developments in this field
Conclusions
Now that many family classifications have been reworked to reflect monophyletic lineages, different types of data, such as geographical distributions, morphological traits and phenologies, can be integrated in an evolutionary framework through a taxon portal. Because systematists have the knowledge and the need to aggregate information around specimens (vouchers) and species names, a reliable, verifiable and connected information system will be created. A legume portal that integrates scientifically validated information on one of the most economically and ecologically important plant families would become an important tool and source of data for different user communities, from researchers to government officials, environmental consultants, and the general public. Also, data cleaning performed by the legume portal could feedback to large international data aggregators, improving data quality in these databases and overcoming redundant-data validation by different researchers. As with many biodiversity data and taxon-centric portals, the biggest challenge for a legume portal will be long-term sustainability and continued relevance. This requires a scalable model that is flexible in terms of sources of data harvested, programming language and informatics tools and, most of all, that remains endorsed and supported by users and by the legume-systematics community.
Conflicts of interest
Daniel J. Murphy is the Editor-in-Chief for Australian Systematic Botany and Ashley N. Egan is a guest editor for this special issue. Despite this relationship, they did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making process for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
This research did not receive any specific funding; however, support for preparation of this manuscript was indirectly provided by the Natural Sciences and Engineering Research Council of Canada.
Acknowledgements
This paper stems from a workshop organised by Yasuhiro Kubota (University of the Ryukyus), Félix Forest (Royal Botanic Gardens, Kew) and Firouzeh Javadi (Kyushu University) during the 7th International Legume Conference held in Sendai, Japan, in August 2018. We thank Danilo Oliveira for discussions on portal development, Luc Brouillet for discussion on taxonomic resources, and Patrick Herendeen, Richard White, Donald Hobern, Jan Wieringa and Colin Hughes for interesting input on an earlier version of the manuscript.
References
Adey ME, Allkin R, Bisby FA, White RJ, Macfarlane TD (1984) The Vicieae database: an experimental taxonomic monograph. In ‘Databases in Systematics’. (Eds R Allkin, FA Bisby) Systematics Association Special Volume 26, pp. 175–188. (Academic Press: London, UK)Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, Saito K (2012) KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant & Cell Physiology 53, e1
| KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research.Crossref | GoogleScholarGoogle Scholar |
Allkin R (1984) Handling taxonomic descriptions by computer. In ‘Databases in Systematics’. (Eds R Allkin, FA Bisby) Systematics Association Special Volume 26, pp. 263–278. (Academic Press: London, UK)
Allkin R, White RJ (1988) Data management models for biological classification. In ‘Classification and related methods of data analysis’. (Ed. HH Bock) pp. 653–402. (Elsevier: Amsterdam, Netherlands)
Allkin R, White RJ (1993) XDF Data exchange format. In ‘Advances in Computer Methods for Systematic Biology: Artificial Intelligence, Databases and Computer Vision’. (Ed. R Fortuner) pp. 474–475. (The John Hopkins University Press: Baltimore, MD, USA)
Allkin R, Winfield PJ (1993) Software development strategies for global plant information systems. In ‘Designs for a Global Plant Information System’. (Eds FA Bisby, GF Russell, RJ Pankhurst) pp. 304–318. (Academic Press: London, UK)
Allkin R, White RJ, Winfield PJ (1992) Handling the taxonomic structure of biological data. Mathematical and Computer Modelling 16, 1–9.
| Handling the taxonomic structure of biological data.Crossref | GoogleScholarGoogle Scholar |
Angiosperm Phylogeny Group (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161, 105–121.
| An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III.Crossref | GoogleScholarGoogle Scholar |
Antonelli A, Hettling H, Condamine FL, Vos K, Nilsson RH, Sanderson MJ, Sauquet H, Scharn R, Silvestro D, Töpel M, Bacon CD, Oxelman B, Vos RA (2017) Toward a self-updating platform for estimating rates of speciation and migration, ages, and relationships of taxa. Systematic Biology 66, 152–166.
| Toward a self-updating platform for estimating rates of speciation and migration, ages, and relationships of taxa.Crossref | GoogleScholarGoogle Scholar | 27616324PubMed |
Banfield R, Lombardo CT, Wax T (2015) ‘Design Sprint: a Practical Guidebook for Building Great Digital Products.’ (O’Reilly Media, Inc.: Sebastopol, CA, USA)
Bennett D, Hettling H, Silvestro D, Zizka A, Bacon C, Faurby S, Vos R, Antonelli A (2018) phylotaR: an automated pipeline for retrieving orthologous DNA sequences from GenBank in R. Life 8, 20
| phylotaR: an automated pipeline for retrieving orthologous DNA sequences from GenBank in R.Crossref | GoogleScholarGoogle Scholar |
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Research 41, D36–D42.
| GenBank.Crossref | GoogleScholarGoogle Scholar | 23193287PubMed |
Berendsohn WG, Güntsch A, Hoffmann N, Kohlbecker A, Luther K, Müller A (2011) Biodiversity information platforms: from standards to interoperability. ZooKeys 150, 71–87.
| Biodiversity information platforms: from standards to interoperability.Crossref | GoogleScholarGoogle Scholar |
Binggeli P (1996) A taxonomic, biogeographical and ecological overview of invasive woody plants. Journal of Vegetation Science 7, 121–124.
| A taxonomic, biogeographical and ecological overview of invasive woody plants.Crossref | GoogleScholarGoogle Scholar |
Bisby FA (1993) Botanical strategies for compiling a global plant checklist. In ‘Designs for a Global Plant Information System’. (Eds FA Bisby, GF Russell, G RJ Pankhurst) pp. 145–157. (Academic Press: London, UK)
Bisby FA (2000) The quiet revolution: biodiversity informatics and the internet. Science 289, 2309–2312.
| The quiet revolution: biodiversity informatics and the internet.Crossref | GoogleScholarGoogle Scholar | 11009408PubMed |
Bisby FA, Buckingham J, Harborne JB (1994) ‘Phytochemical Dictionary of the Leguminosae.’ (Chapman & Hall: London, UK)
Bisby FA, Ruggiero MA, Roskov YR, Cachuela-Palacio M, Kimani SW, Kirk PM, Soulier-Perkins A, van Hertum J (2006) ‘Species 2000 & ITIS Catalogue of Life: 2006 Annual Checklist. CD-ROM, Species 2000.’ (University of Reading: Reading, UK)
Bowser A, Wiggins A, Shanley L, Preece J, Henderson S (2014) Sharing data while protecting privacy in citizen science. Interaction 21, 70–73.
| Sharing data while protecting privacy in citizen science.Crossref | GoogleScholarGoogle Scholar |
Bridge PD, Roberts PJ, Spooner BM, Panchal G (2003) On the unreliability of published DNA sequences. New Phytologist 160, 43–48.
| On the unreliability of published DNA sequences.Crossref | GoogleScholarGoogle Scholar |
Butler D (2006) Mashups mix data into global service. Nature 439, 6–7.
| Mashups mix data into global service.Crossref | GoogleScholarGoogle Scholar | 16397468PubMed |
Buttigieg PL, Pafilis E, Lewis SE, Schildhauer MP, Walls RL, Mungall CJ (2016) The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation. Journal of Biomedical Semantics 7, 57
| The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation.Crossref | GoogleScholarGoogle Scholar | 27664130PubMed |
Cantrill DJ (2018) The Australasian virtual herbarium: tracking data usage and benefits for biological collections. Applications in Plant Sciences 6, e1026
| The Australasian virtual herbarium: tracking data usage and benefits for biological collections.Crossref | GoogleScholarGoogle Scholar | 29732257PubMed |
Cayuela L, Granzow-de la Cerda I, Albuquerque FS, Golicher JD (2012) Taxonstand: an R package for species names standardization in vegetation databases. Methods in Ecology and Evolution 3, 1078–1083.
| Taxonstand: an R package for species names standardization in vegetation databases.Crossref | GoogleScholarGoogle Scholar |
Chard K, Dart E, Foster I, Shifflett D, Tuecke S, Williams J (2018) The modern research data portal: a design pattern for networked, data-intensive science. PeerJ – Computer Science 4, e144
| The modern research data portal: a design pattern for networked, data-intensive science.Crossref | GoogleScholarGoogle Scholar |
Cicero C, Spencer CL, Bloom DA, Guralnick RP, Koo MS, Otegui J, Russell LA, Wieczorek JR (2017) Biodiversity informatics and data quality on a global scale. In ‘The Extended Specimen: Emerging Frontiers in Collections-based Ornithological Research. Studies in Avian Biology, number 50’. (Ed. MS Webster) pp. 201–218. (CRC Press: Boca Raton, FL, USA)
Conte MG, Gaillard S, Lanau N, Rouard M, Périn C (2008) GreenPhylDB: a database for plant comparative genomics. Nucleic Acids Research 36, D991–D998.
| GreenPhylDB: a database for plant comparative genomics.Crossref | GoogleScholarGoogle Scholar | 17986457PubMed |
Costello M, Michener W, Gahegan M, Zhang Z-Q, Bourne P (2013) Biodiversity data should be published, cited, and peer reviewed. Trends in Ecology & Evolution 28, 454–461.
| Biodiversity data should be published, cited, and peer reviewed.Crossref | GoogleScholarGoogle Scholar |
Dallwitz MJ (1993) DELTA and INTKEY. In ‘Advances in Computer Methods for Systematic Biology: Artificial Intelligence, Databases, Computer Vision’. (Ed. R Fortuner) pp. 287–296. (The Johns Hopkins University Press: Baltimore, MD, USA)
Dash S, Campbell J, Cannon E, Cleary A, Huang W, Kalberer S, Karingula V, Rice A, Singh J, Umale P, Weeks N, Wilkey A, Farmer A, Cannon S (2016) Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Research 44, D1181–D1188.
| Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family.Crossref | GoogleScholarGoogle Scholar | 26546515PubMed |
Deans AR, Lewis SE, Huala E, Anzaldo SS, Ashburner M, Balhoff JP, Blackburn DC, Blake JA, Burleigh JG, Chanet B, Cooper LD, Courtot M, Csösz S, Cui H, Dahdul W, Das S, Dececchi TA, Dettai A, Diogo R, Druzinsky RE, Dumontier M, Franz NM, Friedrich F, Gkoutos GV, Haendel M, Harmon LJ, Hayamizu TF, He Y, Hines HM, Ibrahim N, Jackson LM, Jaiswal P, James-Zorn C, Köhler S, Lecointre G, Lapp H, Lawrence CJ, Le Novère N, Lundberg JG, Macklin J, Mast AR, Midford PE, Mikó I, Mungall CJ, Oellrich A, Osumi-Sutherland D, Parkinson H, Ramírez MJ, Richter S, Robinson PN, Ruttenberg A, Schulz KS, Segerdell E, Seltmann KC, Sharkey MJ, Smith AD, Smith B, Specht CD, Squires RB, Thacker RW, Thessen A, Fernandez-Triana J, Vihinen M, Vize PD, Vogt L, Wall CE, Walls RL, Westerfeld M, Wharton RA, Wirkner CS, Woolley JB, Yoder MJ, Zorn AM, Mabee P (2015) Finding our way through phenotypes. PLoS Biology 13, e1002033
| Finding our way through phenotypes.Crossref | GoogleScholarGoogle Scholar | 25562316PubMed |
Delisle F, Lavoie C, Jean M, Lachance D (2003) Reconstructing the spread of invasive plants: taking into account biases associated with herbarium specimens. Journal of Biogeography 30, 1033–1042.
| Reconstructing the spread of invasive plants: taking into account biases associated with herbarium specimens.Crossref | GoogleScholarGoogle Scholar |
Dressler S, Schmidt M, Zizka G (2014) Introducing African plants: a photo guide – an interactive photo data-base and rapid identification tool for continental Africa. Taxon 63, 1159–1161.
| Introducing African plants: a photo guide – an interactive photo data-base and rapid identification tool for continental Africa.Crossref | GoogleScholarGoogle Scholar |
Faria SM, Lewis GP, Sprent JI, Sutherland JM (1989) Occurrence of nodulation in the Leguminosae. New Phytologist 111, 607–619.
| Occurrence of nodulation in the Leguminosae.Crossref | GoogleScholarGoogle Scholar |
Fecher B, Friesike S, Hebing M (2015) What drives academic data sharing? PLoS One 10, e0118053
| What drives academic data sharing?Crossref | GoogleScholarGoogle Scholar | 25714752PubMed |
Gardiner LM, Bachman SP (2016) The role of citizen science in a global assessment of extinction risk in palms (Arecaceae). Botanical Journal of the Linnean Society 182, 543–550.
| The role of citizen science in a global assessment of extinction risk in palms (Arecaceae).Crossref | GoogleScholarGoogle Scholar |
Gewin V (2002) All living things, online. Nature 418, 362–363.
| All living things, online.Crossref | GoogleScholarGoogle Scholar | 12140529PubMed |
Godfray HCJ (2002) Challenges for taxonomy. Nature 417, 17–19.
| Challenges for taxonomy.Crossref | GoogleScholarGoogle Scholar |
Gonzales M, Archuleta E, Farmer A, Gajendran K, Grant D, Shoemaker R, Beavis W, Waugh M (2005) The legume information system (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Research 33, D660–D665.
| The legume information system (LIS): an integrated information resource for comparative legume biology.Crossref | GoogleScholarGoogle Scholar | 15608283PubMed |
Gunn CR (1984) Fruits and seeds of genera in the subfamily Mimosoideae (Fabaceae). Technical bulletin number 1681. USDA Agricultural Research Service, Washington, DC, USA.
Gunn CR (1991) Fruits and seeds of genera in the subfamily Caesalpinioideae (Fabaceae). Technical bulletin number 1755, USDA Agricultural Research Service, Washington, DC, USA.
Heaton L (2018) Introduction. In ‘La reconfiguration du travail scientifique en biodiversité, Pratiques amateurs et technologies numériques’. (Eds L Heaton, F Miller, PD da Silva, S Proulx) pp. 9–29. (Les Presses de l’Université de Montréal: Montréal, QC, Canada)
Hinchliff CE, Smith SA, Allman JF, Burleigh JG, Chaudhary R, Coghill LM, Crandall KA, Deng J, Drew BT, Gazis R, Gude K, Hibbett DS, Katz LA, Laughinghouse HD Hinchliff CE, Smith SA, Allman JF, Burleigh JG, Chaudhary R, Coghill LM, Crandall KA, Deng J, Drew BT, Gazis R, Gude K, Hibbett DS, Katz LA, Laughinghouse HD (2015) Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proceedings of the National Academy of Sciences of the United States of America 112, 12764–12769.
| Synthesis of phylogeny and taxonomy into a comprehensive tree of life.Crossref | GoogleScholarGoogle Scholar | 26385966PubMed |
Hobern D, Apostolico A, Arnaud E, Bello JC, Canhos D, Dubois G, Field D, Alonso Garcia E, Hardisty A, Harrison J, Heidorn B, Krishtalka L, Mata E, Page RDM, Parr C, Price J, Willoughby S (2012) ‘Global Biodiversity Informatics Outlook: Delivering Biodiversity Knowledge in the Information Age.’ (Global Biodiversity Information Facility: Copenhagen, Denmark).
Hobern D, Baptiste B, Copas K, Guralnick R, Hahn A, van Huis E, Kim ES, McGeoch M, Naicker I, Navarro L, Noesgaard D, Price M, Rodrigues A, Schigel D, Sheffield CA, Wieczorek J (2019) Connecting data and expertise: a new alliance for biodiversity knowledge. Biodiversity Data Journal 7, e33679
| Connecting data and expertise: a new alliance for biodiversity knowledge.Crossref | GoogleScholarGoogle Scholar | 30886531PubMed |
Hollis S, Brummitt R (1992) ‘World Geographical Scheme for Recording Plant Distributions. Plant Taxonomic Database Standards Number 2. International Working Group on Taxonomic Databases for Plant Sciences (TDWG).’ (Hunt Institute for Botanical Documentation: Pittsburgh, PA, USA)
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T (2010) MassBank: a public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry 45, 703–714.
| MassBank: a public repository for sharing mass spectral data for life sciences.Crossref | GoogleScholarGoogle Scholar | 20623627PubMed |
Jolley-Rogers G, Varghese T, Harvey P, dos Remedios N, Miller JT (2014) Phylojive: integrating biodiversity data with the tree of life. Bioinformatics 30, 1308–1309.
| Phylojive: integrating biodiversity data with the tree of life.Crossref | GoogleScholarGoogle Scholar | 24443378PubMed |
Joshi HJ, Hirsch-Hoffmann M, Baerenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk KJ, Egelhofer V, Wienkoop S, Weckwerth W, Bruley C, Rolland N, Toyoda T, Nakagami H, Jones AM, Briggs SP, Castleden I, Tanz SK, Millar AH, Heazlewood JL (2011) MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiology 155, 259–270.
| MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data.Crossref | GoogleScholarGoogle Scholar | 21075962PubMed |
Kattge J, Díaz S, Lavorel S, Prentice IC, Leadley P, Bönisch G, Garnier E, Westoby M, Reich PB, Wright IJ, Cornelissen JHC, Violle C, Harrison SP, Van Bodegom PM, Reichstein M, Enquist BJ, Soudzilovskaia NA, Ackerly DD, Anand M, Atkin O, Bahn M, Baker TR, Baldocchi D, Bekker R, Blanco C, Blonder B, Bond WJ, Bradstock R, Bunker DE, Casanoves F, Cavender-Bares J, Chambers JQ, Chapin FS, Chave J, Coomes D, Cornwell WK, Craine JM, Dobrin BH, Duarte L, Durka W, Elser J, Esser G, Estiarte M, Fagan WF, Fang J, Fernández-Méndez F, Fidelis A, Finegan B, Flores O, Ford H, Frank D, Freschet GT, Fyllas NM, Gallagher RV, Green WA, Gutierrez AG, Hickler T, Higgins S, Hodgson JG, Jalili A, Jansen S, Joly C, Kerkhoff AJ, Kirkup D, Kitajima K, Kleyer M, Klotz S, Knops JMH, Kramer K, Kühn I, Kurokawa H, Laughlin D, Lee TD, Leishman M, Lens F, Lenz T, Lewis SL, Lloyd J, Llusià J, Louault F, Ma S, Mahecha MD, Manning P, Massad T, Medlyn B, Messier J, Moles AT, Müller SC, Nadrowski K, Naeem S, Niinemets Ü, Nöllert S, Nüske A, Ogaya R, Oleksyn J, Onipchenko VG, Onoda Y, Ordoñez J, Overbeck G, Ozinga WA, Patiño S, Paula S, Pausas JG, Peñuelas J, Phillips OL, Pillar V, Poorter H, Poorter L, Poschlod P, Prinzing A, Proulx R, Rammig A, Reinsch S, Reu B, Sack L, Salgado-Negret B, Sardans J, Shiodera S, Shipley B, Siefert A, Sosinski E, Soussana J-F, Swaine E, Swenson N, Thompson K, Thornton P, Waldram M, Weiher E, White M, White S, Wright SJ, Yguel B, Zaehle S, Zanne AE, Wirth C (2011) TRY: a global database of plant traits. Global Change Biology 17, 2905–2935.
| TRY: a global database of plant traits.Crossref | GoogleScholarGoogle Scholar |
Kirkbride JH Jr, Gunn CR, Weitzman AL (2003a) Fruits and seeds of genera in the subfamily Faboideae (Fabaceae), Vol. I. Technical bulletin number 1890, USDA Agricultural Research Service, Washington, DC, USA.
Kirkbride JH Jr, Gunn CR, Weitzman AL (2003b) Fruits and seeds of genera in the subfamily Faboideae (Fabaceae), Vol. II. Technical bulletin number 1890, USDA Agricultural Research Service, Washington, DC, USA.
Knapp J, Zeratsky J, Kowitz B (2016) ‘Sprint: How to Solve Big Problems and Test New Ideas in Just Five days.’ (Simon and Schuster: New York, NY, USA)
Kress WJ, Garcia-Robledo C, Soares JVB, Jacobs D, Wilson K, Lopez IC, Belhumeur PN (2018) Citizen science and climate change: mapping the range expansions of native and exotic plants with the mobile app Leafsnap. Bioscience 68, 348–358.
| Citizen science and climate change: mapping the range expansions of native and exotic plants with the mobile app Leafsnap.Crossref | GoogleScholarGoogle Scholar |
Kumar N, Belhumeur PN, Biswas A, Jacobs DW, Kress WJ, Lopez IC, Soares JV (2012) Leafsnap: a computer vision system for automatic plant species identification. In ‘Computer Vision: ECCV 2012’. (Eds A Fitzgibbon, S Lazebnik, P Perona, Y Sato, C Schmid) pp. 502–516. (Springer: Berlin, Germany)
Lang PL, Willems FM, Scheepens JF, Burbano HA, Bossdorf O (2019) Using herbaria to study global environmental change. New Phytologist 221, 110–122.
| Using herbaria to study global environmental change.Crossref | GoogleScholarGoogle Scholar | 30160314PubMed |
Lawler A (2001) Up for the count? Science 294, 769–770.
| Up for the count?Crossref | GoogleScholarGoogle Scholar | 11679647PubMed |
Legume Phylogeny Working Group (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77.
| A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny.Crossref | GoogleScholarGoogle Scholar |
Lewis GP, Schrire B, Mackinder B, Lock M (Eds) (2005) ‘Legumes of the World.’ (Royal Botanic Gardens, Kew: London, UK)
Li J, Dai X, Liu T, Zhao PX (2012) LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Research 40, D1221–D1229.
| LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes.Crossref | GoogleScholarGoogle Scholar | 22110036PubMed |
Lock JM (1989) ‘Legumes of Africa: a Checklist.’ (Royal Botanic Gardens, Kew: London, UK)
Meineke EK, Davies TJ, Daru BH, Davis CC (2018) Biological collections for understanding biodiversity in the Anthropocene. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 374, 20170386
| Biological collections for understanding biodiversity in the Anthropocene.Crossref | GoogleScholarGoogle Scholar | 30455211PubMed |
Michener WK (2015) Ecological data sharing. Ecological Informatics 29, 33–44.
| Ecological data sharing.Crossref | GoogleScholarGoogle Scholar |
Michonneau F, Brown JW, Winter DJ (2016) rotl: an R package to interact with the Open Tree of Life data. Methods in Ecology and Evolution 7, 1476–1481.
| rotl: an R package to interact with the Open Tree of Life data.Crossref | GoogleScholarGoogle Scholar |
Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O’Leary MA (2015) A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evolutionary Bioinformatics Online 11, 43–48.
| A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway.Crossref | GoogleScholarGoogle Scholar | 25861210PubMed |
Miller JT, Pirzl R, Rosauer D, Jolley-Rogers G, Varghese T (2019) Phylolink: phylogenetically based profiling, visualisations and metrics for biodiversity. Bioinformatics 35, 1229–1230.
| Phylolink: phylogenetically based profiling, visualisations and metrics for biodiversity.Crossref | GoogleScholarGoogle Scholar | 30202854PubMed |
Nelson G, Ellis S (2018) The history and impact of digitization and digital data mobilization on biodiversity research. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 374, 20170391
| The history and impact of digitization and digital data mobilization on biodiversity research.Crossref | GoogleScholarGoogle Scholar | 30455209PubMed |
Nelson G, Sweeney P, Gilbert E (2018) Use of globally unique identifiers (GUIDs) to link herbarium specimen records to physical specimens. Applications in Plant Sciences 6, e1027
| Use of globally unique identifiers (GUIDs) to link herbarium specimen records to physical specimens.Crossref | GoogleScholarGoogle Scholar | 29732258PubMed |
O’Leary MA, Kaufman S (2011) MorphoBank: phylophenomics in the ‘cloud’. Cladistics 27, 529–537.
| MorphoBank: phylophenomics in the ‘cloud’.Crossref | GoogleScholarGoogle Scholar |
Parr CL, Dunn RR, Sanders NJ, Weiser MD, Photakis M, Bishop TR, Fitzpatrick MC, Arnan X, Baccaro F, Brandão CR, Chick L, Donoso DA, Fayle TM, Gómez C, Grossman B, Munyai TC, Pacheco R, Retana J, Robinson A, Sagata K, Silva RR, Tista M, Vasconcelos H, Yates M, Gibb H (2017) GlobalAnts: a new database on the geography of ant traits (Hymenoptera: Formicidae). Insect Conservation and Diversity 10, 5–20.
| GlobalAnts: a new database on the geography of ant traits (Hymenoptera: Formicidae).Crossref | GoogleScholarGoogle Scholar |
Penev L, Mietchen D, Chavan V, Hagedorn G, Smith V, Shotton D, Ó Tuama É, Senderov V, Georgiev T, Stoev P, Groom Q, Remsen D, Edmunds S (2017) Strategies and guidelines for scholarly publishing of biodiversity data. Research Ideas and Outcomes 3, e12431
| Strategies and guidelines for scholarly publishing of biodiversity data.Crossref | GoogleScholarGoogle Scholar |
Poisot T, Bruneau A, Gonzalez A, Gravel D, Peres-Neto P (2019) Ecological data should not be so hard to find and reuse. Trends in Ecology & Evolution 34, 494–496.
| Ecological data should not be so hard to find and reuse.Crossref | GoogleScholarGoogle Scholar |
Ratnasingham S, Hebert P (2007) BoLD: the barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes 7, 355–364.
| BoLD: the barcode of life data system (http://www.barcodinglife.org).Crossref | http://www.barcodinglife.org).&journal=Molecular Ecology Notes&volume=7&pages=355-364&publication_year=2007&author=S%20Ratnasingham&hl=en&doi=10.1111/j.1471-8286.2007.01678.x" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | 18784790PubMed |
Rees J, Cranston K (2017) Automated assembly of a reference taxonomy for phylogenetic data synthesis. Biodiversity Data Journal 5, e12581
| Automated assembly of a reference taxonomy for phylogenetic data synthesis.Crossref | GoogleScholarGoogle Scholar |
Rosindell J, Harmon LJ (2012) OneZoom: a fractal explorer for the tree of life. PLoS Biology 10, e1001406
| OneZoom: a fractal explorer for the tree of life.Crossref | GoogleScholarGoogle Scholar | 23091419PubMed |
Sanderson MJ, Boss D, Chen D, Cranston KA, Wehe A (2008) The PhyLoTA browser: processing GenBank for molecular phylogenetics research. Systematic Biology 57, 335–346.
| The PhyLoTA browser: processing GenBank for molecular phylogenetics research.Crossref | GoogleScholarGoogle Scholar | 18570030PubMed |
Schuettpelz E, Frandsen PB, Dikow RB, Brown A, Orli S, Peters M, Metallo A, Funk VA, Dorr LJ (2017) Applications of deep convolutional neural networks to digitized natural history collections. Biodiversity Data Journal 5, e21139
| Applications of deep convolutional neural networks to digitized natural history collections.Crossref | GoogleScholarGoogle Scholar |
Smith S, Walker J (2019) PyPHLAWD: a python tool for phylogenetic dataset construction. Methods in Ecology and Evolution 10, 104–108.
| PyPHLAWD: a python tool for phylogenetic dataset construction.Crossref | GoogleScholarGoogle Scholar |
Soltis PS (2017) Digitization of herbaria enables novel research. American Journal of Botany 104, 1281–1284.
| Digitization of herbaria enables novel research.Crossref | GoogleScholarGoogle Scholar | 29885238PubMed |
Soltis PS, Nelson G, James SA (2018) Green digitization: online botanical collections data answering real-world questions. Applications in Plant Sciences 6, e1028
| Green digitization: online botanical collections data answering real-world questions.Crossref | GoogleScholarGoogle Scholar | 29732255PubMed |
Sprent JI (2001) ‘Nodulation in Legumes.’ (Royal Botanic Gardens, Kew: London, UK)
Stein LD (2003) Integrating biological databases. Nature Reviews – Genetics 4, 337–345.
| Integrating biological databases.Crossref | GoogleScholarGoogle Scholar | 12728276PubMed |
Tedersoo L, Laanisto L, Rahimlou S, Toussaint A, Hallikma T, Pärtel M (2018) Global database of plants with root‐symbiotic nitrogen fixation: Nod DB. Journal of Vegetation Science 29, 560–568.
| Global database of plants with root‐symbiotic nitrogen fixation: Nod DB.Crossref | GoogleScholarGoogle Scholar |
Tenopir C, Allard S, Douglass K, Aydinoglu AU, Wu L, Read E, Manoff M, Frame M (2011) Data sharing by scientists: practices and perceptions. PLoS One 6, e21101
| Data sharing by scientists: practices and perceptions.Crossref | GoogleScholarGoogle Scholar | 21738610PubMed |
Unger J, Merhof D, Renner S (2016) Computer vision applied to herbarium specimens of German trees: testing the future utility of the millions of herbarium specimen images for automated identification. BMC Evolutionary Biology 16, 248
| Computer vision applied to herbarium specimens of German trees: testing the future utility of the millions of herbarium specimen images for automated identification.Crossref | GoogleScholarGoogle Scholar | 27852219PubMed |
van Horn G, Mac Aodha O, Song Y, Shepard A, Adam H, Perona P, Belongie S (2017) The iNaturalist challenge 2017 dataset. Available at http://arxiv.org/abs/1707.06642 [Verified 31 May 2019]
van Kleunen M, Dawson W, Essl F, Pergl J, Winter M, Weber E, Kreft H, Weigelt P, Kartesz J, Nishino M, Antonova LA, Barcelona JF, Cabezas FJ, Cárdenas D, Cárdenas-Toro J, Castano N, Chacón E, Chatelain C, Ebel AL, Figueiredo E, Fuentes N, Groom QJ, Henderson L, Inderjit , Kupriyanov A, Masciadri S, Meerman J, Morozova O, Moser D, Nickrent DL, Patzelt A, Pelser PB, Baptiste MP, Poopath M, Schulze M, Seebens H, Shu WS, Thomas J, Velayos M, Wieringa JJ, Pysek P (2015) Global exchange and accumulation of non-native plants. Nature 525, 100
| Global exchange and accumulation of non-native plants.Crossref | GoogleScholarGoogle Scholar | 26287466PubMed |
Vilgalys R (2003) Taxonomic misidentification in public DNA databases. New Phytologist 160, 4–5.
| Taxonomic misidentification in public DNA databases.Crossref | GoogleScholarGoogle Scholar |
Wäldchen J, Mäder P (2018) Machine learning for image based species identification Methods in Ecology and Evolution 9, 2216–2225.
| Machine learning for image based species identificationCrossref | GoogleScholarGoogle Scholar |
Weber A, Skog LE (2007) The genera of Gesneriaceae. Basic information with illustration of selected species. 2nd edn. Available at http://www.genera-gesneriaceae.at [Verified 31 May 2019]
Wheeler QD, Raven PH, Wilson EO (2004) Taxonomy: impediment or expedient? Science 303, 285
| Taxonomy: impediment or expedient?Crossref | GoogleScholarGoogle Scholar | 14726557PubMed |
White RJ (1984) Implementing small database systems with specialised features. In ‘Databases in Systematics’. (Eds R Allkin, FA Bisby) Systematics Association Special Vol. 26, pp. 291–308. (Academic Press: London, UK)
White RJ, Allkin R (1992) Language for the definition and exchange of biological data sets. Mathematical and Computer Modelling 16, 199–223.
| Language for the definition and exchange of biological data sets.Crossref | GoogleScholarGoogle Scholar |
White RJ, Allkin R, Winfield PJ (1993) Systematic databases: the Baobab design and the Alice system. In ‘Advances in Computer Methods for Systematic Biology: Artificial Intelligence, Databases, Computer Vision’. (Ed. R Fortuner) pp. 297–311. (Johns Hopkins University Press: Baltimore, MD, USA)
Wieczorek J, Döring M, De Giovanni R, Robertson T, Vieglais D (2009) Darwin Core, biodiversity information standards (TDWG). Available at http://rs.tdwg.org/dwc/ [Verified 31 May 2019]
Wilson EO (2000) A global biodiversity map. Science 289, 2279
| A global biodiversity map.Crossref | GoogleScholarGoogle Scholar | 11041790PubMed |
Wilson EO (2003) The encyclopedia of life. Trends in Ecology & Evolution 18, 77–80.
| The encyclopedia of life.Crossref | GoogleScholarGoogle Scholar |
Younis S, Weiland C, Hoehndorf R, Dressler S, Hickler T, Seeger B, Schmidt M (2018) Taxon and trait recognition from digitized herbarium specimens using deep convolutional neural networks. Botany Letters 165, 377–383.
| Taxon and trait recognition from digitized herbarium specimens using deep convolutional neural networks.Crossref | GoogleScholarGoogle Scholar |
Zarucchi JL, Winfield PJ, Polhill RM, Hollis S, Bisby FA, Allkin R (1993) The ILDIS project on the world’s legume species diversity. In ‘Designs for a Global Plant Species Information system’. (Eds FA Bisby, RJ Pankhurst, GR Russell) pp. 131–144. (Oxford University Press: Oxford, UK)
1 The authors compiled this paper as part of the work of the Legume Phylogeny Working Group (LPWG).
Appendix 1. Online resources available for taxonomy, systematics and phylogenetics
This list is not exhaustive and does not include numerous national and regional initiatives. All web sites were accessed between February and May 2019. APIs, application programming interfaces
|