A phylogenetic investigation of the taxonomically problematic Eucalyptus odorata complex (E. section Adnataria series Subbuxeales): evidence for extensive interspecific gene flow and reticulate evolution
Patrick S. Fahey A B * , Frank Udovicic D , David J Cantrill D , Dean Nicolle E , Todd G. B. McLay
A B * , Frank Udovicic D , David J Cantrill D , Dean Nicolle E , Todd G. B. McLay  C E and Michael J. Bayly
C E and Michael J. Bayly  C
C
A Queensland Alliance of Agriculture and Food Innovation, University of Queensland, St Lucia, Qld, Australia.
B Research Centre for Ecosystem Resilience, Australian Institute of Botanical Science, The Royal Botanic Garden, Sydney, NSW, Australia.
C Currency Creek Arboretum, Melrose Park, SA, Australia.
D School of BioSciences, The University of Melbourne, Parkville, Vic., Australia.
E National Herbarium of Victoria, Royal Botanic Gardens Victoria, South Yarra, Vic., Australia.
Australian Systematic Botany 35(5) 403-435 https://doi.org/10.1071/SB21029
Submitted: 30 July 2021 Accepted: 20 September 2022 Published: 20 October 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
To investigate the relationships among species in the taxonomically problematic Eucalyptus odorata species complex, we generated molecular data using double-digest restriction site-associated DNA sequencing (ddRADseq) and Diversity Arrays Technology sequencing (DArTseq). These data were analysed utilising principal-component analysis (PCA), phylogenetic networks, phylogeny reconstruction and hybridisation tests. Twelve species that are variously recognised in the complex were sampled from across their ranges, along with co-occurring members of E. section Adnataria, to allow for patterns of hybridisation and gene flow to be identified. Despite the large genetic datasets generated, many relationships within the E. odorata complex were poorly resolved, and few species were monophyletic, likely owing to both biological factors including recent speciation and extensive hybridisation and introgression, and potential over-splitting of taxa. We show that multiple taxa with limited distributions are the result of reticulate evolutionary events and that typical Eucalyptus viridis R.T.Baker and the possibly con-specific E. aenea K.D.Hill are sister to the rest of the complex. The remaining species appeared to represent a discontinuous crescent-shaped cline running from the Flinders Ranges to the south-western slopes region of New South Wales, with limited support for an east–west split in this cline across the Murray River Basin. Eucalytpus viridis var. latiuscula Blakely, which is not closely related to the typical variety of this species in our data, may represent a northern extension to this cline.
Keywords: biogeography, DArTseq, ddRADseq, Eucalyptus, hybridisation, phylogenetics, reticulate evolution, south-eastern Australia.
Introduction
Species in many plant genera will readily hybridise with closely related and co-occurring species despite being morphologically and ecologically distinct with largely separate evolutionary histories (Abbott et al. 2013). This makes defining species difficult under some of the most widely applied species concepts such as the biological species concept (BSC), which regards species as the widest populations of organisms that can interbreed, while being unable to interbreed with other populations (Mayr 1963, 2000), and the phylogenetic species concept, which includes all individuals descending from a common ancestor as a species (Baum 1992). In Australia, one large and iconic group where this species definition problem is prevalent is the eucalypts, a diverse clade of woody shrubs and trees comprising >750 recognised species and numerous undescribed entities (Council of Heads of Australasian Herbaria 2016) that are the dominant arboreal taxa across much of the continent. Within the largest eucalypt genus, Eucalyptus, infra-sectional hybridisation is a frequently observed phenomenon despite ‘pure’ species dominating most natural populations (Griffin et al. 1988); however, there is a sharp decline in hybridisation frequency and hybrid fitness at the sectional level (Larcombe et al. 2015). It has previously been suggested that hybridisation is particularly frequent in E. subgenus Symphyomyrtus (Jones et al. 2016; Flores-Rentería et al. 2017), the subgenus to which all species sampled in this study belong.
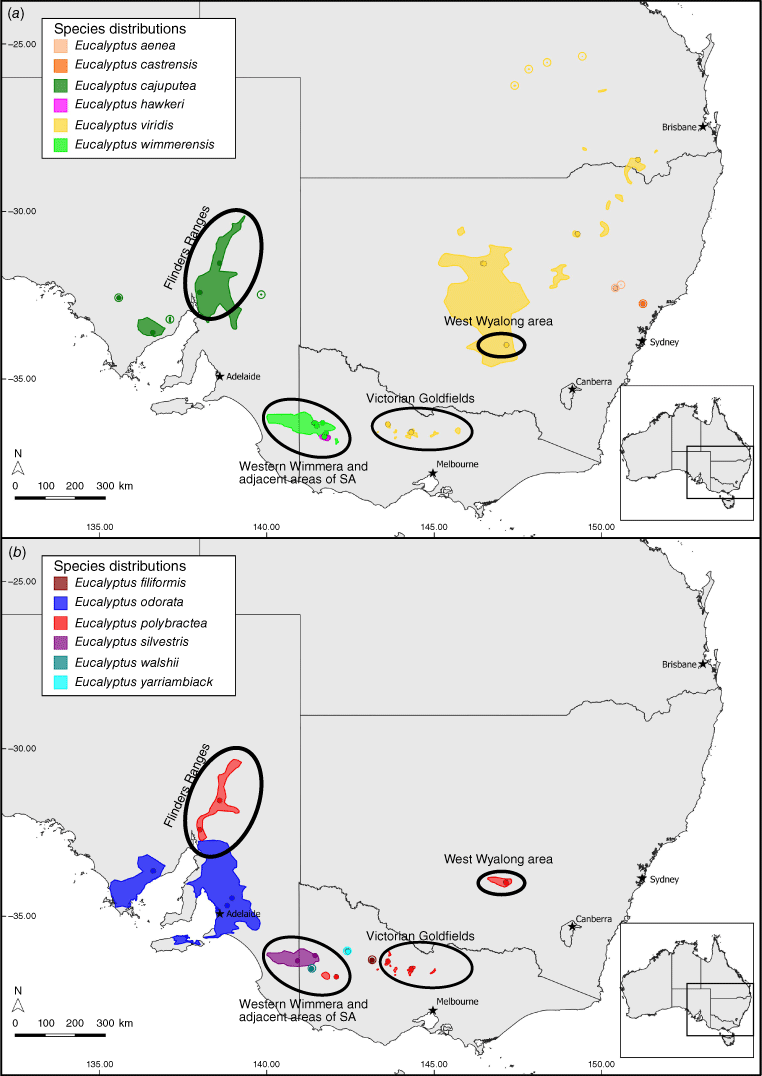
The group investigated in this study, the E. odorata complex, provides an extra layer of complexity to the problem of defining species, in the scattered yet overlapping distribution of several taxa within the complex (Fig. 1), making it difficult to conclude whether morphologically similar entities that occur hundreds of kilometres apart are conspecific. The complex consists of between 3 and 12 species, depending on which classification system is employed (Table 1) and taxa in the complex are united by their mallee to small tree-like habit, lanceolate to narrowly lanceolate juvenile leaves up to 2.5 cm wide, simple axillary inflorescences, buds with outer operculum intact at anthesis, and cup-shaped to barrel-shaped fruit with three or four locules (Rule 2018). The complex is part of E. section Adnataria series Subbuxeales, along with the grey boxes, four species of lignotuberous but generally single-trunked woodland or forest trees (E. albens Benth., E. microcarpa (Maiden) Maiden, E. moluccana Roxb., E. woollsiana R.T.Baker) and two other mallee species that are closely related to the E. odorata complex, E. albopurpurea (Boomsma) D.Nicolle and E. froggattii Blakely (Nicolle 2019). Eucalyptus froggattii is recognisable by its terminal inflorescences, square buds and fruit, and tightly crowded oils glands in the adult leaves (Brooker et al. 2015). Eucalyptus albopurpurea is less distinct from members of the E. odorata complex and may be better placed within the complex because it is known to intergrade with E. odorata Behr on Kangaroo Island (Nicolle 2000), but differs in its wider juvenile (up to 4.7 cm) and adult leaves, apparently terminal inflorescences, marginally longer buds and fruit, and often pink to purple flowers (Brooker et al. 2015).
|
The diversity of the complex is best understood by following its taxonomic history, which starts with the three accepted species described prior to 1910 (Table 1), namely, E. odorata, E. polybractea R.T.Baker and E. viridis R.T.Baker. The first of these, E. odorata, is a mallee or tree with variably larger buds and fruit than for the other two, which occurs in temperate woodlands to the west of the Murray Basin in South Australia (SA), although historically it has also been considered to be present in the western Wimmera of Victoria and adjacent areas of south-eastern SA (Brooker et al. 2015). Eucalyptus polybractea was described to include mallees that have bluish-green leaves and somewhat pruinose buds and fruit, which occur in the following three highly disjunct areas: on sandstone outcrops in the Victoria Goldfields, around the township of West Wyalong in New South Wales (NSW) and in the Flinders Ranges of SA (Brooker et al. 2015). In 2018, the range of E. polybractea was extended into the Victorian Wimmera by the recognition of a new subspecies, E. polybractea subsp. subcerea, with reportedly pruinose seedlings and faintly pruinose buds and fruit in this region (Rule 2018). The final of these three long-standing taxa is E. viridis, which includes mallees with linear green leaves and non-glaucous buds and fruits, and has historically been considered to have a distribution that encompasses much of that of E. polybractea, with a northern extension from West Wyalong to central Queensland along the inland slopes of the Great Dividing Range (GDR), with some small populations in the Hunter Valley (Brooker et al. 2015). The species was also previously considered to occur in the Wimmera and was considered more common in this area than was E. odorata (Chippendale 1988; Brooker et al. 2015). This classification broadly held for 90 years despite several varieties and subspecies of these taxa being proposed and some taxonomic confusion regarding the name E. fruticetorum F.Muell. ex Miq., which Blakely (1934) applied to E. polybractea despite the type material representing E. odorata (Pryor and Johnson 1971).
Starting with the publication of E. wimmerensis Rule in 1990, there has been a significant increase in the number of described taxa in the group, with eight species published in the past 30 years (Table 1). Eucalyptus wimmerensis was described (Rule 1990) to include all populations in the Wimmera region of Victoria and adjacent parts of SA that had previously been considered E. viridis (Fig. 1a). These populations differ from typical E. viridis largely in their juvenile leaf morphology, but also exhibit wider adult leaves and marginally larger fruits; all traits suggesting a closer affinity with E. polybractea and E. odorata than with E. viridis (Rule 1990; Nicolle 2006). Rule (2018) described five subspecies of E. wimmerensis; however, these show minor morphological differences and, given there are many questions regarding species-level taxonomy in the complex, we do not analyse these subspecies in detail in this paper. The next taxonomic change in the complex was again made by Rule, with the separation of all populations east of the Murray Basin (Fig. 1b), formerly considered to be E. odorata, into the new taxon E. silvestris Rule on the basis of their smaller buds and fruit, lustrous adult leaves, and broader juvenile leaves with longer petioles (Rule 1994). Other authors have suggested these traits may result from hybridisation between E. odorata or E. wimmerensis and the grey box, E. microcarpa (Brooker et al. 2015; Nicolle 2019).
Further populations that had previously been regarded as E. viridis were separated into another new taxon, E. aenea K.D.Hill, by Hill (1997). This species includes populations in the Goulburn River National Park (NSW) and nearby areas (Fig. 1a) that grow on exposed slopes among taller eucalypt forest and differ from typical E. viridis in their broader adult and juvenile leaves, bluish-green juvenile leaves and wholly smooth bark (Hill 1997). Currently, this is the only species in the group not recognised on the Australian Plant Name Index (Council of Heads of Australasian Herbaria 2016), being regarded as a synonym of E. viridis. In 2002, a further population of mallee-boxes with affinities to E. viridis and E. aenea discovered on the Singleton Army Base (NSW) was described as E. castrensis K.D.Hill by Hill and Stanberg (2002) (Fig. 1a). Plants from this population have larger adult leaves, buds and fruit, broader juvenile leaves and a stronger medial constriction of their operculum than does E. aenea, while also growing into more robust mallees that retain basal rough bark (Hill and Stanberg 2002). Per Nicolle (2019), the type population of this taxa consists of two forms, one potentially referable to E. aenea and the second that matches the type material that may represent E. aenea × E. microcarpa–E. moluccana hybrids, with the concept of the name as applied by Hill and Stanberg (2002) and Copeland and Hunter (2005), including both the potential E. aenea and E. aenea × E. microcarpa–E. moluccana hybrid forms.
In a 2004 publication, Rule described the following three further taxa in the complex from Victoria: E. filiformis Rule, E. hawkeri Rule and E. walshii Rule. Eucalyptus filiformis is known from only seven individuals that form a copse on Mount Jeffcott in the eastern Wimmera (Fig. 1b) and show morphological affinities with both E. viridis and E. polybractea, differing primarily from the former in having dull bluish-green leaves with clearly visible venation and larger fruits, and the latter in having persistent thin grey, box bark, narrower adult and juvenile leaves and thin-walled fruit (Rule 2004). The seven known individuals of this species show minimal morphological differentiation. A further individual at this locality was subsequently discovered by Rule and provided to us under the informal name ‘E. aff. polybractea (Mount Jeffcott)’ on the basis of its smooth bark, glaucous juvenile leaves and minute, lightly waxy buds, and barrel-shaped fruits (K. Rule, pers. comm., 18 November 2018).
Eucalyptus walshii is also known only from a single population with minor morphological differences from the other species in the complex. This species occurs near Broughtons Waterhole in the Little Desert of western Victoria (Fig. 1b), with key morphological features being pale smooth bark above a basal stocking of box bark, a tree-like habit and broad adult leaves with dense reticulate venation (Rule 2004). It was described as showing greatest morphological affinities with E. odorata and E. albopurpurea (Rule 2004), which both occur only west of the Murray basin under Rule’s classification.
The third of the mallee-box species from Rule’s (2004) publication, E. hawkeri, is somewhat more widespread, occurring in many mallee communities in the southern Wimmera (Rule 2018; Fig. 1b) The primary characters of this species are its slender tree-like habit, lanceolate and often pendulous adult leaves, and substantial stocking of rough bark. This taxon has been suggested by Nicolle (2019) and Brooker et al. (2015) as being a recurring intergrade between E. wimmerensis and E. microcarpa.
The most recently described species in the group was E. yarriambiack Rule from a small population restricted to a narrow roadside strip in an otherwise extensively cleared agricultural area near Yarriambiack Creek, north of the town of Brim in the southern Murray Mallee of Victoria (Fig. 1a). This population lies outside the current distribution of all other species of the E. odorata complex, and differs from them in its tree-like growth habit, box bark coverage of its main branches, and smaller fruit (Rule 2012). Nicolle (2019) noted that the type material may represent a hybrid between E. largiflorens F.Muell. and E. odorata, despite the lack of other E. odorata complex members in the area.
The final member of the group is E. cajuputea Miq., a name considered a synonym of E. odorata as early as 1867 (Council of Heads of Australasian Herbaria 2016) and resurrected by Nicolle and Roberts (2013) to include all populations previously regarded as E. viridis in the Flinders Ranges, where the type material for the species was collected, and northern Eyre Peninsula, with outlying populations in the Gawler Ranges and on Oulnina Hill (Fig. 1a). These populations have wider juvenile and adult leaves that lack the lustrous colouration of E. viridis (Nicolle and Roberts 2013). A detailed overview of the morphological variation in the group is provided by Rule (2018).
In this study, we aim to use and compare genomic data generated from double-digest restriction-associated DNA sequencing (ddRADseq; Peterson et al. 2012) and Diversity Array Technologies sequencing (DArTseq; Sansaloni et al. 2010) to build a systematic understanding of the taxa in the E. odorata complex. The DArTseq approach was designed using eucalypts (Sansaloni et al. 2010) and has been successfully used to investigate both phylogenetic and population genetic relationships in various groups of eucalypts in the past (Steane et al. 2011, 2017; Jones et al. 2016; Rutherford et al. 2016, 2018, 2019, 2020; Jordan et al. 2017), whereas ddRADseq has seen very little use in eucalypts (Aguirre et al. 2019), despite widespread use in phylogenetic studies of other taxa (Yang et al. 2016; Guo et al. 2020). Both techniques use restriction-enzyme digestion to fragment genomic DNA; however, DArTseq is a proprietary method performed on a fee-for-service basis only by Diversity Array Technologies (Canberra, ACT, Australia), whereas ddRADseq can be performed ‘in-house’ by researchers at a lower cost per sample. Thus, by using both, we aim to show the costs and advantages of each approach in phylogenetic studies of eucalypts, as well as exploring the value of combined DArTseq/ddRADseq data. We sample all 12 species within the E. odorata complex from multiple locations throughout their ranges to test monophyly, and resolve relationships and patterns of hybridisation among them. This represents the first molecular systematics investigation focussed on the group and the first direct comparison between ddRADseq and DArTseq data in answering phylogenetic and taxonomic questions in Eucalyptus.
Materials and methods
Sample collection and preparation
The classification system of Nicolle (2019) was followed in this study, with the exception of the consideration of all 12 species in the E. odorata complex outlined in Table 1 as potentially valid. Where possible, samples of each species were collected from a representative set of wild populations, and this was supplemented, when necessary, with cultivated specimens from Currency Creek Arboretum, the Royal Botanic Gardens Victoria, and the private seedling experiments of Kevin Rule (Table 2). All species in the complex were represented by at least two samples except for E. castrensis, which grows only on Department of Defence managed land (Hill and Stanberg 2002) and for which only a single cultivated specimen, which is more referable to the putative E. aenea form than the putative hybrid typical material, was accessible during this study. In addition to two samples of E. hawkeri from Mount Arapilies in the southern Wimmera, we identified an additional specimen from the northern Wimmera as E. hawkeri, on the basis of its single-trunked habit, substantial stocking of rough bark and somewhat weeping foliage, providing the first record of this species north of the Little Desert. Furthermore, two samples associated with the E. odorata complex, but of uncertain identity, were included, namely, a seedling grown from seed of Rule’s ‘E. aff. polybractea (Mount Jeffcott)’, and a sample that could not be positively assigned to either E. dumosa A.Cunn. ex J.Oxley or E. polybractea from Wychitella Nature Conservation Reserve in the Victorian goldfields that was suspected to be a hybrid. In addition to the members of the E. odorata complex, sampling was extended to include many co-occurring species of E. section Adnataria to test for possible hybridisation. A sample each of E. dumosa (E. section Dumaria) and E. oleosa F.Muell. ex Miq. (E. section Bisectae), were collected as outgroups for phylogenetic analysis (Table 2).

|
Approximately 10 leaves were collected from each sampled individual and placed immediately into a coffee filter nested in silica beads to dehydrate before storage at the University of Melbourne. Voucher specimens were collected for at least one individual of each species at each collection site and were lodged at the University of Melbourne Herbarium (MELU; Table 2).
Because DArTseq is performed in 94 sample plates and was undertaken after the generation of preliminary results from the ddRADseq investigation (91 samples), there were some changes in sampling between the two datasets. Namely, as discussed below, the wild population of E. filiformis was found to be clonal, so six of the eight samples included in the ddRADseq dataset were excluded from the DArTseq run, and an extra two samples of E. polybractea, five of E. viridis, one E. aenea and a third outgroup sample of E. leptophylla F.Muell. ex Miq. were included to bring the total sample number in the DArTseq dataset to 94.
Field collections for this study were completed under scientific collecting permits granted by Department of Environment, Land, Water and Planning, Government of Victoria (10008557), NSW National Parks & Wildlife Service, Office of Environment & Heritage, Government of New South Wales (SL102100) and Department for Environment and Water, Government of South Australia (Q26766-1).
DNA isolation, library preparation and sequencing
The modified CTAB protocol of Schuster et al. (2018) was employed for total DNA isolation from 70 to 80 mg of dry leaf material for each of the 100 samples. A Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) was used to check extraction purity and a Qubit fluorometer (ver. 2.0, Thermo Fisher Scientific) to quantify the concentration of extracted DNA. Informed by the results of Yang et al. (2016) in silico tests of digestion of the E. grandis W.Hill genome, the restriction enzymes EcoR1-HF and Msp1 were chosen for genome digestion to create the ddRADseq library. The library preparation and pooling followed the protocol of Fahey et al. (2021). The resulting pool was quantified on a 2200 Tape Station (Agilent) using a D1000 kit and sequencing was undertaken at the Walter and Eliza Hall Institute of Medical Research (WEHI) Genomics Hub, Melbourne, Vic., Australia, on an Illumina NextSeq. 500 using a 300 cycle, 2× paired-end reads kit. For DArTseq, extractions were standardised to a concentration of 50 ng μL−1 and sent to Diversity Array Technologies Pty Ltd (Canberra, ACT, Australia) for high-density sequencing using their proprietary DArTseq pipeline (Kilian et al. 2012). Filtered read data as well as DArT SNP data were returned for analysis.
Dataset filtering, construction, and analysis
The Cutadapt (ver. 2.8) python script (Martin 2011) was used to trim raw reads from ddRADseq to remove restriction-site residues and bases added for adaptor diversity with a maximum error rate of 0.5. The assembly pipeline ipyrad (ver. 0.9.62, see https://ipyrad.readthedocs.io/en/master/, Eaton and Overcast 2020) was used to reconstruct loci by mapping reads to the Eucalyptus grandis reference genome (Myburg et al. 2014); this allowed us to avoid problems related to inclusion of paralogous loci as ipyrad discards any reads that do not map uniquely to the reference genome. Various values for the statistical and majority rule base call minimum (6, 20), clustering threshold (0.85, 0.9, 0.95) and minimum number of samples with data at each locus (four samples, 25, 50, 75% of total samples) were tested to find the most informative dataset. The dataset presented in this paper is that derived using base call minimums of six, clustering threshold of 0.85% and a minimum of four samples returned per locus; however, the loci reconstructed using each of the different input variables showed little variation beyond the increasing number of loci retained when the minimum sample threshold was decreased (Supplementary Table S1).
Principal-component analyses (PCA) and network analyses were performed on both the ddRADseq and DArTseq SNP datasets individually. Because the total size of the ddRADseq alignment produced by ipyrad was too large to feasibly analyse, we converted the single-nucleotide polymorphism (SNP) dataset produced by ipyrad to a NEXUS alignment to perform these analyses. Additionally, using the DArTR R package (ver. 1.9.9.1, see https://CRAN.R-project.org/package=dartR; Gruber et al. 2018) we converted the DArTseq data to an alignment of SNPs for network analysis. Networks were created in Splitstree (ver. 4.16.1, Universität Tübingen, Tübingen, Germany; Huson and Bryant 2006) using the NeighbourNet algorithm and Uncorrected-P distances ignoring ambiguities and with outgroups excluded.
Principal-component analysis is sensitive to missing data and uneven sampling (McVean 2009). So, we created a further SNP dataset from our ddRADseq data utilising VCFtools (ver. 0.1.16, see https://vcftools.github.io/index.html; Danecek et al. 2011), where we allowed maximum missing data per SNP of 10%, filtered for one SNP per 5000-bp window of the E. grandis reference genome, and included only samples in E. series Subbuxeales and excluded seven of the eight clonal E. filiformis samples and two E. wimmerensis samples with significantly above-average missing data, namely, KRule1714 and KRule3012 with 3215 and 1413 loci respectively in the final assembly, against an average of 10 313 loci for all other samples. Similar filtering was applied to the DArTseq SNPs utilising the DArTR package in R (R Foundation for Statistical Computing, Vienna, Austria), including filtering for a single SNP per locus, reproducibility of >0.98, callrate >0.90 and a minor allele frequency of >0.02. PCAs were performed in R utilising the VCFR (ver. 1.12.0, https://CRAN.R-project.org/package=vcfR; Knaus and Grünwald 2017) package to load ddRADseq VCF data, and the dudi.pca function of adegenet (ver. 2.1.5, https://CRAN.R-project.org/package=adegenet; Jombart and Ahmed 2011), before being graphed using ggplot2 (ver. 3.3.5, https://CRAN.R-project.org/package=ggplot2; Wickham 2016).
The PHYLIP alignment produced by ipyrad was used to estimate a maximum-likelihood (ML) phylogeny in RAxML (ver. 8.2.12, see https://cme.h-its.org/exelixis/web/software/raxml/; Stamatakis 2014) by using the rapid hill-climbing tree-search algorithm and performing 100 rapid bootstraps. Because of the anonymous nature of the data, no partitioning was used, along with a GTR substitution matrix. Additionally, a maximum-parsimony (MP) analysis was undertaken using PAUP (ver. 4.0a, see https://paup.phylosolutions.com/; Swofford 2002), with 100 bootstrap replicates and multistate characters interpreted as polymorphisms. Owing to the size and complexity of the dataset, the required computational resources proved prohibitive for Bayesian phylogeny estimation. Trees were visualised in TreeGraph 2 (ver. 2.15, see http://treegraph.bioinfweb.info/; Stöver and Müller 2010) to build figures. The same phylogenetic analyses were performed on an alignment of the DArTseq data generated using the gl2fasta function of the DArTR R package. The resultant phylogenies are available in the supplementary material of this article (Supplementary Fig. S1–S4).
Although minor differences in relationships among samples existed between the two individual data sources, so as to build the most robust phylogeny possible, a further ipyrad run was performed utilising similar settings as for the ddRADseq dataset. The only change to those settings was to treat the data as RAD reads rather than ddRAD and this was undertaken on the combined DArT reads and the forward reads from ddRADseq to reconstruct the largest alignment possible while not incidentally including any loci twice. Although previous studies have highlighted that different techniques to generate molecular data for phylogenetic studies can generate different results (Collins and Hrbek 2015; Kirschner et al. 2021), in this case, both techniques are enzymatic reduced-representation library-generation techniques and therefore there should be limited causes of bias in the representation of the genome between them. Both MP and ML phylogenetic analyses were performed on the combined dataset, as they were on the individual datasets. Additionally, a tetrad analysis with 100 bootstrapping replicates was performed in the ipyrad toolbox (Eaton and Overcast 2020) on the combined dataset and is presented in the supplementary material (Supplementary Fig. S5).
To investigate the patterns of introgression in samples that showed unexpected or unstable placement in the phylogenetic analyses and PCAs, we undertook ABBA-BABA tests utilising the combined ddRAD and DArT data. ABBA-BABA tests were performed using the ipyrad analysis toolbox. In all tests, species and samples of interest were treated as P2 individuals and the sample PSF41B (E. oleosa) was used as the outgroup. For those samples where the ABBA-BABA tests indicated hybridisation between parental species we had sampled, we also performed NewHybrid tests (ver. 1.1, see https://github.com/eriqande/newhybrids; Anderson and Thompson 2002) to determine the hybrid generations. NewHybrids was performed using the gl.nhybrids function from the the DArTR R package. A filtered version of the combined SNP dataset with a minimum of 75% of samples represented at each locus and one SNP per 5000-bp window of the E. grandis reference genome was used for NewHybrids analyses.
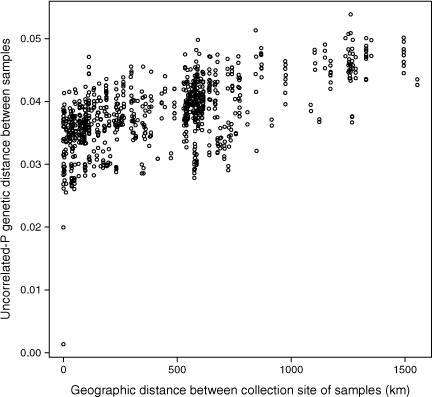
The full dataset included several suspected hybrids and introgressed samples that caused reduced support values throughout the phylogeny; therefore, we completed additional ML and MP analyses on both the individual ddRADseq and DArTseq datasets, and on the combined dataset, with 10 samples being removed on the basis of the findings of earlier analyses and hybridisation tests. Seven samples that were suspected to be introgressed a priori, showing unexpected placement in the initial tree and support for introgression in ABBA-BABA tests, were removed, including the suspected E. dumosa × E. polybractea intersectional hybrid PSF96A, the suspected E. filiformis × E. leucoxylon F.Muell. interserial hybrid KRule3912, and the five samples of E. hawkeri and E. silvestris (DN445, KRule5370, PSF54A, PSF61A and PSF105E). Additionally, three samples that were oddly placed in the initial analyses and networks, but for which patterns of introgression could not be established in ABBA-BABA tests, were also excluded, including E. cajuputea PSF50P, E. polybractea PSF50B and E. aenea PSF90J. Because of a lack of supported relationships in the reduced tree within the core of the E. odorata complex, which is defined as including, from this point forward, E. odorata, E. cajuputea, E. wimmerensis, E. walshii, E. yarriambiack, E. filiformis, E. polybractea, and E. viridis samples from south-eastern Queensland, an isolation by distance test was undertaken in R using the mantel function from the vegan package (ver. 2.5-6, J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, and H. Wagner, see https://CRAN.R-project.org/package=vegan) on uncorrelated-P distances calculated from the SNP alignment used in MP analyses.
Results
Datasets
The final ddRAD dataset used in network analyses contained 1 032 139 SNPs across 89 samples, with a missing data proportion of 65.85% and was reduced to 2287 SNPs across 59 samples for PCA analysis. The DArTseq network dataset contained 60 991 SNPs, with 43.6% missing data across 90 samples, whereas the PCA version contained 68 samples and 2681 SNPs. The combined alignment used for ML phylogeny reconstruction contained 119 489 loci and was a total of 12 430 010 bp in length, with the 975 539 SNPs present in the dataset being used for MP phylogeny reconstruction.
ddRADseq and DArTseq networks and PCAs
In both the ddRAD and DArT networks (Fig. 2), the clustering of all samples other than the mallee species in E. series Subbuxeales (the E. odorata complex, E. albopurpurea, and E. froggattii) was identical and matched the relationships established in subsequent phylogenetic analyses and are discussed in regard to them. Relationships within the E. odorata complex are not particularly clear and varied marginally between the datasets (Table 3); however, the clustering of the samples in the core E. odorata complex is much clearer in the DArT network than the ddRAD network. A comparison between the relationships of these samples is provided in Table 3; however, PCAs, phylogenies and ABBA-BABA tests provided greater insight into the relationships in this group.
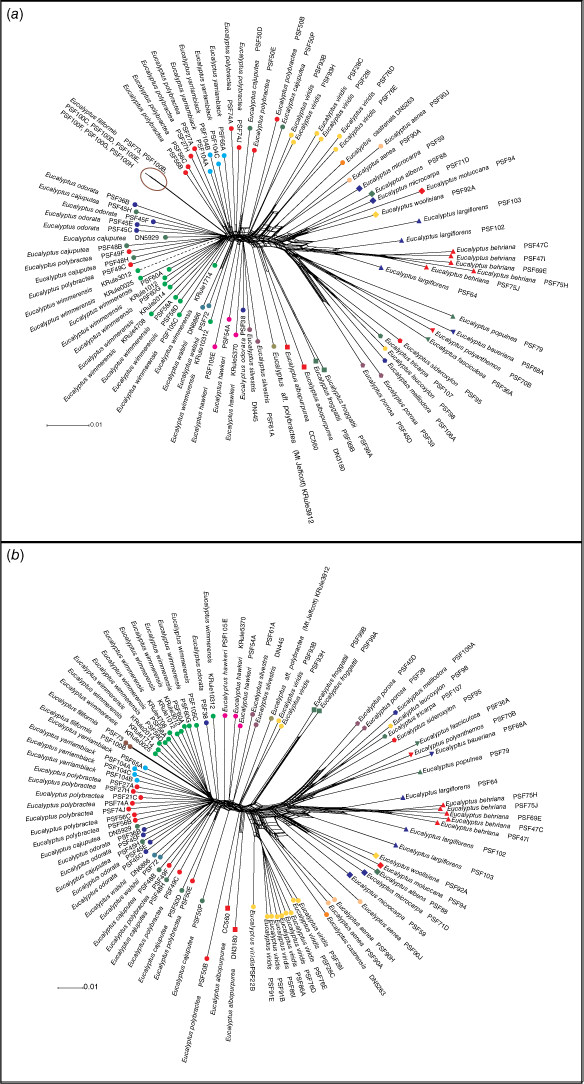
|

|
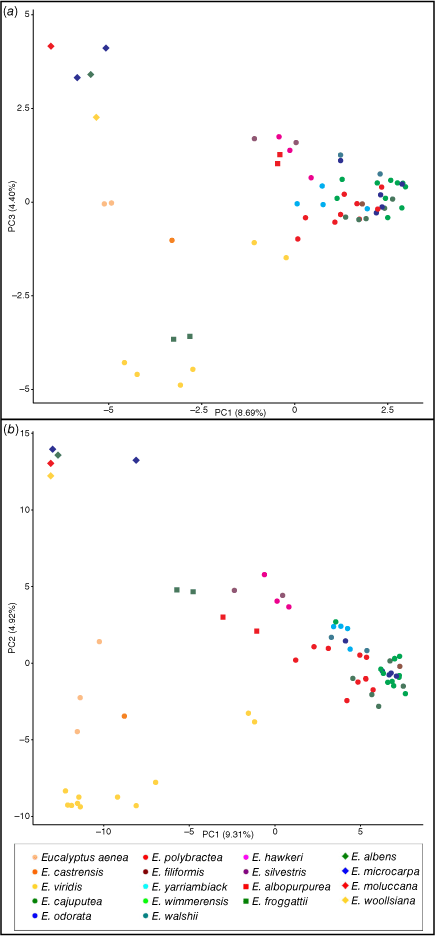
In Fig. 3, we see that although PC3 in the ddRAD analysis matches PC2 from the DArT one, overall, the PCA plots produced are very similar and fit with the patterns observed in the networks and phylogenies. PC2 in the ddRADseq analysis predominately separates the two E. froggattii samples from the other samples and has only a marginally higher eigenvalue than does PC3 (3.80 v. 3.35%). It is likely that the difference in sampling between the two datasets, namely the extra six E. viridis samples in the DArTseq analysis, is responsible for this difference. In both analyses, the following three main groupings form: the grey boxes, E. viridis and allied species, and the remainder of the E. odorata complex. Both E. froggattii and E. albopurpurea fall towards the centre of the plot using the two selected PCs, although the former segregates on PC2 and PC3 in the ddRAD and DArT analyses respectively (not shown). In both plots, the E. viridis segregates E. aenea and E. castrensis sits between E. viridis and the grey boxes, and the most northerly population of E. viridis sampled sits between the other E. viridis samples and the rest of the E. odorata complex. Additionally, the two suspected E. wimmerensis–E. microcarpa intergrades, E. hawkeri and E. silvestris, sit between the E. odorata complex and the main E. odorata complex cluster, albeit closer to the former. One sample of the grey-box E. microcarpa (PSF59) collected at a locality where E. wimmerensis is also present, separates from the other grey-box samples towards the E. odorata complex cluster in the DArT plot. All these results suggest possible hybridisation and genetic introgression between these taxa that we investigate using ABBA-BABA tests. There is no separation of the remaining seven species in the E. odorata complex that form the main cluster, although E. yarriambiack and E. walshii sit towards the end of the cluster closest to the grey boxes, which may suggest some introgression into these populations as has previously been hypothesised, a finding we test in our ABBA-BABA tests.
|
Combined data phylogeny
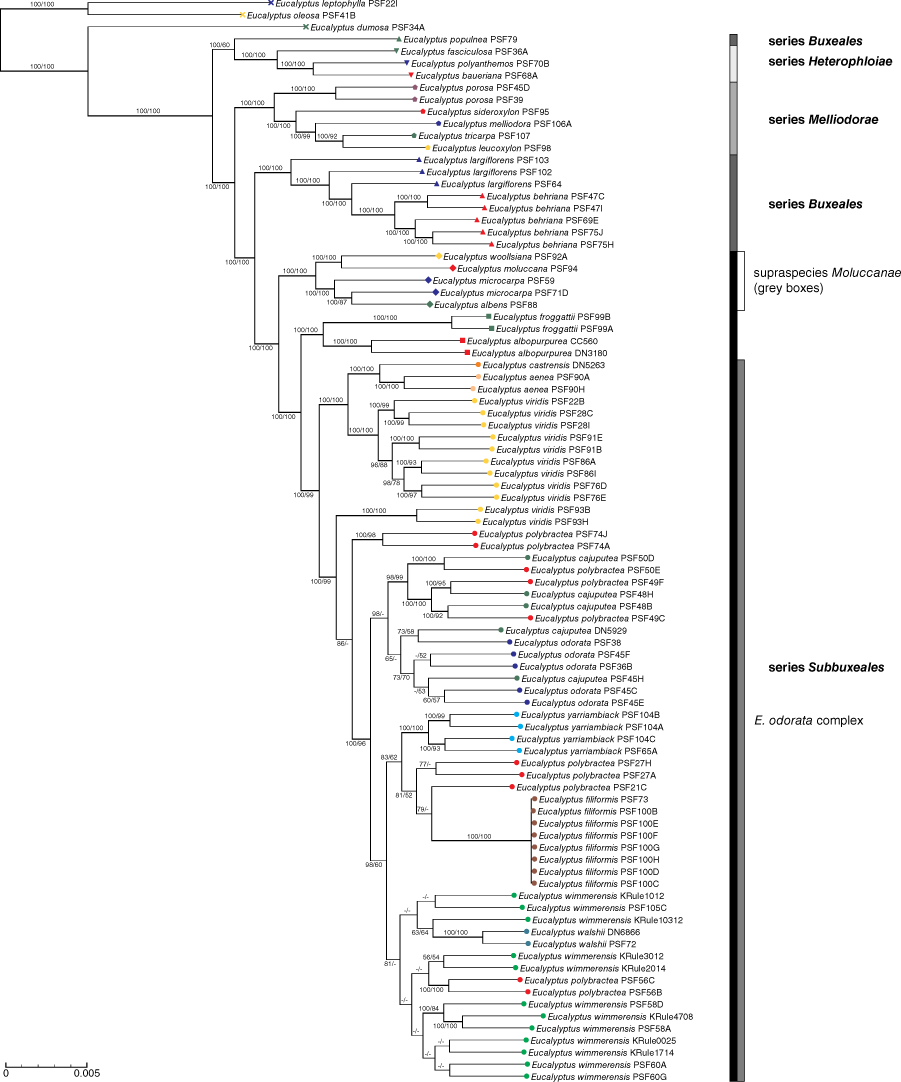
There were no supported differences between ML and MP phylogenies in either the full-sample (Fig. 4) or reduced-sample (Fig. 5) analyses at an 80% bootstrap threshold, with the reduced sampling analyses including more nodes that met this threshold (50 in the complete dataset, 55 in the reduced sampling dataset). The following discussion focusses primarily on the reduced sample phylogeny, unless otherwise specified.

|
|
As expected, the outgroups formed two clades, a basal one containing species of E. section Bisectae and a further one containing the sample of E. dumosa (E. section Dumaria) and the putative E. polybractea × E. dumosa hybrid sample (PSF96A), which was included only in the full dataset analysis. The first clade to diverge in E. section Adnataria included the three sampled species from E. series Heterophloiae (E. baueriana Schauer, E. polyanthemos Schauer and E. fasciculosa F.Muell.) and E. populnea from E. series Buxeales, although the inclusion of the latter in this clade is not supported in the MP analysis. A relationship between E. populnea F.Muell. and E. ser. Heterophloiae makes E. series Buxeales polyphyletic in our phylogeny. The next clades to diverge are E. series Melliodorae, which contains both samples of E. porosa F.Muell. ex Miq. as sister to the rest of the series, and the ‘E. aff. polybractea (Mount Jeffcott)’ seedling, followed by a clade containing E. behriana and E. largiflorens from the polyphyletic E. series Buxeales, which is resolved as sister to a monophyletic E. series Subbuxeales. Within the main E. series Buxeales clade, E. largiflorens formed a grade that subtends a monophyletic E. behriana clade. Within E. series Subbuxeales, the four grey-box species were supported as a monophyletic clade sister to the mallee species (E. odorata complex, E. albopurpurea and E. froggattii). Within the grey boxes, the only species represented by more than one sample, E. microcarpa, was not monophyletic, with one sample, PSF59, being the earliest diverging member of the clade matching the PCA and network findings. Eucalyptus albopurpurea and E. froggattii were supported as sister species and, in turn, as the sister clade to the E. odorata complex. Although the E. odorata complex was supported as a monophyletic group, there were very few resolved relationships within the complex. None of the widespread species (E. cajuputea, E. odorata, E. polybractea, E. viridis and E. wimmerensis) was found to be monophyletic, although, in most cases, where multiple samples from the same site were included, they were supported as each other’s closest relatives.
The majority of E. viridis samples, including all from Victoria and the central west of NSW, were supported as a monophyletic lineage, which was sister to a clade containing the two species from the Hunter Valley, E. aenea and E. castrensis. Within this main E. viridis clade, the three Goldfields samples were supported as a sister lineage to the samples from north of the Murray River, with the samples from each of the three locations in the latter group forming sister pairs and the Pilliga population diverging first from a West Wyalong and Candelego clade. The two samples of E. viridis from the Inglewood area on the Qld–NSW border were not supported as part of this clade; rather, these samples were the next clade to diverge with respect to the rest of the E. odorata complex, followed by a clade containing the two samples of E. polybractea from the West Wyalong area, rendering the latter species polyphyletic also.
Within the rest of the E. odorata complex, there was support in the ML analysis for an east–west divide across the Murray River Basin, further rendering E. polybractea polymorphic, although the MP analysis did not corroborate this. In the western clade, all samples from the Flinders Ranges, both of E. polybractea and E. cajuputea (but excluding the two samples removed because of suspected introgression from other taxa, namely PSF50B and PSF50P) formed a monophyletic lineage, with the two sampling locations (Wilpena Pound and Devils Peak) being represented by supported clades. There were no supported relationships between any of the other E. cajuputea and E. odorata samples in the western clade. Additionally, there was also support for a division between samples from the western Wimmera and those from the eastern Wimmera and Victorian goldfields in the eastern clade, although, again, this was supported only in the ML analysis. There were minimal supported relationships in the western Wimmera clade, with only samples from a single collecting locality being supported as monophyletic groups, including the samples of E. walshii and E. polybractea subsp. subcerea. In the eastern Wimmera and Goldfields clade, the four samples of E. yarriambiack were supported as a clade, with the ML analysis placing them sister to the samples of E. polybractea from this region and the clonal E. filiformis. Our test showed significant IBD in this core clade (Mantel statistic r: 0.597, significance: 1 × 10−4, Fig. 6).
|
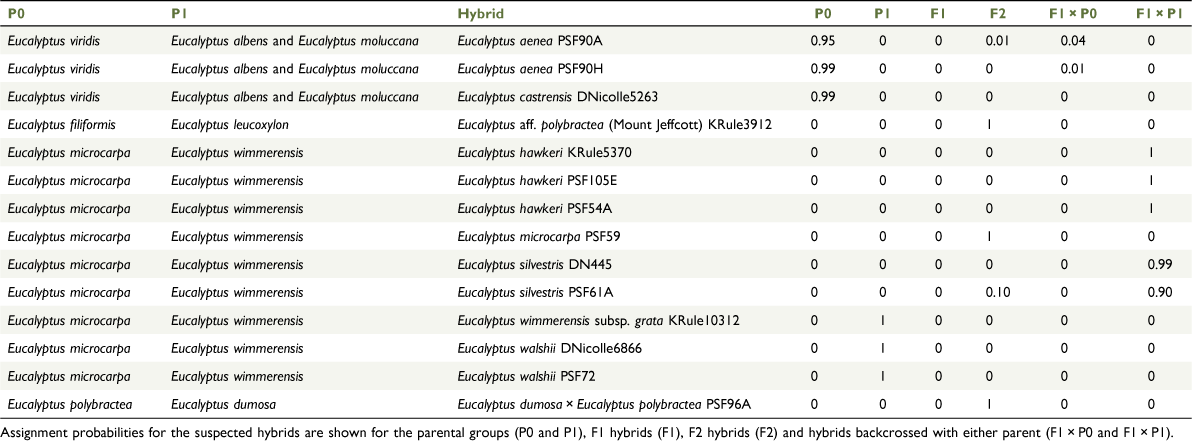
Hybridisation tests
There was a number of ABBA-BABA tests on samples with unexpected and unstable placement in other analyses that showed significant introgression at the commonly used threshold Z-score of three (Zheng and Janke 2018; Table 4). Two tests showed very high Z-scores and D-statistics, corresponding to the two samples not being assigned to existing species, namely the E. dumosa × E. polybractea individual (PSF96A) and the seedling of E. aff. polybractea (Mount Jeffcott) (KRule3912), which is an apparent E. filiformis × E. leucoxylon hybrid. For all tests for introgression from E. microcarpa into E. hawkeri, E. silvestris, E. walshii and E. wimmerensis subsp. grata with respect to E. wimmerensis, the Z-scores were greater than three. The individual tests of the former two returned higher D-statistics, ranging from 0.147 to 0.209, matching their more intermediary placement in phylogenetic analyses and PCAs, whereas E. walshii and E. wimmerensis subsp. grata returned lower D-statistics of 0.131 and 0.110 respectively, as expected on the basis of their more consistent placement with E. wimmerensis samples in other analyses. Support for introgression of E. wimmerensis into the E. microcarpa sample PSF59 was also found with a lower D-statistic of 0.111. No support for introgression of E. largiflorens nor E. microcarpa into E. yarriambiack, when compared with E. polybractea from the Victorian goldfields, was found.

|
Although reaching the threshold Z-score of three, D-statistics for E. aenea (excluding PSF90J) and E. castrensis being introgressed from the co-occurring grey boxes E. albens and E. moluccana were low (0.071–0.089). There was no support for our sample of E. castrensis being more introgressed from the grey-box species than is E. aenea. Tests of the aberrant E. aenea sample PSF90J being more introgressed by the grey boxes than other E. aenea samples returned the opposite result, with negative D-statistics suggesting that both E. aenea and E. viridis share more derived alleles with E. albens and E. moluccana than this sample. A similar situation is observed for the two samples from the Flinders Ranges that show aberrant placement (PSF50B and PSF50P), which showed negative D-statistics, some with a Z-score greater than three, when compared with all other taxa (not shown) except E. populnea, which has a positive D-statistic but fails to reach the threshold Z-score of three. This suggests that a species we have not sampled, potentially one genetically related to E. populnea, is the source of introgression into these samples.
The NewHybrid tests (Table 5) suggested that none of the samples that showed evidence of hybridisation and introgression was an F1 hybrid. The seedling of E. aff. polybractea (Mount Jeffcott; KRule3912), the E. dumosa × E. polybractea individual (PSF96A) and the E. microcarpa sample with E. wimmerensis introgression (PSF59) were all assigned F2 status. The last of these was an unexpected result, given its consistent placement with the grey-box samples in other analyses and a low D-statistic in the ABBA-BABA analysis. As expected, all E. hawkeri and E. silvestris samples were hybrids backcrossed with E. wimmerensis, although both E. walshii samples and the E. wimmerensis subsp. grata sample were assigned to the parental E. wimmerensis. The two E. aenea and one E. castrensis sample tested were both assigned to E. viridis rather than to a hybrid generation, supporting there being only historic introgression into these populations from one or both grey-box species with which they co-occur.
|
Discussion
Comparison of ddRADseq and DArTseq
In the context of phylogenetic studies of Eucalyptus, we have shown that there is minimal difference in the phylogenetic signal in the data generated through ddRADseq (a hands-on, in-house reduced-representation genetic-library approach) and DArTseq (a proprietary service gaining popularity in genetic studies of eucalypts and other plants), with each having benefits over the other. Unlike ddRADseq, DArTseq is designed to deal with highly repetitive genomes, such as those of many eucalypts (Sansaloni et al. 2010). Additionally, the data generated by DArTseq have the benefits of reproducibility and this approach offers the ability to expand and combine datasets generated at different times as part of separate sequencing runs because of the targeted nature of the loci genotyped (Sansaloni et al. 2010). Reproducibility and combinability are more problematic in ddRADseq datasets developed at different times, where there is a degree of stochasticity in which loci are returned each run owing to slight differences in library preparation and sequencing (O’Leary et al. 2018). In addition, we had significantly more even coverage across our samples in the DArTseq than in the ddRADseq data, which allows more confidence in the results of analyses, despite the fact there were fewer overall loci and SNPs. Where ddRADseq comes out ahead is the benefits of performing the library preparation in house, which gives flexibility in the number of samples included, the length of the loci in the library and the sequencing platform used, all for a lower cost. Ultimately, these two techniques may be best suited to different situations, DArTseq is preferable if you will want to combine data sequenced at different times, or across different studies, but ddRADseq might provide a cheaper alternative for a one-off dataset, where all samples are processed at once. Both of our datasets appear to be at the point where simply adding more genetic data does not improve resolution and support of phylogenetic relationships, given they show similar patterns individually (Fig. 2, 3) and we do not increase nor change the signal by combining them (Fig. 4, 5). This is in line with the findings of Luo et al. (2018) who performed case studies of molecular species delimitation techniques and found simply adding more loci to datasets often did not increase the discriminatory power of the analyses.
Phylogeny of E. section Adnataria
Although this study was focussed on the E. odorata complex, we have also produced one of the most resolved phylogenies for section Adnataria (Fig. 5). Previous phylogenetic studies of section Adnataria have either relied on plastid DNA (Flores-Rentería et al. 2017; Alwadani et al. 2019), which does not resolve phylogenetic relationships in the eucalypts because of a large cyto-nuclear discrepancy (Steane et al. 1998; Jackson et al. 1999), internal transcribed spacer sequences, which do not provide enough resolution to reconstruct relationships within the section (Steane et al. 2002, 2007; Thornhill et al. 2019), or have not resolved any relationships between series and species with support (Woodhams et al. 2013). However, because our sampling was comprehensive only for taxa that commonly co-occur with members of the E. odorata complex, we sampled only four of the nine series in E. section Adnataria that are recognised in the classification of Nicolle (2019). Three of the sampled series (Heterophloiae, Melliodorae and Subbuxeales) were monophyletic, with the placement of E. populnea as sister to E. series Heterophloiae, rendering E. series Buxeales polyphyletic (Fig. 5). A possible explanation for the placement of this sample is introgression from E. conica H.Deane & Maiden, a member of E. series Heterophloiae, which occurs at the collection locality and that we did not sample in this study. As we sampled only 3 of the 15 species placed in E. series Buxeales by Nicolle (2019), including a single E. populnea individual, and as the group shares several morphological traits (Brooker 2000), we recommend further phylogenetic investigation of the series to test its monophyly.
Brooker (2000) placed E. series Melliodorae in E. subsection Terminales along with E. series Heterophloiae and several series not sampled in our study, although he placed E. series Buxeales and Subbuxeales in E. subsection Apicales. This does not match our phylogeny, which shows E. series Melliodorae as more closely related to the last two series than to E. series Heterophloiae (Fig. 5). No previous phylogenetic studies have resolved these subsections to be reciprocally monophyletic (Woodhams et al. 2013; Flores-Rentería et al. 2017), suggesting a re-assessment of the subsectional classification within E. section Adnataria may be needed. Brooker’s E. subsection Terminales was defined as including species with inflexed stamens and an outer ring of staminodes (Brooker 2000), which, if our phylogeny is a true representation of evolutionary relationships, suggests that these traits may either be the ancestral state of E. section Adnataria or independently derived in both series, especially given the placement of E. porosa, which lacks staminodes, in E. series Melliodorae. The close relationship between E. series Buxeales and Subbuxeales is consistent with established classifications, with some authors not recognising E. series Subbuxeales as distinct from E. series Buxeales (Brooker 2000; Brooker et al. 2015).
We are able to confirm that E. porosa is best placed in E. series Melliodorae (Fig. 5), despite morphological similarities to members of the E. odorata complex (E. series Subbuxeales), including its typically mallee growth habit, lack of an operculum scar and axillary inflorescence arrangement (Brooker et al. 2015). This aligns with the classification of Nicolle (2019), but not that of Brooker (2000) who considered this species a member of E. series Buxeales, which included the E. odorata complex in their classification. Eucalyptus series Melliodorae is united by several key morphological traits that E. porosa also shares, namely, the outer operculum being held to anthesis, axillary inflorescences, an intramarginal vein remote from the leaf edge, and a tardily deciduous, broad staminal ring. Only the first two of these characteristics are shared with members of E. series Subbuxeales (Brooker et al. 2015), whereas E. froggattii is the only member of E. series Subbuxeales with a remote intramarginal vein (Brooker and Nicolle 2013). A final trait shared by the other members of E. series Melliodorae we sampled but lacked by E. porosa and two other members of the series we have not sampled (E. bosistoana F.Muell. and E. argophloia Blakely) is a sterile outer ring of stamens, which fits with the placement of E. porosa in this study as sister to the rest of the series (Fig. 5). Additional support for the placement of E. porosa in E. series Melliodorae comes from hybrids of E. porosa × E. leucoxylon being common (Nicolle 2014), but no E. porosa × E. odorata complex hybrids having been recorded.
The seedling of ‘E. aff. polybractea (Mount Jeffcott)’ was also part of the E. series Melliodorae clade in our full dataset phylogeny (Fig. 4). The maternal parent of this seedling is found at Mount Jeffcott within 100 m of the only known stand of E. filiformis, from which it differs primarily in its glaucous juvenile foliage and smooth bark (K. Rule, pers. comm., 22 November 2018). Of the other E. section Adnataria taxa that occur at the site (E. microcarpa, E. largiflorens and E. leucoxylon), E. leucoxylon is by far the most abundant species perhaps explaining why, despite the extensive gene-flow between E. microcarpa and members of the E. odorata complex in the Wimmera, our results, both the phylogeny (Fig. 4) and ABBA-BABA tests (Table 4), showed that this seedling is a E. filiformis × E leucoxylon hybrid. This finding fits with the observations of its morphology (K. Rule, pers. comm., 3 February 2021), although, because we did not sample the maternal parent tree, we cannot say if it is also a E. leucoxylon hybrid, potentially explaining its unique morphology.
The two clades within E. series Subbuxeales (Fig. 5) are supported by morphology, as the typically single-trunked grey-box species and mallee box species form reciprocally monophyletic clades. This is in line with previous studies that have shown that the mallee growth form may be tied to genetic lineages in other eucalypt groups (Hines and Byrne 2001). Although grey boxes are very morphologically similar and establishing geographic boundaries among the taxa are difficult because they intergrade with one another (Bean 2009; Flores-Rentería et al. 2017), E. albopurpurea and E. froggattii have not previously been hypothesised to be closely related in the literature. Eucalyptus albopurpurea has often been regarded as more morphologically similar to members of the E. odorata complex, in particular E. odorata itself, with which it intergrades with on Kangaroo Island (Nicolle 2000, 2014), than to E. froggattii, which is set apart morphologically by its square buds and fruits (Brooker et al. 2015). However, these two species do have morphological similarities not shared by the members of the E. odorata complex, including apparently terminal compound inflorescences, generally broader leaves, and slightly larger buds and fruit (Brooker et al. 2015). More work is needed to clarify the relationship between these two species and the E. odorata complex.
Relationships within the E. odorata complex
Relationships within the E. odorata complex are largely unresolved in our phylogeny, with very little bootstrap support in both the MP and ML analyses, and no species with multiple populations sampled being resolved as monophyletic. However, our PCA (Fig. 3) and hybridisation tests (Tables 4, 5) have shed some light on the possible patterns of relatedness and introgression that explain this lack of resolution. There is strong support for the idea that E. viridis and the two segregate species from the Hunter Valley, E. aenea and E. castrensis, form a clade sister to the rest of the complex if northern (Queensland) populations currently regarded as E. viridis are excluded. Although E. viridis co-occurs with E. polybractea at multiple locations and the two may hybridise on occasion (but no evidence of this was seen in this study), the former is the most morphologically distinct species in the complex, given its linear juvenile leaves and its narrow, green adult leaves, and, at most sites, the two are easily recognisable and morphologically distinct. The two Hunter Valley segregates of E. viridis, E. aenea and E. castrensis, form a sister lineage to E. viridis in our phylogeny. However, the PCA (Fig. 3) and hybridisation tests (Tables 4, 5) give weight to the hypothesis that these populations have experienced introgression from a grey-box species, which may account for their morphological distinctness, although given the results of our ABBA-BABA tests (Table 4), E. albens appears the probable parent rather than E. moluccana as was hypothesised by Nicolle (2019). As the NewHybrid analysis assigns these samples to E. viridis rather than to a hybrid generation, this finding of introgression from a grey box is likely to be the result of historic introgression. The genetically distinct sample of E. aenea (PSF90J) was recognised as differing from smooth-barked typical E. aenea in the field because of its stocking of rough bark that reached ~3 m up the trunks. Although genetically distinct from the other E. aenea samples in our dataset, this is not due to more substantial genetic input from a grey box as hypothesised. Although grey boxes are the most closely related species to occur in the vicinity of E. aenea, ironbark species of E. section Adnataria (E. crebra F.Muell., and E. fibrosa F.Muell.) dominate the site and may be the culprit for this genetic distinctness, although our dataset does not allow us to test this. As our E. castrensis sample is more referable to the broader application of the name per Hill and Stanberg (2002) that may represent E. aenea, and we have not sampled material from the putative E. aenea × E. microcarpa–E. moluccana hybrids, which match the type material of the species (Nicolle 2019), we cannot say anything further regarding the distinction of E. castrensis from E. aenea.
Previous authors have noted that Queensland populations in the vicinity of Inglewood, where the two northern E. viridis samples included in this study were collected, and Durikai State Forest have broader leaves than typical for E. viridis, with Blakely (1934) classifying these populations as E. viridis var. latiuscula Blakely. Chippendale (1988) suggested that these populations may be hybrids of E. viridis and the grey-box E. woollsiana, whereas Brooker et al. (2015) believed that these populations show greater morphological similarities to E. wimmerensis, which they regard as a subspecies of E. viridis, than to the typical form of E. viridis found in the Victorian goldfields and scattered populations in NSW. In the more resolved reduced sampling phylogeny (Fig. 5), the northern samples are placed as the next clade to diverge in the complex after the main E. viridis clade, with the samples of E. polybractea from the most northerly population diverging next, although the node between these clades is supported only in the ML analysis. The ABBA-BABA tests also do not support introgression from the co-occurring grey-box E. woollsiana into these samples compared with E. viridis and E. polybractea from West Wyalong (Table 4), as was previously hypothesised (Chippendale 1988). Along with the placement of these samples in the PCA analyses (Fig. 3), this raises the question of whether rather than an E. viridis × E. woolsiana hybrid, they may be E. viridis × E. polybractea hybrids, although given the lack of E. polybractea populations nearby (the nearest known population at West Wyalong being over 820 km away), they would have to be phantom hybrids, which have been observed in eucalypts previously, albeit without such a large geographic distance to the phantom parent (Kirkpatrick et al. 1973; Hopper and Wardell-Johnson 2004). However, our ABBA-BABA tests showed no support for this hypothesis (Table 4), with the northern E. viridis samples showing a similar number of shared alleles with typical E. viridis as our samples of E. polybractea from the most northerly population of this species at West Wyalong. This leaves the simplest explanation as the most probable, in that these northern populations previously regarded as E. viridis var. latiuscula represent a currently unrecognised distinct entity that is sister to the core E. odorata complex rather than being closely related to typical E. viridis, which fits with the observation of morphological similarities to E. wimmerensis of seedlings from populations previously ascribed to this variety at Inglewood and Durakai in Queensland by Brooker et al. (2015).
Relationships between the few supported clades and the majority of samples in the core E. odorata complex are unsupported, despite the topology returned broadly reflecting geography. Only the taxa known from a single site, namely E. yarriambiack and E. filiformis, are supported as monophyletic (Fig. 5), and because we have sampled all known wild individuals of E. filiformis, we can say with confidence on the basis of the lack of genetic differences between them, that the species represents a single clonal colony. Previously it has been noted that E. filiformis does not appear to not readily reproduce (Rule 2004), although specimens grown at the Royal Botanic Gardens Victoria from seed show differences in adult morphology from the wild individuals (Rule 2018). We have sampled one of these cultivated individuals (PSF73) and shown it to also be clonal, suggesting there may be occasional pollination occurring in the wild population and, for this individual at least, any morphological differences from the wild plants are not due to genetic differences.
Eucalyptus polybractea is polyphyletic in the phylogeny, although relationships among populations are not fully resolved, especially between the type population at West Wyalong (samples PSF74A and PSF74J) and the Victorian goldfields populations (samples PSF21C, PSF27A and PSF27H). Although our phylogenetic analyses suggest that the two samples from the Wimmera (samples PSF56B and PSF56C) are potentially conspecific with E. wimmerensis and the West Wyalong population is sister to the rest of the core E. odorata complex (Fig. 5), in our network analyses collections from the Victorian goldfields and West Wyalong were more genetically similar to one another, and to E. yarriambiack and E. filiformis, than to the Wimmera samples (Fig. 2). On the basis of the supported Flinders Ranges clade, which contains both E. polybractea and E. cajuputea samples (Fig. 5), we suggest that E. polybractea should not be considered to occur west of the Murray Basin.
Although relationships among E. wimmerensis samples in the phylogeny remain unclear, they show little genetic differentiation, although the species’ boundaries remain unclear, given the uncertainty regarding the identity of the E. polybractea populations in the Wimmera. Although E. wimmerensis subsp. grata and Eucalyptus walshii were not genetically distinct from E. wimmerensis in our analyses, the PCA (Fig. 3) and ABBA-BABA tests (Table 4) suggested a low level of introgression from E. microcarpa, which may be responsible for the more robust stature of plants in these populations. The other species found only in the Wimmera and adjacent areas of SA, namely E. silvestris and E. hawkeri, both appear to represent more recent geneflow between E. wimmerensis and the co-occurring E. microcarpa on the basis of our ABBA-BABA and NewHybrid tests (Table 4). This largely fits with the classifications of both Nicolle (2019) and Brooker et al. (2015), although those authors suggested E. odorata as being the E. odorata complex parent in the case of E. silvestris. We have some hesitancy ruling out this hypothesis, given our sampling, because we have shown that there is little genetic distinction between E. odorata and E. wimmerensis, and we cannot rule out there being populations in the Wimmera or adjacent areas of SA that better fit in E. odorata that we have not sampled.
The distinctions between the western taxa, E. cajuputea, E. odorata, and South Australian populations of E. polybractea, are unresolved in our study and require further investigation; however, it is clear what we have called E. polybractea in the Flinders Ranges has minimal genetic links to the typical E. polybractea of Victoria and NSW and may be best considered conspecific with E. cajuputea. Samples from the Flinders Ranges, identified as both E. cajuputea and E. polybractea, form a single clade in our phylogeny when the two aberrant samples are excluded (Fig. 5), suggesting that there is a single lineage in this region that is distinct from other populations from west of the Murray Basin. The two aberrant samples, one each identified as E. polybractea and E. cajuputea, showed significant negative results for most comparisons in our ABBA-BABA tests (Table 4), suggesting that introgression from a species we have not sampled is possible. The only other E. section Adnataria species that occur at Wilpena Pound and with which hybridisation may be occurring are E. porosa, which we have sampled and can therefore rule out, and E. intertexta R.T.Baker, which we have not sampled as part of this study. Eucalyptus intertexta (E. series Buxeales) is related to E. populnea and E. largiflorens, potentially explaining the single positive D-statistic with E. populnea in our ABBA-BABA tests, although the unresolved placement of the E. populnea sample in our phylogenies confounds this because we are unable to establish the relationship between this species and other members of E. series Buxeales. The mallee species E. leptophylla also co-occurs with the population these samples were sourced from and is superficially morphologically similar to members of the E. odorata complex, despite being in E. section Bisectae. This species may be the unknown parent, with its comparatively distant relationship to the E. odorata complex potentially explaining the significant negative D-statistics. Although we have a single sample of this species in our dataset, as part of the most divergent outgroup clade, we were not able to use it as an ingroup in ABBA-BABA tests to test this hypothesis because these require the inclusion of an outgroup with an evolutionary divergence point prior to the divergence of the three ingroup samples (Durand et al. 2011). The other samples of E. cajuputea and E. odorata formed a polytomy along with the Flinders Ranges clade in the ML analysis, which may support a lack of distinctness of these two species outside the Flinders Ranges.
Genetic variation within the core E. odorata complex as a cline
Our findings of extensive introgression among members of the E. odorata complex and co-occurring box species is not unexpected given that previous studies on E. section Adnataria have shown extensive hybridisation leading to morphological taxa not forming genetic clades (Flores-Rentería et al. 2017). However, in regard to the taxonomy of the E. odorata complex, the nature of the core of the clade as a discontinuous cline of morphological and genetic variation running from the Flinders Ranges, south and then east through south-eastern SA, east through the Wimmera and then the Goldfields of Victoria and then north to West Wyalong in NSW also plays a significant role in disagreements among authorities over where taxonomic boundaries should be drawn. This clinal genetic variation has been taxonomically divided by different authors at different points on the basis of different factors they consider most important for classification and, in this paper, we have, a priori, broken it into the seven largely geographically distinct species (E. odorata, E. cajuputea, E. wimmerensis, E. polybractea, E. filiformis, E. walshii and E. yarriambiack).
We see evidence for this cline in the most distal populations (Flinders Range and West Wyalong) being the most distinct, and in relationships between neighbouring populations fluctuate between analyses. This includes, for instance, E. filiformis clustering with E. wimmerensis in one network (Fig. 2b) but being more closely related to E. polybractea from the Victorian goldfields in other analyses (Fig. 2a, 5), and the swapping of relationships between the three main groups (samples from west of the Murray River Basin, E. wimmerensis and allied samples, and eastern E. polybractea and allied samples) in the core group between analyses, even if the alternate relationships are not supported (Fig. 4). This may be indicative of recent rapid diversification of a widespread ancestral population that has undergone vicariance at multiple locations at approximately congruent times. Clines along which diversification has occurred, often resulting in morphologically distinct species with a hybrid zone between them, have previously been observed in other eucalypt groups, including E. populnea and E. brownii Maiden & Cambage (Holman et al. 2003), E. melanophloia F.Muell. and E. whitei Maiden & Blakely (Holman et al. 2011), and the green ashes (E. sect. Eucalyptus; Rutherford et al. 2018). The strong support for IBD (Fig. 6, r: 0.597) in our data is congruent with the existence of a genetic cline.
Taxonomy within the E. odorata complex
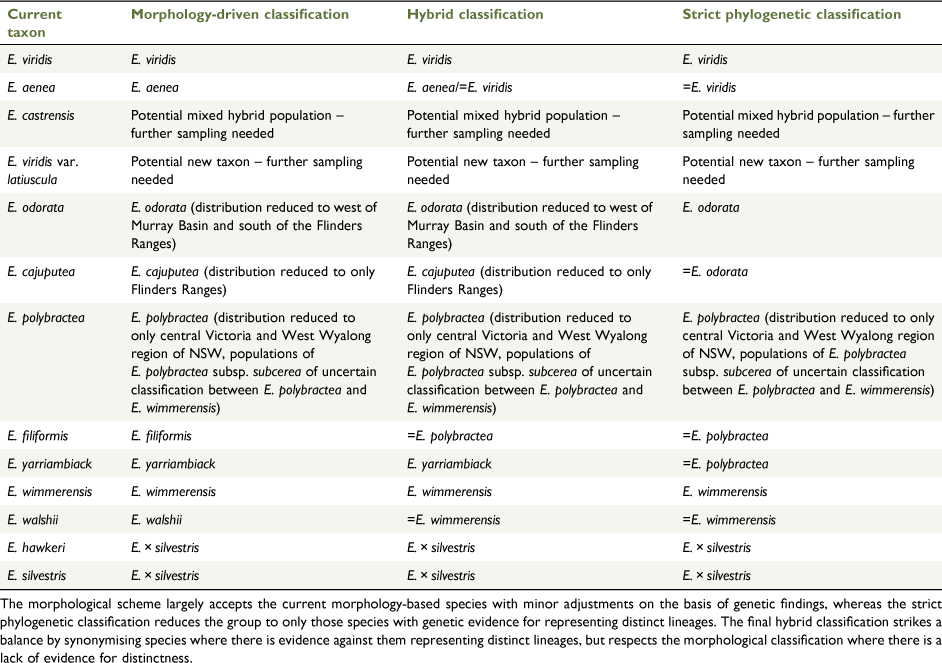
With the extensive level of interspecific geneflow, our dataset has provided evidence for, and the lack of resolved relationships among populations in the E. odorata complex; we do not feel that it is appropriate to make major taxonomic changes on the basis of our study without further work to investigate patterns of diversity in finer detail. However, we see several approaches that could be taken to clarify the taxonomy of the group as outlined in Table 6. The simplest is to uphold the current morphology-based species classification, with adjustments to species distributions where necessary to match resolved phylogenetic relationships. This approach, essentially applying the morphological species concept, would require accepting the recognition of hybrid entities as species, which is being realised as a major driver of plant evolution (Mayr 2000). The one clear change that is well supported in our data is that populations currently regarded as E. polybractea in the Flinders Ranges are not related to the eastern populations of that species and the circumscription of E. cajuputea should be expanded to include these populations.
|
The second approach is to greatly reduce the number of species in the complex to only those that are supported as monophyletic, strictly applying the phylogenetic species concept (Baum 1992). This approach would likely reduce the complex back to two or three species, depending on what future study reveals regarding the distinctness of the northern populations of E. viridis. Eucalyptus aenea and E. castrensis would be synonymised with E. viridis and the cline of the rest of the complex broken up as follows: E. odorata to include populations west of the Murray Basin, E. wimmerensis to cover all populations in the western Wimmera of Victoria and adjacent areas of SA, and E. polybractea to accommodate populations from the eastern Wimmera, Victorian goldfields and around West Wyalong. Additionally, the name E. viridis var. latiuscula may need to be resurrected and its taxonomic rank re-assessed to accommodate the northern E. viridis populations that may represent a further extension of this cline. We would advocate that this approach is less than optimal, because each of these taxa would cover a large range of morphological variation that is somewhat correlated with geography and there are outstanding questions regarding the monophylly of these taxa. For these reasons, at this point, we would advocate a looser application of the phylogenetic concept and using the framework of integrative taxonomy to consider both the morphological and molecular evidence, synonymising species with molecular evidence against them being distinct entities, while maintaining morphological taxa with inconclusive molecular support for their status as distinct populations.
Following this reasoning, E. hawkeri and E. silvestris may need to be synonymised because both represent intergrades between E. wimmerensis and E. microcarpa. Although also showing introgression from E. microcarpa, E. walshii is perhaps better synonymised with E. wimmerensis, given its much closer genetic ties to this species. Recognition of E. filiformis is problematic, because it is clonal and, given its close placement to E. polybractea in our phylogeny and networks (Fig. 2, 5), possibly an outlying population of E. polybractea that has unique morphology owing to the small population size causing bottlenecking and genetic drift. A similar problem exists for E. yarriambiack because our data suggest that it is not experiencing introgression from the co-occurring E. largiflorens, but does not represent a distinct lineage from E. polybractea, rather being another potential small, isolated population undergoing genetic drift or a genetic bottleneck that should be synonymised with this species. However, in the case of E. yarriambiack, the population is not clonal, holds greater genetic diversity than does the clonal E. filiformis and is far enough outside the range of E. polybractea that there is likely to be no ongoing gene flow with populations of that species, all suggesting that the species status for E. yarriambiack is not unreasonable. For E. viridis, E. aenea and E. castrensis, we recommend that further phylogenetic studies are undertaken before taxonomy is re-assessed, because, although we have shown these three taxa are each other’s closest relatives, we have not sampled widely enough to determine whether E. aenea and E. castrensis are distinct lineages from E. viridis or isolated populations experiencing introgression from the co-occurring and more locally abundant grey-box species E. albens. In addition, further work is needed to investigate the relationships of the Queensland populations currently regarded as E. viridis to that species and to E. polybractea, or whether the name E. viridis var. latiuscula needs to be resurrected and given the rank of species.
Future directions
In the case of the E. odorata complex, this study has seemingly reached the extent of informativeness for data generated using restriction site-associated DNA sequencing. To get a clearer view of the evolutionary history of the group, other data types are likely to be needed to increase resolution, such as target capture data for gene sequences that have the potential to be better correlated with the observed morphological variation. Conversely, given the lack of divergence among species, population genetic approaches such as denser sampling at each site and Structure analyses (Pritchard et al. 2000), or its spatially explicit cousin, ConStruct (Bradburd et al. 2018), may help resolve species boundaries in the complex. However, it may not be possible to establish a clear and widely accepted species taxonomy in the E. odorata complex because speciation is incomplete and ongoing, and we can access only a single point in time without knowing the future trajectory of diverging populations. Classifying organisms into discrete groups is a very human task and such discreet groups may not reflect the processes of the natural world, which are much more continuous in nature and, thus, this task will always be problematic.
Conclusions
Overall, we have been able to confirm the boundaries of the E. odorata complex, clarified the classification of some taxa and resolved the relationships among a limited number of species. We have also shown that ABBA-BABA tests are a valuable tool for understanding relationships in the eucalypts, and these results together with our NewHybrids analysis have shown that there is clearly a large amount of historic and recent hybridisation happening in this group, especially with the closely related grey boxes. We showed that E. viridis, E. aenea and E. castrensis represent a distinct lineage within the complex, and E. viridis var. latiuscula from southern Qld may represent a valid taxon. Given the substantial amount of genetic data we have employed in this study, the lack of resolution within the core E. odorata complex may reflect the evolutionary history of the group rather than insufficient phylogenetic signal. The core group of species, namely E. cajuputea, E. odorata, E. wimmerensis, E. yarriambiack, E. filiformis, E. walshii and E. polybractea, possibly represents a semi-circular-shaped genetic cline that runs from the Flinders Ranges south-east through south-eastern SA, east through inland Victoria before turning north toward the West Wyalong area of NSW, which different authors have divided into separate taxa at different points. Further work on this group is required to answer the questions that our results have raised; however, it seems likely that finding suitable taxonomic solutions may remain challenging.
Supplementary material
Supplementary material is available online.
Data availability
The data that support this study are available in Dryad at https://doi.org/10.5061/dryad.prr4xgxmp.
Conflicts of interest
David Cantrill and Mike Bayly are both editors for Australian Systematic Botany. Despite this relationship, they did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
Research costs were financially supported by a grant from Eucalypt Australia (grant number 2016‐42) and the Jim Ross Studentship from the Cybec Foundation. Patrick Fahey was supported by the Sophie Ducker Postgraduate Scholarship from the University of Melbourne Botany Foundation in 2019 and 2020.
Acknowledgements
We thank Kevin Rule for provision of samples, determination of specimens, and advice on the project, and Harvey Orel for assisting with field collections. We are grateful to Peter Symes for granting permission for and accommodating sampling from The Royal Botanic Gardens Victoria’s living collection.
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW, Zinner D (2013) Hybridization and speciation. Journal of Evolutionary Biology 26, 229–246.| Hybridization and speciation.Crossref | GoogleScholarGoogle Scholar |
Aguirre NC, Filippi CV, Zaina G, Rivas JG, Acuña CV, Villalba PV, García MN, González S, Rivarola M, Martínez MC, Puebla AF, Morgante M, Hopp HE, Paniego NB, Marcucci Poltri SN (2019) Optimizing ddRADseq in non-model species: a case study in Eucalyptus dunnii Maiden. Agronomy 9, 484
| Optimizing ddRADseq in non-model species: a case study in Eucalyptus dunnii Maiden.Crossref | GoogleScholarGoogle Scholar |
Alwadani KG, Janes JK, Andrew RL (2019) Chloroplast genome analysis of box-ironbark Eucalyptus. Molecular Phylogenetics and Evolution 136, 76–86.
| Chloroplast genome analysis of box-ironbark Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160, 1217–1229.
| A model-based method for identifying species hybrids using multilocus genetic data.Crossref | GoogleScholarGoogle Scholar |
Baum D (1992) Phylogenetic species concepts. Trends in Ecology & Evolution 7, 1–2.
| Phylogenetic species concepts.Crossref | GoogleScholarGoogle Scholar |
Bean AR (2009) Taxonomic and nomenclatural notes on the Eastern grey boxes (Eucalyptus ser. Moluccanae Chippendale, Myrtaceae) and the reinstatement of Eucalyptus woollsiana R.T.Baker. Austrobaileya 8, 25–34.
Blakely WF (1934) ‘A key to the eucalypts: with descriptions of 500 species and 138 varieties, and a companion to J. H. Maiden’s critical revision of the genus Eucalyptus.’ (The Worker Trustees: Sydney, NSW, Australia)
Bradburd GS, Coop GM, Ralph PL (2018) Inferring continuous and discrete population genetic structure across space. Genetics 210, 33–52.
| Inferring continuous and discrete population genetic structure across space.Crossref | GoogleScholarGoogle Scholar |
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Hér. (Myrtaceae). Australian Systematic Botany 13, 79–148.
| A new classification of the genus Eucalyptus L’Hér. (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Brooker MIH, Nicolle D (2013) ‘Atlas of leaf venation and oil gland patterns in the eucalypts.’ (CSIRO Publishing: Melbourne, Vic., Australia)
Brooker MIH, Slee AV, Connors JR, Duffy S, West J (2015) ‘EUCLID: eucalypts of Australia’, 4th edn. (CSIRO: Canberra, ACT, Australia)
Chippendale GM (1988) ‘Flora of Australia. Vol. 19: Myrtaceae – Eucalyptus, Angophora.’ (Australian Government Publishing Service: Canberra, ACT, Australia)
Collins RA, Hrbek T (2015) An in silico comparison of reduced-representation and sequence-capture protocols for phylogenomics. bioRxiv 2015, 032565
| An in silico comparison of reduced-representation and sequence-capture protocols for phylogenomics.Crossref | GoogleScholarGoogle Scholar |
Copeland LM, Hunter JT (2005) Range extension, habitat and conservation status of three rare mallees, Eucalyptus castrensis, Eucalyptus fracta and Eucalyptus pumila from the Hunter Valley, NSW. Cunninghamia 9, 307–309.
Council of Heads of Australasian Herbaria (2016) ‘Australian Plant Census, IBIS database. Centre for Australian National Biodiversity Research.’ (Australian Government: Canberra, ACT, Australia)
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27, 2156–2158.
| The variant call format and VCFtools.Crossref | GoogleScholarGoogle Scholar |
Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Molecular Biology And Evolution 28, 2239–2252.
| Testing for ancient admixture between closely related populations.Crossref | GoogleScholarGoogle Scholar |
Eaton DAR, Overcast I (2020) ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594.
| ipyrad: interactive assembly and analysis of RADseq datasets.Crossref | GoogleScholarGoogle Scholar |
Fahey PS, Fowler RM, McLay TGB, Udovicic F, Cantrill DJ, Bayly MJ (2021) Divergent lineages in a semi-arid mallee species, Eucalyptus behriana, correspond to a major geographic break in southeastern Australia. Ecology and Evolution 11, 664–678.
| Divergent lineages in a semi-arid mallee species, Eucalyptus behriana, correspond to a major geographic break in southeastern Australia.Crossref | GoogleScholarGoogle Scholar |
Flores-Rentería L, Rymer PD, Riegler M (2017) Unpacking boxes: integration of molecular, morphological and ecological approaches reveals extensive patterns of reticulate evolution in box eucalypts. Molecular Phylogenetics and Evolution 108, 70–87.
| Unpacking boxes: integration of molecular, morphological and ecological approaches reveals extensive patterns of reticulate evolution in box eucalypts.Crossref | GoogleScholarGoogle Scholar |
Griffin AR, Burgess IP, Wolf L (1988) Patterns of natural and manipulated hybridisation in the genus Eucalyptus L’Hérit. – 1. A review. Australian Journal of Botany 36, 41–66.
| Patterns of natural and manipulated hybridisation in the genus Eucalyptus L’Hérit. – 1. A review.Crossref | GoogleScholarGoogle Scholar |
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691–699.
| dartR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing.Crossref | GoogleScholarGoogle Scholar |
Guo C, Ma P-F, Yang G-Q, Ye X-Y, Guo Y, Liu J-X, Liu Y-L, Eaton DAR, Guo Z-H, Li D-Z (2020) Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos. Systematic Biology 70, 756–773.
| Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos.Crossref | GoogleScholarGoogle Scholar |
Hill KD (1997) New species in Angophora and Eucalyptus (Myrtaceae) from New South Wales. Telopea 7, 97–109.
| New species in Angophora and Eucalyptus (Myrtaceae) from New South Wales.Crossref | GoogleScholarGoogle Scholar |
Hill KD, Stanberg LC (2002) Eucalyptus castrensis (Myrtaceae), a new species from New South Wales. Telopea 9, 773–776.
| Eucalyptus castrensis (Myrtaceae), a new species from New South Wales.Crossref | GoogleScholarGoogle Scholar |
Hines B, Byrne M (2001) Genetic differentiation between mallee and tree forms in the Eucalyptus loxophleba complex. Heredity 87, 566–572.
| Genetic differentiation between mallee and tree forms in the Eucalyptus loxophleba complex.Crossref | GoogleScholarGoogle Scholar |
Holman JE, Hughes JM, Fensham RJ (2003) A morphological cline in Eucalyptus: a genetic perspective. Molecular Ecology 12, 3013–3025.
| A morphological cline in Eucalyptus: a genetic perspective.Crossref | GoogleScholarGoogle Scholar |
Holman JE, Hughes JM, Fensham RJ (2011) Origins of a morphological cline between Eucalyptus melanophloia and Eucalyptus whitei. Australian Journal of Botany 59, 244–252.
| Origins of a morphological cline between Eucalyptus melanophloia and Eucalyptus whitei.Crossref | GoogleScholarGoogle Scholar |
Hopper S, Wardell-Johnson G (2004) Eucalyptus virginea and E. relicta (Myrtaceae), two new rare forest trees from south-western Australia allied to E. lanepoolei, and a new phantom hybrid. Nuytsia 15, 71–84.
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology And Evolution 23, 254–267.
| Application of phylogenetic networks in evolutionary studies.Crossref | GoogleScholarGoogle Scholar |
Jackson HD, Steane DA, Potts BM, Vaillancourt RE (1999) Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae). Molecular Ecology 8, 739–751.
| Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071.
| adegenet 1.3-1: new tools for the analysis of genome-wide SNP data.Crossref | GoogleScholarGoogle Scholar |
Jones RC, Nicolle D, Steane DA, Vaillancourt RE, Potts BM (2016) High density, genome-wide markers and intra-specific replication yield an unprecedented phylogenetic reconstruction of a globally significant, speciose lineage of Eucalyptus. Molecular Phylogenetics and Evolution 105, 63–85.
| High density, genome-wide markers and intra-specific replication yield an unprecedented phylogenetic reconstruction of a globally significant, speciose lineage of Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Jordan R, Hoffmann AA, Dillon SK, Prober SM (2017) Evidence of genomic adaptation to climate in Eucalyptus microcarpa: implications for adaptive potential to projected climate change. Molecular Ecology 26, 6002–6020.
| Evidence of genomic adaptation to climate in Eucalyptus microcarpa: implications for adaptive potential to projected climate change.Crossref | GoogleScholarGoogle Scholar |
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In ‘Data production and analysis in population genomics. Vol. 888’. (Eds Pompanon F, Bonin A) pp. 67–89. (Humana Press: Totowa, NJ, USA)
Kirkpatrick JB, Simmons D, Parsons RF (1973) The relationship of some populations involving Eucalyptus cypellocarpa and E. globulus to the problem of phantom hybrids. New Phytologist 72, 867–876.
| The relationship of some populations involving Eucalyptus cypellocarpa and E. globulus to the problem of phantom hybrids.Crossref | GoogleScholarGoogle Scholar |
Kirschner P, Arthofer W, Pfeifenberger S, Záveská E, Schönswetter P, Steiner FM, Schlick-Steiner BC, The STEPPE Consortium (2021) Performance comparison of two reduced-representation based genome-wide marker-discovery strategies in a multi-taxon phylogeographic framework. Scientific Reports 11, 3978
| Performance comparison of two reduced-representation based genome-wide marker-discovery strategies in a multi-taxon phylogeographic framework.Crossref | GoogleScholarGoogle Scholar |
Knaus BJ, Grünwald NJ (2017) vcfr: a package to manipulate and visualize variant call format data in R. Molecular Ecology Resources 17, 44–53.
| vcfr: a package to manipulate and visualize variant call format data in R.Crossref | GoogleScholarGoogle Scholar |
Larcombe MJ, Holland B, Steane DA, Jones RC, Nicolle D, Vaillancourt RE, Potts BM (2015) Patterns of reproductive isolation in Eucalyptus: a phylogenetic perspective. Molecular Biology And Evolution 32, 1833–1846.
| Patterns of reproductive isolation in Eucalyptus: a phylogenetic perspective.Crossref | GoogleScholarGoogle Scholar |
Luo A, Ling C, Ho SYW, Zhu C-D (2018) Comparison of methods for molecular species delimitation across a range of speciation scenarios. Systematic Biology 67, 830–846.
| Comparison of methods for molecular species delimitation across a range of speciation scenarios.Crossref | GoogleScholarGoogle Scholar |
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10
| Cutadapt removes adapter sequences from high-throughput sequencing reads.Crossref | GoogleScholarGoogle Scholar |
Mayr E (1963) ‘Animal species and evolution.’ (Belknap Press of Harvard University Press: Cambridge, MA, USA)
Mayr E (2000) The biological species concept. In ‘Species concepts and phylogenetic theory: a debate’. (Eds QD Wheeler, R Meier) pp. 17–29. (Columbia University Press: New York, NY, USA)
McVean G (2009) A genealogical interpretation of principal components analysis. PLoS genetics 5, e1000686
| A genealogical interpretation of principal components analysis.Crossref | GoogleScholarGoogle Scholar |
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, Goodstein DM, Dubchak I, Poliakov A, Mizrachi E, Kullan ARK, Hussey SG, Pinard D, van der Merwe K, Singh P, van Jaarsveld I, Silva-Junior OB, Togawa RC, Pappas MR, Faria DA, Sansaloni CP, Petroli CD, Yang X, Ranjan P, Tschaplinski TJ, Ye C-Y, Li T, Sterck L, Vanneste K, Murat F, Soler M, Clemente HS, Saidi N, Cassan-Wang H, Dunand C, Hefer CA, Bornberg-Bauer E, Kersting AR, Vining K, Amarasinghe V, Ranik M, Naithani S, Elser J, Boyd AE, Liston A, Spatafora JW, Dharmwardhana P, Raja R, Sullivan C, Romanel E, Alves-Ferreira M, Külheim C, Foley W, Carocha V, Paiva J, Kudrna D, Brommonschenkel SH, Pasquali G, Byrne M, Rigault P, Tibbits J, Spokevicius A, Jones RC, Steane DA, Vaillancourt RE, Potts BM, Joubert F, Barry K, Pappas GJ, Strauss SH, Jaiswal P, Grima-Pettenati J, Salse J, Van de Peer Y, Rokhsar DS, Schmutz J (2014) The genome of Eucalyptus grandis. Nature 510, 356–362.
| The genome of Eucalyptus grandis.Crossref | GoogleScholarGoogle Scholar |
Nicolle D (2000) New taxa of Eucalyptus informal subgenus Symphyomyrtus (Myrtaceae), endemic to South Australia. Journal of the Adelaide Botanic Garden 19, 83–94.
Nicolle D (2006) ‘Eucalypts of Victoria and Tasmania.’ (Bloomings Books: Melbourne, Vic., Australia)
Nicolle D (2014) Myrtaceae (partly) (version 1). In ‘Flora of South Australia. Vol. 5’. (Ed. J Kellermann) (State Herbarium of South Australia: Adelaide, SA, Australia)
Nicolle D (2019) Classification of the eucalypts (Angophora, Corymbia and Eucalyptus) Version 4. Available at https://www.dn.com.au/Classification-Of-The-Eucalypts.pdf
Nicolle D, Roberts I (2013) ‘Native eucalypts of South Australia.’ (Dean Nicolle: Adelaide, SA, Australia)
O’Leary SJ, Puritz JB, Willis SC, Hollenbeck CM, Portnoy DS (2018) These aren’t the loci you’e looking for: principles of effective SNP filtering for molecular ecologists. Molecular Ecology 27, 3193–3206.
| These aren’t the loci you’e looking for: principles of effective SNP filtering for molecular ecologists.Crossref | GoogleScholarGoogle Scholar |
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and geotyping in model and non-model species. PLoS One 7, e37135
| Double digest RADseq: an inexpensive method for de novo SNP discovery and geotyping in model and non-model species.Crossref | GoogleScholarGoogle Scholar |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
| Inference of population structure using multilocus genotype data.Crossref | GoogleScholarGoogle Scholar |
Pryor LD, Johnson LAS (1971) ‘A classification of the eucalypts.’ (The Australian National University: Canberra, ACT, Australia)
Rule K (1990) Eucalyptus wimmerensis, a new species of Eucalyptus (Myrtaceae) from Victoria and South Australia. Muelleria 7, 193–201.
| Eucalyptus wimmerensis, a new species of Eucalyptus (Myrtaceae) from Victoria and South Australia.Crossref | GoogleScholarGoogle Scholar |
Rule K (1994) Eucalyptus silvestris, a new species of Eucalyptus (Myrtaceae) for Victoria and South Australia and notes on Victorian occurrences of Eucalyptus odorata. Muelleria 8, 193–199.
| Eucalyptus silvestris, a new species of Eucalyptus (Myrtaceae) for Victoria and South Australia and notes on Victorian occurrences of Eucalyptus odorata.Crossref | GoogleScholarGoogle Scholar |
Rule K (2004) New taxa in Eucalyptus (Myrtaceae) for Victoria and notes on Victorian populations of Eucalyptus calycogona. Muelleria 20, 9–32.
| New taxa in Eucalyptus (Myrtaceae) for Victoria and notes on Victorian populations of Eucalyptus calycogona.Crossref | GoogleScholarGoogle Scholar |
Rule K (2012) Five new endemic eucalypts for Victoria. Muelleria 30, 83–105.
| Five new endemic eucalypts for Victoria.Crossref | GoogleScholarGoogle Scholar |
Rule K (2018) Eucalyptus wimmerensis revisited and notes on the morphologies and taxonomies of five Victorian mallee-boxes. Muelleria 37, 39–64.
| Eucalyptus wimmerensis revisited and notes on the morphologies and taxonomies of five Victorian mallee-boxes.Crossref | GoogleScholarGoogle Scholar |
Rutherford S, Wilson PG, Rossetto M, Bonser SP (2016) Phylogenomics of the green ash eucalypts (Myrtaceae): a tale of reticulate evolution and misidentification. Australian Systematic Botany 28, 326–354.
| Phylogenomics of the green ash eucalypts (Myrtaceae): a tale of reticulate evolution and misidentification.Crossref | GoogleScholarGoogle Scholar |
Rutherford S, Rossetto M, Bragg JG, McPherson H, Benson D, Bonser SP, Wilson PG (2018) Speciation in the presence of gene flow: population genomics of closely related and diverging Eucalyptus species. Heredity 121, 126–141.
| Speciation in the presence of gene flow: population genomics of closely related and diverging Eucalyptus species.Crossref | GoogleScholarGoogle Scholar |
Rutherford S, van der Merwe M, Wilson PG, Kooyman RM, Rossetto M (2019) Managing the risk of genetic swamping of a rare and restricted tree. Conservation Genetics 20, 1113–1131.
| Managing the risk of genetic swamping of a rare and restricted tree.Crossref | GoogleScholarGoogle Scholar |
Rutherford S, Wan JSH, Cohen JM, Benson D, Rossetto M (2020) Looks can be deceiving: speciation dynamics of co-distributed Angophora (Myrtaceae) species in a varying landscape. Evolution 75, 310–329.
| Looks can be deceiving: speciation dynamics of co-distributed Angophora (Myrtaceae) species in a varying landscape.Crossref | GoogleScholarGoogle Scholar |
Sansaloni CP, Petroli CD, Carling J, Hudson CJ, Steane DA, Myburg AA, Grattapaglia D, Vaillancourt RE, Kilian A (2010) A high-density diversity arrays technology (DArT) microarray for genome-wide genotyping in Eucalyptus. Plant Methods 6, 16
| A high-density diversity arrays technology (DArT) microarray for genome-wide genotyping in Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS One 13, e0195034
| Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
| RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Byrne M, Vaillancourt RE, Potts BM (1998) Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae). Australian Systematic Botany 11, 25–40.
| Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Steane DA, Nicolle D, McKinnon GE, Vaillancourt RE, Potts BM (2002) Higher-level relationships among the eucalypts are resolved by ITS-sequence data. Australian Systematic Botany 15, 49–62.
| Higher-level relationships among the eucalypts are resolved by ITS-sequence data.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Nicolle D, Potts BM (2007) Phylogenetic positioning of anomalous eucalypts by using ITS sequence data. Australian Systematic Botany 20, 402–408.
| Phylogenetic positioning of anomalous eucalypts by using ITS sequence data.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Nicolle D, Sansaloni CP, Petroli CD, Carling J, Kilian A, Myburg AA, Grattapaglia D, Vaillancourt RE (2011) Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Molecular Phylogenetics and Evolution 59, 206–224.
| Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping.Crossref | GoogleScholarGoogle Scholar |
Steane DA, Potts BM, McLean EH, Collins L, Holland BR, Prober SM, Stock WD, Vaillancourt RE, Byrne M (2017) Genomic scans across three eucalypts suggest that adaptation to aridity is a genome-wide phenomenon. Genome Biology and Evolution 9, 253–265.
| Genomic scans across three eucalypts suggest that adaptation to aridity is a genome-wide phenomenon.Crossref | GoogleScholarGoogle Scholar |
Stöver BC, Müller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11, 7
| TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |
Swofford D (2002) ‘PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0.’ (Sinauer Associates: Sunderland, MA, USA)
Thornhill AH, Crisp MD, Külheim C, Lam KE, Nelson LA, Yeates DK, Miller JT (2019) A dated molecular perspective of eucalypt taxonomy, evolution and diversification. Australian Systematic Botany 32, 29–48.
| A dated molecular perspective of eucalypt taxonomy, evolution and diversification.Crossref | GoogleScholarGoogle Scholar |
Wickham H (2016) ‘ggplot2: Elegant Graphics for Data Analysis.’ (Springer-Verlag: New York, NY, USA)
Woodhams M, Steane DA, Jones RC, Nicolle D, Moulton V, Holland BR (2013) Novel distances for dollo data. Systematic Biology 62, 62–77.
| Novel distances for dollo data.Crossref | GoogleScholarGoogle Scholar |
Yang G-Q, Chen Y-M, Wang J-P, Guo C, Zhao L, Wang X-Y, Guo Y, Li L, Li D-Z, Guo Z-H (2016) Development of a universal and simplified ddRAD library preparation approach for SNP discovery and genotyping in angiosperm plants. Plant Methods 12, 39
| Development of a universal and simplified ddRAD library preparation approach for SNP discovery and genotyping in angiosperm plants.Crossref | GoogleScholarGoogle Scholar |
Zheng Y, Janke A (2018) Gene flow analysis method, the D-statistic, is robust in a wide parameter space. BMC Bioinformatics 19, 10
| Gene flow analysis method, the D-statistic, is robust in a wide parameter space.Crossref | GoogleScholarGoogle Scholar |