Phylogeny, classification and biogeography of Philotheca sect. Erionema (Rutaceae) based on nrDNA sequences
Erin L. Batty A B , Gareth D. Holmes A B , Daniel J. Murphy B , Paul I. Forster C , Will C. Neal A and Michael J. Bayly
A B , Daniel J. Murphy B , Paul I. Forster C , Will C. Neal A and Michael J. Bayly  A *
A *
A School of BioSciences, The University of Melbourne, Vic. 3010, Australia.
B Royal Botanic Gardens Victoria, Birdwood Avenue, Melbourne, Vic. 3004, Australia.
C Department of Environment and Science, Queensland Herbarium, Brisbane Botanic Gardens, Mt Coot-tha Road, Toowong, Qld 4066, Australia.
Australian Systematic Botany 35(4) 326-338 https://doi.org/10.1071/SB22003
Submitted: 12 January 2022 Accepted: 18 July 2022 Published: 16 September 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Philotheca sect. Erionema includes 14 species from eastern Australia and one from south-western Australia. We conducted a phylogenetic analysis of the section, including samples of all species, using sequences of the ITS and ETS regions of nuclear ribosomal DNA. Results were broadly congruent with a previous analysis based on morphological and flavonoid data. The analysis is consistent with the monophyly of the section and supports the monophyly of six species represented by multiple samples. Philotheca verrucosa (A. Rich.) Paul G. Wilson was resolved as paraphyletic with respect to P. freyciana Rozefelds but with poor support. Philotheca glasshousiensis, P. myoporoides and P. myoporoides subsp. myoporoides were clearly polyphyletic, including separate geographic clades and the classification of each of these taxa requires revision. In particular, disjunct northern populations of P. glasshousiensis probably represent a distinct species, the five subspecies of P. myoporoides could be treated as separate species and at least two other distinct groups that are currently included under the circumscription of subsp. myoporoides could be treated as species. The phylogeny revealed deeply divergent, geographically overlapping clades in eastern Australia and substantial distances (up to 900 km) between sister taxa. We infer that biogeography of the group has been shaped largely by vicariant differentiation of taxa.
Keywords: Australia, biogeography, nrDNA, Philotheca freyciana, Philotheca glasshousiensis, Philotheca myoporoides, phylogeny, Rutaceae, taxonomy.
Introduction
Philotheca sect. Erionema (F.Muell.) Paul G.Wilson is one of four sections recognised in the Australian genus Philotheca Rudge (Wilson 2013). The section currently includes 14 species (21 taxa, including subspecies) from eastern Australia and 1 species, P. brucei1 (with 3 subspecies) from south-western Australia (Fig. 1). This is distinguished from other sections of the genus primarily on the basis of the that have two (or rarely several) glands at the base of the white anther apiculum and by the seeds that are ellipsoid, laterally flattened, have a linear hilum on the adaxial face and have a basal chalazal region (Wilson 1998, 2013). Members of the section are insect pollinated (Armstrong 1991) shrubs or small trees that occur in open eucalypt forests or woodlands or in heathlands, sometimes associated with rock outcrops (Wilson 2013).
|

|
Classification of sect. Erionema has seen several changes in the last two decades. The section was transferred from Eriostemon Sm. (sensu Wilson 1970) to Philotheca (Wilson 1998) based on Armstrong’s (1991) morphological phylogenetic analyses of Australian Rutaceae. Circumscription of the section was amended by Wilson (1998) to include the Western Australian (WA) species, P. brucei (previously included in a monotypic section, Eriostemon sect. Osmanthos Paul G.Wilson), on the basis of shared anther and seed morphology. New taxa have also been assigned to sect. Erionema including: a new species, P. freyciana that was segregated from P. verrucosa by Rozefelds (2001a); a new subspecies of P. buxifolia described by Wilson (1998); and a number of new taxa or combinations that were published in a complex of species related to P. myoporoides (Bayly 1998; Rozefelds 2001b; Forster 2005). This last group, in particular, has been taxonomically problematic, with up to 10 largely allopatric subspecies being recognised under a widespread and broadly circumscribed P. myoporoides (Rozefelds 2001b). The five Queensland (Qld) subspecies of this group were elevated to species rank by Forster (2005) but that treatment did not consider the status of the remaining subspecies, resulting in markedly different species concepts for taxa on either side of the New South Wales (NSW)–Qld state border.
Relationships in sect. Erionema have been assessed using datasets based on morphology and leaf flavonoids (Bayly 2001). Those analyses supported monophyly of the section, including P. brucei but many relationships were not well resolved. In particular, most members of the P. myoporoides group (including currently accepted subspecies and recent segregate species) formed a large polytomy that also included P. verrucosa, P. scabra, P. hispidula and P. buxifolia. Relationships inferred among that group were based largely on homoplasious morphological characters.
A small number of Philotheca species have been included in molecular phylogenetic studies assessing higher-level relationships of Australian Rutaceae (Chase et al. 1999; Scott et al. 2000; Groppo et al. 2008; Salvo et al. 2010; Bayly et al. 2013; Morton and Telmer 2014; Appelhans et al. 2021) or as outgroups in analyses of other genera (Othman et al. 2010; French et al. 2016). At most, these studies have included one species of sect. Erionema. As such, there are currently no molecular data for assessing the circumscription of this section or the relationships of taxa within this.
The current study uses newly generated sequences of the internal transcribed spacers (ITS) and external transcribed spacer (ETS) of nuclear ribosomal DNA to assess relationships among members of Philotheca sect. Erionema. The aims of this study were: (1) to test the monophyly of the section, in particular the inclusion of P. brucei; (2) to test the classification of species and subspecies, especially those associated with P. myoporoides; and (3) to gain insight into the biogeographic history of this group in eastern and south-western Australia.
Materials and methods
Taxon sampling and DNA sequencing
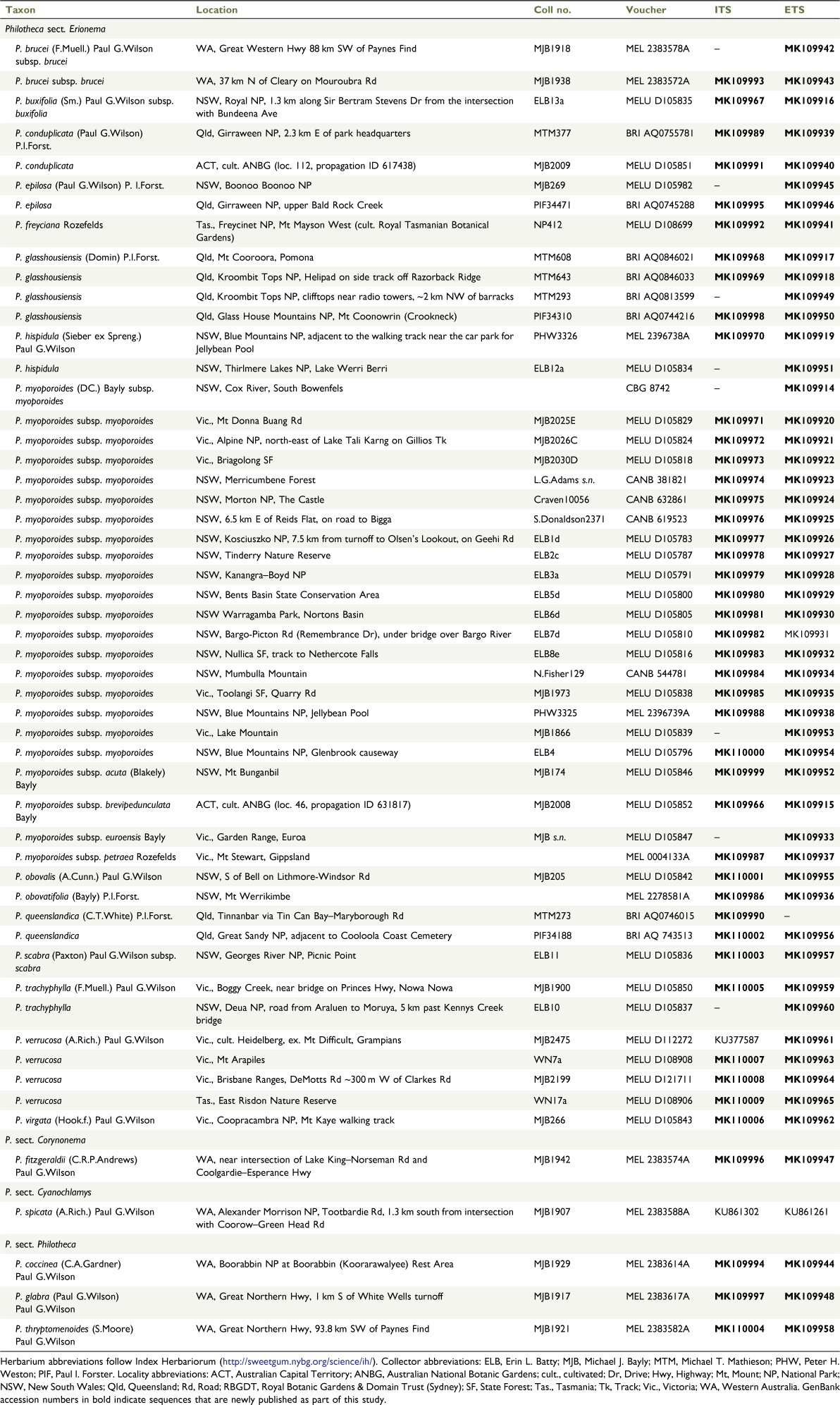
Plant material was obtained from field collections, cultivated plants of known provenance, and in a few cases from existing herbarium specimens (Table 1).
In total, 49 ingroup samples were used, representing all 15 species of sect. Erionema (19 of the 24 currently accepted taxa including segregate subspecies; the 5 subspecies not sampled were Philotheca brucei subsp. brevifolia and subsp. cinerea, P. buxifolia subsp. falcata and subsp. obovata, and P. scabra subsp. latifolia). Initial results indicated that P. myoporoides subsp. myoporoides, the most geographically widespread subspecies in the section, was genetically heterogeneous. This taxon was therefore sampled more intensively using material from 19 different locations. Representatives of the three other sections of Philotheca were used as outgroups: P. coccinea, P. glabra and P. thryptomenoides from sect. Philotheca; P. fitzgeraldii from sect. Corynonema (Paul G.Wilson) Paul G.Wilson; P. spicata from sect. Cyanochlamys (Bartl. ex F.Muell.) Paul G.Wilson.
DNA isolation, amplification and sequencing of ITS and ETS followed the methods described by Bayly et al. (2015).
Sequence editing and alignment
Contiguous sequences for each region were assembled and edited using Sequencher (ver. 4.8, Gene Codes Corporation, Ann Arbor, MI, USA) or Geneious (ver. 9, see https://www.geneious.com/; Kearse et al. 2012). Sequencing chromatograms were carefully scrutinised for double signal peaks that could indicate variation among the many copies of ITS and ETS regions that are present in the genome of each plant. Standard IUPAC nucleotide ambiguity codes were used for any positions in the sequence showing evidence of such polymorphisms. Most sequences included some polymorphic sites (fewer than ten across the entire span of the ITS + ETS regions) but two samples had very high numbers, namely Philotheca myoporoides subsp. brevipedunculata and P. buxifolia, with 29 and 27 polymorphic sites respectively across the combined rDNA sequences. These two sequences were retained in the dataset so that these taxa could be represented in analyses, although we recognise that the large number of polymorphic nucleotide sites in these sequences could create uncertainty about their phylogenetic placement. In both of these samples, the 5.8S rDNA region was highly conserved when compared with the flanking ITS-1 and ITS-2 regions (showing no differences in P. buxifolia and only one ambiguous base in P. myoporoides subsp. brevipedunculata), making the recovered sequences unlikely to include rDNA pseudogenes (see Bailey et al. 2003).
Sequences were aligned in Geneious with some manual adjustment. Insertion–deletion events (INDELs) were coded using the ‘simple INDEL coding’ approach of Simmons and Ochoterena (2000), with a single character appended to the end of the data matrix, representing each INDEL, whether single- or multi-base. The final alignment, including INDEL characters (identified as distinct ITS and ETS INDEL CHARSETs in the nexus file), is deposited in TreeBase (see http://treebase.org/; study accession number 26368).
Phylogenetic analyses
Combined ITS and ETS sequences were analysed using maximum parsimony (MP) as implemented in PAUP* 4.0 beta (ver. 10, D. L. Swofford, see https://paup.phylosolutions.com/) and by Bayesian inference (BI) using MrBayes (ver. 3.1.2, see https://nbisweden.github.io/MrBayes/; Ronquist and Huelsenbeck 2003), with and without the presence of INDEL characters. MP analyses were performed using heuristic tree searches, a CLOSEST addition sequence, TBR branch swapping, MAXTREES set at 30 000, all characters equally weighted and gaps treated as missing data. MP Bootstrap analyses used 1000 ‘full heuristic’ replicates with MAXTREES set at 3000 per replicate. Models for BI analyses were selected using the Akaike Information Criterion as implemented in MrModeltest (ver. 2.3, J. Nylander, see https://github.com/nylander/MrModeltest2). The chosen models were GTR + Г + I for ITS and GTR + Г for ETS; INDELs were analysed under the ‘restriction’ model. BI analyses used the default settings of MrBayes and each included two runs of four chains, each run for 2 000 000 generations. Trees were sampled every 1000 generations and a majority rule consensus was computed (with trees from the first 500 000 generations discarded as burn-in). To determine that the runs had converged on a stationary distribution and that the burn-in period was adequate, the distribution of likelihood values in Tracer (ver. 1.5, A. Rambaut, see https://beast.community/tracer.html) and the standard deviation of split frequencies (that was <0.01 at the end of the runs) were evaluated.
Results
The combined nrDNA dataset included 1279 characters, of which 365 were variable and 237 were parsimony informative (119 parsimony informative characters from ITS, including 4 INDEL characters and 119 from ETS, including 6 INDEL characters). BI and MP analyses of the combined ITS and ETS dataset produced congruent results, as did analyses with and without INDEL characters included, therefore only the BI tree based on the analysis including INDEL characters is presented here (Fig. 2), showing both Bayesian posterior probabilities (PP) and parsimony bootstrap support (BS) values. The MP analysis (not shown) produced 30 000 equally parsimonious trees with a length 650 steps and a consistency index of 0.66.
The analysis (Fig. 2) is consistent with the monophyly of sect. Erionema (PP 0.99, BS 84%) as currently circumscribed, including the WA species P. brucei. The basal nodes in the BI tree are not well supported but both P. brucei and a clade of three species (P. obovalis, P. virgata and P. trachyphylla) clearly sit outside a large, well-supported clade (PP 1, BS 92%) that includes all other taxa. That large clade is referred to here as the ‘pedunculate clade’ because the members are characterised by inflorescences with pedicels (one to many) borne on a peduncle; in contrast, pedicels in P. brucei, P. obovalis, P. virgata and P. trachyphylla arise directly from the subtending leaf axil and lack a distinct peduncle. Pedunculate, cymose, axillary inflorescences are inferred to be apomorphic within sect. Erionema and within the genus based on the morphological analysis of Bayly (2001).
Within the pedunculate clade, a well-supported basal dichotomy separates a clade of P. queenslandica sister to two samples of P. glasshousiensis from all other samples.
Ten taxa in the analysis were represented by multiple accessions that allowed their monophyly to be tested. Among these, six were supported as monophyletic, namely P. trachyphylla (PP 1, BS 87%), P. brucei (PP 1, BS 100%), P. queenslandica (PP 1, BS 100%), P. conduplicata (PP 1, BS 100%), P. hispidula (PP 1, BS 92%), and P. epilosa (PP 1, BS 100%). Philotheca verrucosa was resolved as paraphyletic with respect to P. freyciana but support for that relationship was lacking with PP of 0.79 and BS <50%. Three taxa were notably resolved as polyphyletic, namely P. myoporoides (with the currently recognised subspecies spread across multiple clades), P. myoporoides subsp. myoporoides and P. glasshousiensis.
Samples of P. myoporoides subsp. myoporoides fell into two distinct parts of the nrDNA tree, separated by well-supported nodes. These two clusters of samples have a geographic basis. Neither cluster was resolved as monophyletic, with samples being placed at polytomous nodes that also included other taxa. The two clusters are identified here (Fig. 2) as the ‘southern’ group that includes samples from the montane highlands of eastern Victoria (Vic.) and southern NSW, and the ‘northern’ group that includes samples from near Sydney to the South Coast of NSW, generally at lower altitudes.
The samples of P. glasshousiensis were also resolved in two distinct geographic clades. These are identified here as the ‘southern’ samples (Fig. 2) from the Glasshouse Mountains and Mount Cooroora, and the ‘northern’ samples from Kroombit Tops National Park.
Discussion
Comparison of nrDNA phylogeny with previous analyses based on morphological and flavonoid data
The nrDNA phylogeny for sect. Erionema is largely congruent with that of Bayly (2001) that was based on morphological and leaf flavonoid characters. The degree of congruence between the two studies using different datasets is highly unlikely to result from chance and adds confidence to interpretations of relationships. Here we briefly summarise some of the key points of congruence and incongruence between results of the two studies, with the latter largely relating to nodes that are poorly supported in one study or the other.
Both the current molecular and the earlier morphological and flavonoid studies resolved the pedunculate clade as a group, outside of which Philotheca brucei from WA, and P. obovalis, P. trachyphylla and P. virgata from south-east Australia, were resolved as early diverging lineages. Relationships among those branches were slightly different in the two studies. In the morphological and flavonoid study (Bayly 2001), P. obovalis and P. virgata formed a clade that was sister to the rest of the section, one node below P. trachyphylla in the tree; Bayly (2001) incorrectly indicated that P. obovalis and P. virgata were united by having 4-merous flowers (a state found consistently only in P. virgata) but the two do share the trait, unique in the genus, of having a reduced number of carpels (<5) per flower (Wilson 1970, 2013). In the nrDNA tree, P. obovalis and P. virgata were placed with P. trachyphylla in a well-supported clade (PP 1, BS 78%) in which P. trachyphylla was resolved, but not well supported (PP 0.92, BS <50%), as sister to P. virgata.
Within the pedunculate clade, the nrDNA tree provided better resolution than Bayly’s (2001) tree based on morphological and flavonoid data. Also, by having multiple accessions for many taxa in the present study, the nrDNA tree has highlighted relationships that could not be investigated in the morphological and flavonoid analysis, in which each taxon was represented by a single aggregate unit. Within the pedunculate clade, key points of congruence between the two studies are: (1) the placement of P. queenslandica + southern populations of P. glasshousiensis together as a clade sister to all other taxa; (2) the nesting of P. verrucosa, P. buxifolia, P. scabra and P. hispidula within P. myoporoides. A difference between the studies is that the morphological and flavonoid tree resolved P. buxifolia, P. scabra and P. hispidula as a clade, whereas the nrDNA tree showed P. hispidula as well separated from a P. buxifolia + P. scabra clade, with strong support. In this case, the relationships in the morphological and flavonoid tree were based entirely on homoplasious characters (see fig. 5.15 in Bayly 2001).
Implications for taxonomy
Polyphyly of Philotheca myoporoides
A significant result from the phylogenetic study presented here is the strong evidence that Philotheca myoporoides is not monophyletic either in the current circumscription (Forster 2005; Wilson 2013) or the previously broader circumscriptions of Wilson (1970), Bayly (1998) or Rozefelds (2001b). The current circumscription of P. myoporoides should not be maintained and our results add weight to the argument for elevating the remaining subspecific taxa to the rank of species, as Forster (2005) did with the subspecies from south-eastern Qld. This would entail raising the segregate subspecies (subsp. acuta, subsp. brevipedunculata, subsp. euroensis and subsp. petraea) to species rank. Such an elevation in the rank of these taxa is supported by their generally allopatric distributions (Fig. 1) and generally distinct morphology from each other (albeit largely in leaf shape and size; Bayly 1998; Rozefelds 2001b), and from other species in the genus. Although the monophyly of each of these taxa was not explicitly tested in the current study, each being represented by a single accession, they are all resolved in distinct positions in the phylogeny. For subsp. acuta, an earlier name at species rank, Eriostemon affinis Sprague, is available (Bayly 1998; Wilson 2013) but there are no existing names at species rank for the other segregate subspecies and therefore new combinations or new names are required.
An alternative response to the polyphyly of P. myoporoides would be to retain a broad circumscription of the species and to subsume into this, as synonyms or subspecies, all other members of the pedunculate clade (as identified on Fig. 2). That would require uniting 18 currently accepted taxa under one species, including long-accepted and morphologically distinct species such as P. verrucosa, P. hispidula, P. scabra and P. buxifolia. We consider this option as untenable, given the degree of morphological, ecological and genetic differentiation within the pedunculate clade, and such a broad species concept is inconsistent with other species in the family and would seem unparalleled in current classifications of any other group in the Australian vascular flora.
Polyphyly of Philotheca myoporoides subsp. myoporoides
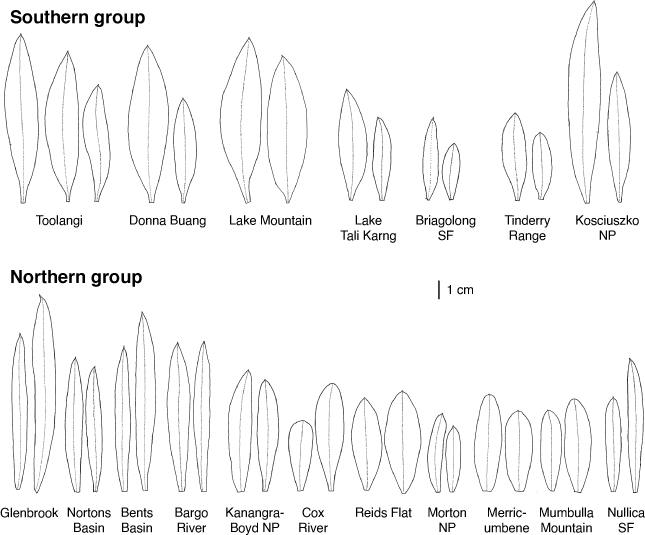
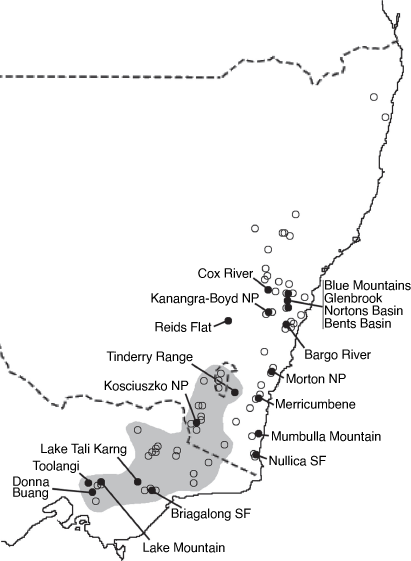
Philotheca myoporoides subsp. myoporoides is clearly polyphyletic, with samples falling in two well-separated parts of the nrDNA tree (Fig. 2). These two genetic groups are geographically and ecologically separated and align, at least partly, with morphological variants previously discussed by Wilson (1970, 2013) and Bayly (1998). In Fig. 3 we have mapped the distribution of the two genetic groups and have also indicated, based on preliminary examination of herbarium material, what we infer to be the geographical ranges of these groups.
|
The ‘southern’ genetic group includes the samples from the highlands of Vic., southern NSW and the Australian Capital Territory (ACT). These typically grow in montane or subalpine forests, commonly in Vic. in forests dominated by Mountain Ash (Eucalyptus regnans F.Muell.) or Alpine Ash (Eucalyptus delegatensis R.T. Baker), often around granite outcrops but also on other substrates. This group equates to the ‘mountain form’ of subsp. myoporoides discussed by Wilson (1970, 2013) and Bayly (1998). The earliest species name relating to this group is Eriostemon lancifolius F.Muell. Plants of this group often have leaves that are broader, relative to their length, than those of the ‘northern’ genetic group but are variable in leaf shape and size (e.g. Fig. 4).
The ‘northern’ nrDNA group in subsp. myoporoides is restricted to NSW and is disjunct from populations of the ‘southern group’ (Fig. 3). It occurs in a range of habitats, mostly at lower altitudes than populations of the ‘southern group’ and is morphologically variable. The samples in our dataset from the Blue Mountains, Glenbrook, Kanangra–Boyd National Park, Bargo River, Bents Basin and Nortons Basin (all from the Central Coast and Central Tablelands of NSW, along water courses, mostly at low altitudes), morphologically match the type of P. myoporoides and have relatively long, linear leaves (Fig. 4). Other samples in the ‘northern’ nrDNA group have different leaf tips and the leaf shapes are relatively short and broad. These include the sample from Cox River that morphologically resembles the type of Eriostemon cuspidatus A.Cunn. from the same locality. Other samples with shorter or broader leaves include that from Reids Flat near Bigga in the Central Tablelands and samples from rocky areas in hills and escarpments of the South Coast or Southern Tablelands regions (Merricumbene Forest in Deua National Park, Morton National Park and Mumbulla Mountain). The last group of specimens occurs in localities close to those of P. myoporoides subsp. brevipedunculata (in Deua National Park and nearby areas) in similar upland habitats; although approaching that taxon in leaf dimensions they have larger leaves, longer peduncles and distinct nrDNA from the single sample of subsp. brevipedunculata included here. Nonetheless, the morphological, ecological and genetic distinctiveness of subsp. brevipedunculata in this area could be worthy of further detailed investigation based on more intensive sampling.
The two distinct genetic groups in subsp. myoporoides should be recognised as at least two different species. However, further work is required to clearly circumscribe them. Although we assign herbarium samples to morphological groups (as in the maps in Fig. 3), there is substantial morphological variation within these groups, especially the northern one, and despite the morphological extremes being fairly distinct, there is a lack of clear separation in leaf attributes (or reproductive features) when all specimens are considered. Also, in the analysis here, neither the ‘southern’ nor ‘northern’ nrDNA groups in subsp. myoporoides was resolved as monophyletic. This could be due to a lack of signal in the ITS and ETS markers or, amongst other hypotheses, it could reflect the presence of multiple taxa in these nrDNA groups. Other species that group closely with the genetic groups of subsp. myoporoides in the nrDNA tree (e.g. P. scabra, P. buxifolia and P. obovatifolia with the ‘northern’ nrDNA group, and P. verrucosa with the ‘southern group’) are morphologically distinct, to the extent that they could not reasonably be treated as conspecific. Further resolution of both genetic relationships and morphological variation in the nrDNA groups of subsp. myoporoides is needed to inform the delimitation of taxa.
Polyphyly of Philotheca glasshousiensis
The polyphyly of P. glasshousiensis provides strong evidence that northern and southern populations should be recognised as two distinct species. The southern populations, from the Glasshouse Mountains and Mount Cooroora, represent P. glasshousiensis sensu stricto, the type being from the Glasshouse Mountains (Mount Coonowrin). These southern populations are most closely related to but clearly distinct from P. queenslandica, being separated by long branches on the phylogenetic tree (Fig. 2). Although these two species both occur in south-eastern Qld, there is a clear ecological separation between them, with P. glasshousiensis growing in rocky areas towards the summits of the mountains and P. queenslandica restricted to lowland heaths in wallum vegetation that is periodically inundated.
Herbarium specimens from the northern populations, at Cania Gorge (not sequenced here) and Kroombit Tops, were included in the circumscription of P. glasshousiensis (or P. myoporoides subsp. leichhardtii (Benth.) Paul G. Wilson) by Bayly (1998) and Forster (2005) on the basis of morphological resemblance and their occurrence in similar cliff line habitat. Our recent, preliminary comparisons indicate that, although the northern populations resemble P. glasshousiensis sensu stricto in most qualitative features (many of which are highly conserved across sect. Erionema), they usually have larger leaves than those of P. glasshousiensis sensu stricto but there is overlap in leaf sizes. More detailed study is needed to clarify the extent to which the two genetic groups can be distinguished morphologically.
The status of Philotheca freyciana warrants further investigation
Philotheca freyciana was described by Rozefelds (2001a) for populations from Freycinet Peninsula, Tasmania (Tas.), that were previously included in the more widespread species P. verrucosa (e.g. Wilson 1970; as Eriostemon verrucosus A.Rich.). Rozefelds (2001a) distinguished P. freyciana from P. verrucosa on the basis of habit, leaf size and anther apex shape. Because of its limited distribution and small population sizes, P. freyciana is listed as Endangered under both Australia’s Environment Protection and Biodiversity Conservation Act 1999 and Tasmania’s Threatened Species Protection Act 1995.
Our analysis placed the single sample of P. freyciana in a clade with weak–moderate support (PP 0.90, BS 74%) with samples of P. verrucosa from Vic. and Tas. There is low sequence variation among samples in this clade and the BI consensus tree suggests that P. freyciana could be nested within P. verrucosa, although with little support (PP 74, BS <50%). Given this result, and that morphological differences between the two species are slight (Duretto 2009), their relationships and genetic distinctiveness are worthy of further investigation. This seems especially worthwhile given the conservation listings of P. freyciana and the potential conservation funding that might be spent to preserve it. Such a study, using additional samples and genetic markers, is currently underway (W. Neal in prep.).
Implementation of taxonomic changes
Although our results highlight the need for several taxonomic changes in sect. Erionema, these changes are not formally implemented here. One reason for this is that further morphological study is needed to assist with the delimitation of some taxa that are identified here on genetic grounds; this particularly applies to the two genetic groups within P. myoporoides subsp. myoporoides and to recognising the northern populations of P. glasshousiensis as a distinct species. The other reason is that the generic circumscription of Philotheca is uncertain and we would prefer not to create new names or combinations for taxa until that is resolved. Our unpublished data, and previous studies (Bayly et al. 2013), strongly suggest that Philotheca is not monophyletic but relationships of the four sections of Philotheca to each other and to related genera are yet to be clarified. It is possible that sect. Erionema might be recognised as a genus distinct from Philotheca. Some of us (MJB and collaborators) are working to resolve relationships of this group and to propose a revised generic classification. Our intention is that new names or combinations for members of sect. Erionema (e.g. raising the segregate subspecies of P. myoporoides to species rank) would be published as part of that work, rather than creating additional, potentially briefly used names in the interim.
Implications for biogeography
The relationships resolved here in sect. Erionema indicate some interesting biogeographic patterns. These include: (1) the disjunction between western and eastern Australia (between Philotheca brucei and all other taxa); (2) the presence of potential deep geographical overlaps of two eastern clades (i.e. the pedunculate clade and the P. obovalis + P. trachyphylla + P. virgata clade); (3) early and substantial divergence of a south-eastern Qld lineage (P. glasshousiensis + P. queenslandica) within the pedunculate clade; (4) two distinct connections between the mainland and Tas. (i.e. P. verrucosa and P. virgata, both in separate clades in the phylogeny); and (5) striking geographic disjunction between the sister taxa P. myoporoides subsp. petraea and P. epilosa (~900 km; Fig. 1c, j), and between the northern populations of P. glasshousiensis s.l. and P. myoporoides subsp. acuta (~850 km; Fig. 1d, f).
The seeds from members of sect. Erionema usually fall within a few metres of the parent and, unlike those from other sections of Philotheca, have no distinctive features to promote dispersal by animals (Armstrong 1991; Bayly 2001). It thus seems likely that long-distance dispersal has not been of major importance in the history of the group and that biogeographic patterns have mostly been shaped by vegetation shifts associated with past climatic and geological changes and a history of differentiation of allopatric taxa through vicariance.
The ages of divergences within sect. Erionema have not been estimated via molecular dating but some inferences can be made from previous work. We have not attempted molecular divergence dating here because fossils most suitable for calibration sit outside the family Rutaceae (Pfeil and Crisp 2008; Bayly et al. 2013) in taxa to which alignment of ITS and ETS markers is problematic and therefore likely to lead to spurious results. Nonetheless, the previous dating study of Bayly et al. (2013), based on chloroplast markers that display a more conservative rate of change (rcbL and atpB), did include one member of sect. Erionema (P. buxifolia) and representatives of the other three subgenera of Philotheca (including two samples of subg. Philotheca). The genus was not resolved as monophyletic in that study and relationships within the group were poorly supported, which, apart from other uncertainties regarding calibrations and mutation rate estimation, clouds understanding of divergence times. The branch connecting P. buxifolia to other taxa in the tree, i.e. a possible stem age for sect. Erionema, was 18–34 Ma (mean 28 Ma). The crown age for the section could be much younger than this, but this estimate at least allows the possibility that sect. Erionema dates to the Paleogene. A Paleogene age would be consistent with a vicariant separation of western and eastern Australia, potentially in the mid Miocene, as inferred for a range of other plant groups of the temperate mesic zone (Crisp and Cook 2007). A history on that timescale could also help to account for the presence of highly disjunct, potentially relictual lineages in south-eastern Australia, and the presence of multiple, deeply diverged but geographically overlapping clades.
The suggested relationship between P. myoporoides subsp. petraea and P. epilosa is particularly remarkable, given not just the distance between them, but also the presence of other members of the section in intervening areas. Despite the substantial distance, a relationship between the two taxa is morphologically plausible as they show a strong resemblance in leaf shape and size, as also noted by I. C. Clarke in annotations on one of the only two herbarium specimens of subsp. petraea (MEL 2030756A). However, if this resemblance is taken as evidence of relationship, a high level of conservation in leaf shape is implied despite substantial isolation in distance and presumably time, and across differing climates.
Data availability
The data supporting this study are publicly available. DNA sequences are available in GenBank (see https://www.ncbi.nlm.nih.gov/genbank/) with accession numbers as indicated in Table 1. The final DNA alignment, including INDEL characters, is available in TreeBase (see http://treebase.org/; study accession number 26368).
Conflicts of interest
Dr Murphy and Dr Bayly are both editors for Australian Systematic Botany. Despite this relationship, these authors did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages editors to publish in the journal and has protocols that keep editors completely separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
This research was supported by an Early Career Researcher Grant from The University of Melbourne.
Acknowledgements
We thank Alison Kellow, Gill Brown, Adele Gibbs, Mike Mathieson, Peter Neish, Daniel Ohlsen, Ian Thompson, Anthony Vadala, Mike Weston, Ruby Wilson, Tundra Morscheck, Paul Carmen, Todd McLay and Rose Barrett for assistance with fieldwork or provision of material. The Directors and staff of MEL, MELU, PERTH, BRI, NSW, AD, CBG, CANB, K, BM and UNE provided loan material or access to collections; MEL, CANB and MELU gave permission to destructively sample from some specimens. The former Queensland Department of Environment and Heritage, New South Wales National Parks and Wildlife Service, former Victorian Department of Conservation and Natural Resources, former Victorian Department of Sustainability and Environment, South Australian Department for Environment and Heritage, and the Western Australian Department of Conservation and Land Management (later DEC) provided permission to collect material in parks and reserves.
References
Appelhans MS, Bayly MJ, Heslewood MM, Groppo M, Verboom GA, Forster PI, Kallunki JA, Duretto MF (2021) A new subfamily classification of the Citrus family (Rutaceae) based on six nuclear and plastid markers. Taxon 70, 1035–1061.| A new subfamily classification of the Citrus family (Rutaceae) based on six nuclear and plastid markers.Crossref | GoogleScholarGoogle Scholar |
Armstrong JA (1991) Studies on pollination and systematics in the Australian Rutaceae. PhD thesis, University of New South Wales, Sydney, NSW, Australia.
Bailey CD, Carr TG, Harris SA, Hughes CE (2003) Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution 29, 435–455.
| Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes.Crossref | GoogleScholarGoogle Scholar |
Bayly MJ (1998) Notes on the Eriostemon myoporoides (Rutaceae) species complex, including new names and a new generic placement in Philotheca. Muelleria 11, 113–126.
Bayly MJ (2001) A cladistic and biogeographic analysis of Philotheca (Rutaceae) and allied genera. PhD thesis, The University of Melbourne, Vic., Australia. Available at https://minerva-access.unimelb.edu.au/handle/11343/37369
Bayly MJ, Holmes GD, Forster PI, Cantrill DJ, Ladiges PY (2013) Major clades of Australasian Rutoideae (Rutaceae) based on rbcL and atpB sequences. PLoS One 8, e72493
| Major clades of Australasian Rutoideae (Rutaceae) based on rbcL and atpB sequences.Crossref | GoogleScholarGoogle Scholar |
Bayly MJ, Duretto MF, Holmes GD, Forster PI, Cantrill DJ, Ladiges PY (2015) Transfer of the New Caledonian genus Boronella to Boronia (Rutaceae) based on analyses of cpDNA and nrDNA. Australian Systematic Botany 28, 111–123.
| Transfer of the New Caledonian genus Boronella to Boronia (Rutaceae) based on analyses of cpDNA and nrDNA.Crossref | GoogleScholarGoogle Scholar |
Chase MW, Morton CM, Kallunki JA (1999) Phylogenetic relationships of Rutaceae: a cladistic analysis of the subfamilies using evidence from RBC and ATP sequence variation. American Journal of Botany 86, 1191–1199.
| Phylogenetic relationships of Rutaceae: a cladistic analysis of the subfamilies using evidence from RBC and ATP sequence variation.Crossref | GoogleScholarGoogle Scholar |
Crisp MD, Cook LG (2007) A congruent molecular signature of vicariance across multiple plant lineages. Molecular Phylogenetics and Evolution 43, 1106–117.
| A congruent molecular signature of vicariance across multiple plant lineages.Crossref | GoogleScholarGoogle Scholar |
Duretto MF (2009) Rutaceae, version 2019:1. In ‘Flora of Tasmania online’. (Ed. MF de Salas) (Tasmanian Herbarium, Tasmanian Museum and Art Gallery: Hobart, Tas., Australia) Available at https://flora.tmag.tas.gov.au/treatments/rutaceae/
Forster PI (2005) New species of Philotheca Rudge (Rutaceae) from Queensland. Austrobaileya 7, 175–181.
French PA, Brown GK, Bayly MJ (2016) Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia. Plant Systematics and Evolution 302, 447–468.
| Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Groppo M, Pirani JR, Salatino MLF, Blanco SR, Kallunki JA (2008) Phylogeny of Rutaceae based on two noncoding regions from cpDNA. American Journal of Botany 95, 985–1005.
| Phylogeny of Rutaceae based on two noncoding regions from cpDNA.Crossref | GoogleScholarGoogle Scholar |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Morton CM, Telmer C (2014) New subfamily classification for the Rutaceae. Annals of the Missouri Botanical Garden 99, 620–641.
| New subfamily classification for the Rutaceae.Crossref | GoogleScholarGoogle Scholar |
Othman RNA, Jordan GJ, Worth JRP, Steane DA, Duretto MF (2010) Phylogeny and infrageneric classification of Correa Andrews (Rutaceae) on the basis of nuclear and chloroplast DNA. Plant Systematics and Evolution 288, 127–138.
| Phylogeny and infrageneric classification of Correa Andrews (Rutaceae) on the basis of nuclear and chloroplast DNA.Crossref | GoogleScholarGoogle Scholar |
Pfeil BE, Crisp MD (2008) The age and biogeography of Citrus and the orange subfamily (Rutaceae: Aurantioideae) in Australasia and New Caledonia. American Journal of Botany 95, 1621–1631.
| The age and biogeography of Citrus and the orange subfamily (Rutaceae: Aurantioideae) in Australasia and New Caledonia.Crossref | GoogleScholarGoogle Scholar |
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
| MRBAYES 3: Bayesian phylogenetic inference under mixed models.Crossref | GoogleScholarGoogle Scholar |
Rozefelds AC (2001a) The Tasmanian species of Philotheca. Muelleria 15, 19–26.
Rozefelds AC (2001b) Notes on the Philotheca myoporoides complex in Victoria. Muelleria 15, 15–18.
Salvo G, Ho SYW, Rosenbaum G, Ree R, Conti E (2010) Tracing the temporal and spatial origins of island endemics in the mediterranean region: a case study from the citrus family (Ruta L., Rutaceae). Systematic Biology 59, 705–722.
| Tracing the temporal and spatial origins of island endemics in the mediterranean region: a case study from the citrus family (Ruta L., Rutaceae).Crossref | GoogleScholarGoogle Scholar |
Scott KD, McIntyre CL, Playford J (2000) Molecular analyses suggest a need for a significant rearrangement of Rutaceae subfamilies and a minor reassessment of species relationships within Flindersia. Plant Systematics and Evolution 223, 15–27.
| Molecular analyses suggest a need for a significant rearrangement of Rutaceae subfamilies and a minor reassessment of species relationships within Flindersia.Crossref | GoogleScholarGoogle Scholar |
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49, 369–381.
| Gaps as characters in sequence-based phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar |
Wilson PG (1970) A taxonomic revision of the genera Crowea, Eriostemon and Phebalium (Rutaceae). Nuytsia 1, 3–155.
Wilson PG (1998) A taxonomic review of the genera Eriostemon and Philotheca (Boronieae: Rutaceae). Nuytsia 12, 239–265.
Wilson PG (2013) Philotheca. Flora of Australia 26, 366–415.
1 Author names for species and subspecies of Philotheca are given in Table 1.