Eucalyptus cryptica (Myrtaceae): a critically endangered new species
Trevor C. Wilson A B * , Susan Rutherford
A B * , Susan Rutherford  B C F , Jia-Yee S. Yap
B C F , Jia-Yee S. Yap  B , Steven M. Douglas D , Enhua Lee E and Maurizio Rossetto
B , Steven M. Douglas D , Enhua Lee E and Maurizio Rossetto  B
B
A Plant Discovery and Evolution, Botanic Gardens of Sydney, Mount Annan, NSW 2567, Australia.
B Research Centre for Ecosystem Resilience, Botanic Gardens of Sydney, Sydney, NSW 2000, Australia.
C Institute of Environment and Ecology, School of the Environment and Safety Engineering, Jiangsu University, Zhenjiang, 212013, PR China.
D Ecological Surveys & Planning, Bundanoon, NSW 2578, Australia.
E Biodiversity and Conservation Division, New South Wales Department of Planning and Environment, Sydney, NSW, Australia.
F Present address: Department of Environmental Science, College of Science Mathematics and Technology, Wenzhou-Kean University, Wenzhou, 325060, PR China.
Australian Systematic Botany 36(5) 386-400 https://doi.org/10.1071/SB22031
Submitted: 5 December 2022 Accepted: 9 August 2023 Published: 5 September 2023
Abstract
Recognition that the critically endangered mallee Eucalyptus sp. Cattai (Gregson s.n. 28 Aug 1954) is a distinct species has been complicated by close morphological similarity between it and other members of E. subgenus Symphyomyrtus section Latoangulatae series Annulares. Recent genomic evidence has demonstrated that it is distinct from other species. In this study, we provide E. sp. Cattai with the new species name, E. cryptica T.C.Wilson, S.Rutherf. & S.M.Douglas, and use genomic scans of adults and seedlings to assist in its description and support its conservation by identifying hybrids. Accompanying the description of E. cryptica are images, diagnostic illustrations and an updated part of the Eucalyptus key for the Flora of New South Wales.
Keywords: conservation, conservation genetics, Eucalyptus, genome‐wide sequencing, hybridisation, Latoangulatae, Symphyomyrtus, taxonomy.
Introduction
The iconic and economically important genus Eucalyptus L’Hér. (Myrtaceae) consists of over 700 species of shrubs and trees, nearly all of which are restricted to Australia and found in a highly diverse range of habitats (Slee et al. 2006; Nicolle 2019). The number of species is an underestimate since numerous unpublished-manuscript names await testing and formal description (Nicolle and Jones 2018; Nicolle 2019).
Progress towards acquiring a stable taxonomy for Eucalyptus has been in part slowed by the capacity of members of the genus to hybridise (e.g. Flores-Rentería et al. 2017; Rutherford et al. 2018). Hybridisation can confound delineation of species because it creates genetic intergrades between taxa (Griffin et al. 1988; Field et al. 2009; Le et al. 2009; Grattapaglia et al. 2012). Even though hybrid intergrades might in time lead to new plant species, hybridisation can also contribute to extinction through genetic swamping or the transfer of maladaptive genes (Rieseberg 1997; Mallet 2005; Abbott et al. 2013; Goulet et al. 2017; Draper et al. 2021). Furthermore, if the survival of a species hinges on conservation management, then the impediment of hybridisation to taxonomy elevates the threat of extinction where conservation activities are not appropriately prioritised because of inaccurate taxonomic understanding (Levin et al. 1996; Rossetto et al. 2021; Wilson et al. 2022).
Misconceptions about hybridisation and its detrimental effect on taxonomic progress are exemplified by the taxon identified as Eucalyptus sp. Cattai (Gregson s.n. 28 Aug 1954) in the Flora of New South Wales (PlantNET 2023), a critically endangered species under both the New South Wales Biodiversity Conservation Act 2016 and the Commonwealth Environment Protection and Biodiversity Conservation Act 1999. It is endemic to the north-western portion of the greater Sydney region, New South Wales (see map in Rutherford et al. 2022). In the late 1990s, Steve Douglas and Ian Brooker informally recognised that populations of E. sp. Cattai were morphologically distinct from other species in the genus. Bark and fruit morphology look similar between E. sp. Cattai and members E. subgenus Symphyomyrtus section Latoangulatae series Annulares (Blakely) Chippend. (Slee et al. 2006; Klaphake 2012), which are commonly known as the ‘red mahoganies’. This group includes three widespread species found in the Sydney region (E. notabilis Maiden, E. resinifera J.White and E. scias L.A.S.Johnson & K.D.Hill) and another species (E. macta L.A.S.Johnson & K.D.Hill) from northern Queensland (Jones et al. 2016; Nicolle 2019). The barely exserted valves of its fruit distinguish E. sp. Cattai from all red mahogany species (with strongly exserted valves). However, the close morphological similarity between E. sp. Cattai and E. notabilis led to the speculation that the former was the result of an intergrade between the latter and nearby populations of E. resinifera (Klaphake 2012), especially given that hybridisation is well documented in other red mahoganies (Le et al. 2009; Steane et al. 2011).
A recent study by Rutherford et al. (2022) tested the species hypothesis for Eucalyptus sp. Cattai by examining genetic variation acquired from genome-wide scans. It concluded that E. sp. Cattai is a distinct species and that it is sister to all other members of the red mahogany clade. The red mahoganies, in turn, were sister to a clade consisting of E. deanei and E. subgenus Symphyomyrtus section Latoangulatae series Transversae Blakely (i.e. the ‘blue gums’), of which some, such as the red mahoganies, occur naturally in the Sydney region (e.g. E. botryoides Sm., E. saligna Sm.). However, low phylogenetic branch support reduced confidence about whether E. sp. Cattai is more closely related to the red mahoganies or the blue gums.
A report on E. sp. Cattai was originally created under the ‘Saving Our Species’ initiative (of the New South Wales Government, led initially by the then Office of Environment & Heritage), which required empirically based evidence to substantiate proposed translocation actions. The genomes of ex situ seedling recruits were scanned for the purpose of establishing new and resilient translocation populations (Bragg et al. 2021), which demonstrated high genetic introgression by other species (Rutherford et al. 2022). In this paper, we assess the genetic profile of new ex situ and in situ specimens in combination with the dataset of Rutherford et al. (2022) and then use an assessment of hybrids to inform our description, illustrations and additional notes for this species that is newly named Eucalyptus cryptica T.C.Wilson, S.Rutherf. & S.M.Douglas. We follow the classification of species by Nicolle (2019), which is, in part, informed by results from the broad phylogenetic study of Eucalyptus (Jones et al. 2016). Measurements of seedlings, derived from both Rutherford et al. (2022) and our own results, have been used to assist with the preparation of the taxonomic description for E. cryptica.
Materials and methods
We acquired specimens of Eucalyptus sp. Cattai for genome-wide scanning and morphological measurements to assess the provenance of adult individuals and the hybrid status of ex situ seedlings. Following the methods of Rutherford et al. (2022), 89 new seedlings of this taxon were germinated and grown at the Australian Botanic Garden, Mount Annan (Royal Botanic Gardens & Domain Trust) from the ‘Clarke’, ‘Saltwater’, ‘Shoplands’ and ‘Logie’ populations (Supplementary Table S1). Maternal lines were kept separate for each accession to provide a more balanced estimate of hybridisation across the distribution of E. sp. Cattai.
A single-nucleotide polymorphism (SNP) dataset used for the analysis of genetic similarity and relationship was constructed following the extraction and DArTseq sequencing methods described by Rutherford et al. (2022). Leaf tissue was collected from 89 ex situ seedlings and 4 adult specimens of E. sp. Cattai, including 1 ex situ specimen at the Australian Botanic Garden (NSW 1079061) and 3 from the recently discovered ‘Larapinta’ site (NSW 1078548, NSW 1078553, NSW 1078549). Genomic scans of these new samples were analysed together with data in the Dryad Digital Repository (https://doi.org/10.5061/dryad.zpc866tbv) produced by Rutherford et al. (2022). The final SNP dataset consisted of data from 109 adult E. sp. Cattai specimens, 105 outgroup specimens (E. subgenus Symphyomyrtus (Schauer) Brooker and more distantly related species of E. L’Hér. subgenus Eucalyptus), and 172 seedlings derived from E. sp. Cattai (n = 386, see Supplementary Table S2). The outgroup sample was provided from the dataset of Rutherford et al. (2022), who sampled replicates of all naturally occurring species of Eucalyptus in the vicinity of E. sp. Cattai to capture evidence of genetic introgression. All collecting operated under a scientific licence (SL101766).
The SNP dataset was analysed to visualise genetic relationship and hybridisation by using the same methods as in Rutherford et al. (2022). The NeighborNet method was employed in Splitstree (ver. 4.14.6, see https://github.com/husonlab/splitstree4; Huson et al. 2008) to generate a network, which is useful for visualising evolutionary histories in groups with substantial reticulation arising from incomplete lineage sorting and hybridisation (Huson and Bryant 2006). Principal-component analysis (PCA) in adegenet (ver. 2.1.1, see https://github.com/thibautjombart/adegenet; Jombart 2008) assessed genetic similarity at the species, individual and population level. NewHybrids (ver. 1.1, see https://github.com/eriqande/newhybrids; Anderson and Thompson 2002) was used to determine hybrids, estimating the posterior probability that an individual belongs to one of the following six genotypic classes: Species A, Species B, F1 hybrid, F2 hybrid, backcross to Species A, or backcross to Species B.
Seventeen (17) morphological characters (Table 1) were measured for all non-hybrid seedlings (i.e. genotypic classes Species A and Species B, as assessed by NewHybrids) according to the methods of Rutherford et al. (2022). The total morphological dataset included 131 specimens (Supplementary Table S3) when combined with seedling measurements of Rutherford et al. (2022).
| Character | Description | |
|---|---|---|
| Plant height | Cotyledon node to the base of the most recently produced leaf node (mm) | |
| Internode length | Distance between the first and second juvenile leaf node (mm) | |
| Stem diameter | Diameter at second node from base (mm) | |
| Smallest leaf length | Length of the smallest leaf lamina (mm) | |
| Smallest leaf width | Width of the smallest leaf lamina (mm) | |
| Smallest leaf petiole length | Length of the smallest leaf petiole (mm) | |
| Smallest leaf petiole width | Width of the smallest leaf petiole (mm) | |
| Largest leaf length | Length of the largest leaf lamina (mm) | |
| Largest leaf width | Width of the largest leaf lamina (mm) | |
| Largest leaf petiole length | Length of the largest leaf petiole (mm) | |
| Largest leaf petiole width | Width of the largest leaf petiole (mm) | |
| Minimum leaf thickness | Smallest thickness of lamina (mm) | |
| Maximum leaf thickness | Greatest thickness of lamina (mm) | |
| Cotyledon lamina length | Length of the cotyledon lamina (mm) | |
| Cotyledon lamina width | Width of the cotyledon lamina (mm) | |
| Cotyledon petiole length | Length of the cotyledon petiole (mm) | |
| Cotyledon petiole width | Width of the cotyledon petiole (mm) |
Measurements for the taxonomy section follow a framework similar to that in other contemporary eucalypt descriptions (e.g. Collins et al. 2019; Bell and Nicolle 2020) and the leaf-vein network was scored using states defined by Slee et al. (2006).
Results and discussion
Our results are consistent with the conclusions of Rutherford et al. (2022) and support recognition of Eucalyptus sp. Cattai as a distinct species closely related to other members of E. subgenus Symphyomyrtus section Latoangulatae series Annulares (red mahoganies) and E. subgenus Symphyomyrtus section Latoangulatae series Transversae (blue gums).
The total dataset comprised 15 168 SNPs (97.6% had a reproducibility score of ≥96%). The proportion of missing data for the samples was between 12.84 and 86.74%, with a mean of 33.81%.
The Splitstree relationship network and principal-component analysis (PCA) indicated that all specimens of E. sp. Cattai form an exclusive cluster that is most genetically similar to a cluster of red mahoganies and a cluster of blue gums and E. deanei (Fig. 1, 2). Although it remains unclear from phenetic results whether E. sp. Cattai belongs to either series, the phylogenetic tree produced by Rutherford et al. (2022) recovers it with the red mahogany clade, albeit with low support. The closer relationship between E. sp. Cattai and red mahoganies corresponds with the shared character of having rough bark over the entire trunk and branches. Eucalyptus deanei and the rest of the blue gum clade have smooth bark at least on their branches, if not over most of the trunk (Brooker 2000; Slee et al. 2006). However, rough bark over the trunk and branches is also exhibited by all species in E. subgenus Symphyomyrtus section Latoangulatae series Robustae (sensu Nicolle 2019), which means that the ‘full bark’ character is unlikely to be synapomorphy for the red mahogany clade.
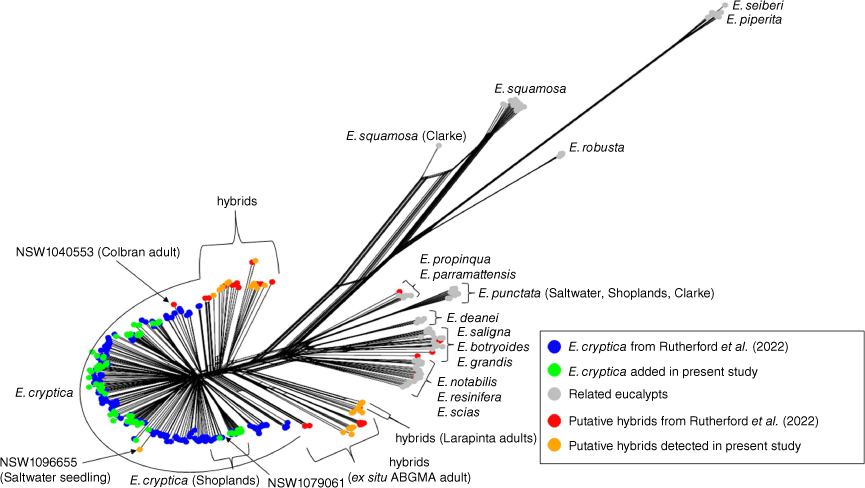
Relationship among Eucalyptus cryptica and other eucalypts from Splitstree analysis of SNP data (15 168 SNPs total) acquired from genome-wide scans including 384 samples. The majority of ‘putative hybrids’ determined by New Hybrids analysis in this study (red and orange) are located along the main stem between E. cryptica and other eucalypts and are associated with a high amount of webbing that can be indicative of genomic reticulation. The majority of these hybrids are also seedlings sourced in situ from mother trees at Clarke, Logie, Saltwater and Shoplands sites expressing typical morphology of E. sp. Cattai. Including previous data from Rutherford et al. (2022), we identified the Larapinta site (three specimens) and six other adults as hybrids. The adult ex situ specimen in the living collection at Australian Botanic Gardens Mount Annan (ABGMA) is recovered within the Shoplands site cluster. Table S2 provides detail for identification, voucher number and site for all specimens.
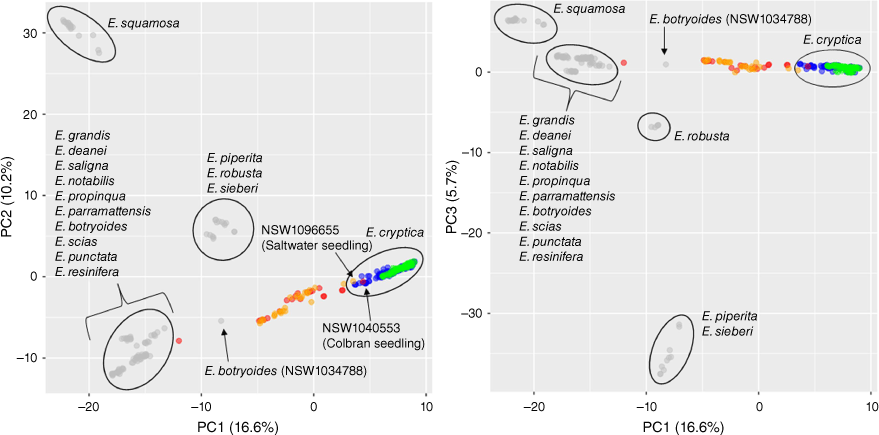
Principal component analyses (PC1 v. PC2, PC1 v. PC3) showing groupings of Eucalyptus cryptica populations and outgroup species based on genome-wide scans (15 168 SNPs total) for 384 samples. Circle colours correspond with the legend of Fig. 1: ‘pure’ E. cryptica samples are coloured blue (originally sampled in Rutherford et al. 2022) and green (new data sampled in this study); putative hybrids are coloured red (Rutherford et al. 2022) and orange (this study); related eucalypt species are coloured grey. Table S2 provides detail for identification, voucher number and site for all specimens.
The cluster of E. sp. Cattai samples that excludes other species also provides support for species recognition and corresponds with results of Rutherford et al. (2022), whose phylogenetic analysis demonstrated that all other red mahoganies were more closely related to each other than to E. sp. Cattai. This evidence is also consistent with our own observations of morphology and those made by Klaphake (2012), which found that E. sp. Cattai has weakly exserted to shallowly enclosed valves in the fruit, whereas all other red mahoganies have strongly exserted valves. In the Splitstree relationship network, wide or extensive ‘webbing’ or ‘cycling’ can be indicative of higher genetic reticulation. Although present, this result was not extensive between the E. sp. Cattai cluster and other species (Fig. 1) and suggests that E. sp. Cattai is not a recent hybrid. Further examination of clade-specific characters for delimiting series will be more useful once our understanding of the relationship between series is improved, such as through broadening the sampling of our dataset to include populations of E. subgenus Symphyomyrtus section Latoangulatae series Robustae. Providing additional outgroups used by Jones et al. (2016), in addition to E. subgenus Symphyomyrtus section Maidenaria, might also concomitantly allow for a more rigorous study on the speculated influence of ancestral hybridisation on the evolution of E. subgenus Symphyomyrtus section Latoangulatae (Jones et al. 2016).
Our results support the view that recent introgression has occurred across the distribution of E. sp. Cattai, and we have shown hybrids additional to those reported by Rutherford et al. (2022) for both seedlings and adults. In total, 36 seedlings were recovered in an intermediate position, situated outside the cluster of their respective population as well as being located between the E. sp. Cattai cluster and other eucalypts (Fig. 1). The little ‘webbing’ or ‘cycling’ reported in the network was associated with some of these seedlings plus three adult members of the Larapinta population. Subsequent testing by NewHybrids confirmed that these specimens were hybrids (Table 2), and, similar to the results by Rutherford et al. (2022), the analysis also assigned the hybrid category to an adult (NSW 1040553) recovered in the pure Colbran cluster and a newly sequenced seedling cluster (NSW 1096655) in the Saltwater cluster. However, these specimens were identified as backcrosses (with E. resinifera) in at least some analyses, which is the likely reason why they were provided a distinctively longer branch but were still grouped within the ‘pure’ population clusters of the phylogenetic network (Fig. 1).
| Sample | Species | Site | Rutherford et al. (2022) | Age | Genetic identity | n admixed | Assigned | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Backcross to Species A | Backcross to Species B | F1 hybrid | F2 hybrid | Total | ||||||||
| NSW1045675 | E. cryptica | Bannerman | Yes | Adult | Hybrid | 3 | 1 | 4 | 2 | 7 | ||
| NSW1045676 | E. cryptica | Bannerman | Yes | Adult | Hybrid | 3 | 1 | 4 | 2 | 7 | ||
| NSW1046200 | E. cryptica | Clarke | Yes | Adult | Hybrid | 2 | 1 | 2 | 6 | 8 | ||
| NSW1040553 | E. cryptica | Colbran | Yes | Adult | Hybrid | 2 | 1 | 1 | 8 | |||
| NSW1041658 | E. cryptica | Georgia | Yes | Adult | Hybrid | 3 | 3 | 2 | 7 | |||
| NSW1041659 | E. cryptica | Georgia | Yes | Adult | Hybrid | 4 | 3 | 2 | 6 | |||
| NSW1078548 | E. cryptica | Larapinta | Adult | Hybrid | 2 | 1 | 3 | 5 | 8 | |||
| NSW1078549 | E. cryptica | Larapinta | Adult | Hybrid | 2 | 1 | 2 | 6 | 8 | |||
| NSW1078553 | E. cryptica | Larapinta | Adult | Hybrid | 1 | 1 | 3 | 6 | 9 | |||
| NSW1043137 | E. cryptica | Clarke | Yes | Seedling | Hybrid | 2 | 1 | 1 | 7 | 8 | ||
| NSW1043421 | E. cryptica | Logie | Yes | Seedling | Hybrid | 1 | 1 | 1 | 8 | 9 | ||
| NSW1043422 | E. cryptica | Logie | Yes | Seedling | Hybrid | 3 | 1 | 1 | 6 | 7 | ||
| NSW1043425 | E. cryptica | Logie | Yes | Seedling | Hybrid | 2 | 1 | 1 | 1 | 6 | 8 | |
| NSW1047890 | E. cryptica | Logie | Yes | Seedling | Hybrid | 4 | 1 | 1 | 5 | 6 | ||
| NSW1047891 | E. cryptica | Logie | Yes | Seedling | Hybrid | 3 | 1 | 1 | 5 | 7 | ||
| NSW1096562 | E. cryptica | Logie | Seedling | Hybrid | 1 | 2 | 6 | 2 | 9 | |||
| NSW1096567 | E. cryptica | Logie | Seedling | Hybrid | 3 | 1 | 1 | 6 | 7 | |||
| NSW1096577 | E. cryptica | Logie | Seedling | Hybrid | 2 | 1 | 8 | 8 | ||||
| NSW1096592 | E. cryptica | Logie | Seedling | Hybrid | 1 | 1 | 9 | 10 | ||||
| NSW1096602 | E. cryptica | Logie | Seedling | Hybrid | 3 | 2 | 1 | 1 | 4 | 7 | ||
| NSW1096607 | E. cryptica | Logie | Seedling | Hybrid | 1 | 1 | 3 | 6 | 9 | |||
| NSW1096650 | E. cryptica | Logie | Seedling | Hybrid | 5 | 6 | 5 | |||||
| NSW1043112 | E. cryptica | Saltwater | Yes | Seedling | Hybrid | 2 | 2 | 7 | 8 | |||
| NSW1043114 | E. cryptica | Saltwater | Yes | Seedling | Hybrid | 3 | 3 | 3 | 2 | 7 | ||
| NSW1043221 | E. cryptica | Saltwater | Yes | Seedling | Hybrid | 3 | 2 | 4 | 2 | 7 | ||
| NSW1047857 | E. cryptica | Saltwater | Yes | Seedling | Hybrid | 2 | 2 | 3 | 1 | 3 | 8 | |
| NSW1082696 | E. cryptica | Saltwater | Seedling | Hybrid | 3 | 2 | 3 | 3 | 7 | |||
| NSW1082714 | E. cryptica | Saltwater | Seedling | Hybrid | 2 | 2 | 7 | 8 | ||||
| NSW1082725 | E. cryptica | Saltwater | Seedling | Hybrid | 4 | 2 | 2 | 3 | 6 | |||
| NSW1082730 | E. cryptica | Saltwater | Seedling | Hybrid | 1 | 3 | 3 | 4 | 9 | |||
| NSW1096528 | E. cryptica | Saltwater | Seedling | Hybrid | 1 | 2 | 6 | 2 | 9 | |||
| NSW1096563 | E. cryptica | Saltwater | Seedling | Hybrid | 2 | 2 | 3 | 4 | 8 | |||
| NSW1096613 | E. cryptica | Saltwater | Seedling | Hybrid | 4 | 1 | 3 | 3 | 6 | |||
| NSW1096620 | E. cryptica | Saltwater | Seedling | Hybrid | 7 | 2 | 2 | 3 | ||||
| NSW1096655 | E. cryptica | Saltwater | Seedling | Hybrid | 2 | 1 | 5 | 1 | 8 | |||
| NSW1096665 | E. cryptica | Saltwater | Seedling | Hybrid | 2 | 2 | 5 | 2 | 8 | |||
| NSW1096690 | E. cryptica | Saltwater | Seedling | Hybrid | 3 | 2 | 3 | 3 | 7 | |||
| NSW1043428 | E. cryptica | Shoplands | Yes | Seedling | Hybrid | 3 | 1 | 3 | 3 | 7 | ||
| NSW1043431 | E. cryptica | Shoplands | Yes | Seedling | Hybrid | 4 | 1 | 1 | 4 | 6 | ||
| NSW1043437 | E. cryptica | Shoplands | Yes | Seedling | Hybrid | 4 | 1 | 1 | 5 | 6 | ||
| NSW1096525 | E. cryptica | Shoplands | Seedling | Hybrid | 2 | 1 | 2 | 5 | 8 | |||
| NSW1096628 | E. cryptica | Shoplands | Seedling | Hybrid | 2 | 2 | 2 | 4 | 8 | |||
| NSW1096633 | E. cryptica | Shoplands | Seedling | Hybrid | 3 | 1 | 1 | 5 | 7 | |||
| NSW1096645 | E. cryptica | Shoplands | Seedling | Hybrid | 4 | 1 | 1 | 4 | 6 | |||
| NSW1096658 | E. cryptica | Shoplands | Seedling | Hybrid | 5 | 2 | 1 | 2 | 5 | |||
| NSW1034790 | E. botryoides | Marie | Yes | Adult | Hybrid | 1 | 10 | |||||
| NSW1046495 | E. botryoides | Etta | Yes | Adult | Hybrid | 1 | 10 | |||||
| NSW1024274 | E. grandis | Kalphalim | Yes | Adult | Hybrid | 1 | 10 | |||||
| NSW1042479 | E. sp. aff. robusta | Woodlands | Yes | Adult | Hybrid | 3 | 3 | 7 | ||||
| NSW1043439 | E. sp. aff. notabilis × E. resinifera | Foxal | Yes | Adult | Hybrid | 1 | 3 | 7 | 9 | |||
| NSW827889 | E. parramattensis | Ulan | Yes | Adult | Hybrid | 1 | 10 | |||||
| NSW1047934 | E. propinqua | Cowarra | Yes | Adult | Hybrid | 1 | 10 | |||||
| NSW1042511 | E. resinifera | Caterson | Yes | Adult | Hybrid | 1 | 10 | |||||
The NewHybrids software tested for hybridisation between Eucalyptus cryptica and other eucalypt species in subgenus Symphyomyrtus. This involved performing multiple independent analyses on specific subsets of data, as outlined in Table S1, following methodology in Rutherford et al. (2022). The table omits individuals that were identified as pure across all NewHybrids analyses. The column ‘Rutherford et al. (2022)’ indicates whether a hybrid was detected by the previous genomic study of E. cryptica (‘yes’) or individuals that are newly sequenced in this study (blank cell). The column ‘n admixed’ indicates the number of times a NewHybrids analysis could not assign an individual to any of the six hybrid classes (i.e. Species A, Species B, F1, F2, backcross to Species A and backcross to Species B). In these cases, a class could not be assigned because the posterior probability of assignment threshold was less than 0.9 (see Table S1 for more details).
Given the prevalence of hybridisation in our results, and because hybrids were detected from most sites (8 of 10), there is a threat of genetic swamping in E. sp. Cattai. This includes all sites sourced for seed (Clarke, Logie, Saltwater, Shoplands) and sites where hybrid adults were detected (Bannerman, Foxal, Larapinta and Georgia). Our results are limited to seed gathered from a single year and, hence, we cannot determine whether the genetic swamping has been constant. However, many sites have adult hybrids, which suggests that introgression has occurred in the past. Fewer of these adult hybrids and a higher number of ‘pure’ individuals in the seedling data (such as in Clarke, Shoplands and Saltwater) are indicative that the northern populations of E. sp. Cattai are of greatest value for conserving the species. The PCA of genetic diversity highlighted their value further, showing that the northern populations encapsulate most of the genetic diversity of E. sp. Cattai (Fig. 3). The Saltwater site has the greatest share of this diversity, and although it is likely that this has been dramatically reduced in situ because of recent land clearing, the ex situ seedling cohort appears to encapsulate nearly all of Saltwater’s genetic diversity.
Principal-component analysis of SNP data summarising genetic variability across Eucalyptus cryptica without hybrids, including adults collected in situ and all available ex situ collections at the Australian Botanic Gardens Mount Annan. The ex situ specimens were 171 seedlings grown from seed collected in situ from the Clarke, Logie, Saltwater and Shoplands sites (diamonds), and one adult tree (triangle). The Clarke, Shoplands and Saltwater populations are more widely distributed across the graph than other populations, demonstrating that they represent most of the genetic diversity of E. cryptica. Table S2 provides detail for identification, voucher number and site for all specimens.
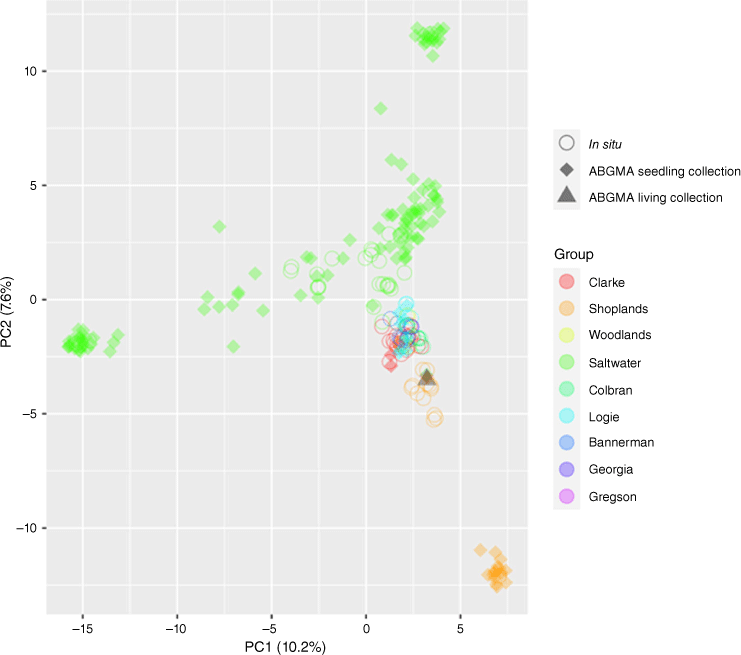
The Shoplands site plus the ex situ adult tree at the Australian Botanic Garden demonstrate unique genetic variation, given their sole occupancy at the opposite end of the PCA cluster from the Saltwater site (Fig. 3). Although the provenance of the ex situ specimen (NSW 1079061) had not been previously specified with more detail than ‘the Kellyville and Annangrove area’, it is assigned to the Shoplands cluster in the Splitstree network (Fig. 1) and PCA (not shown), which suggests that it originates from around the Shoplands site. In addition to establishing its provenance, the conservation value of this individual is further raised, given the finding about its genetic ‘purity’. Future construction of an ex situ collection should now be directed on acquiring individuals with a genetic profile different from that of plants from the Shoplands site.
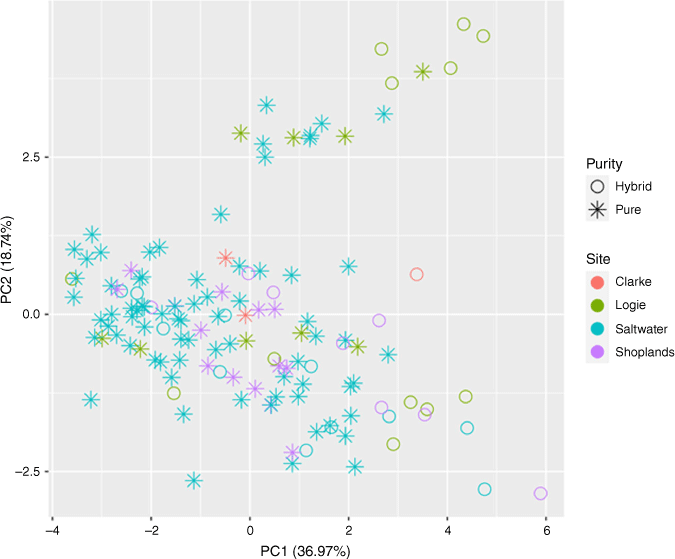
Given the evidence supporting that E. sp. Cattai is a distinct species, we provide the description, illustration and notes to describe it as E. cryptica T.C.Wilson, S.Rutherf. & S.M.Douglas. Our description incorporates the minimum and maximum values of the corresponding morphological measurements (Table 1) for genotyped non-hybrid seedlings in the description. However, a PCA of morphological variation (Fig. 4) does not indicate that there is any difference between pure and hybrid individuals in these characters. Thus, even though the description might be useful for assisting species recognition of unknown seedlings, our results indicated that, at present, the successful detection of hybridisation in seedlings can be confidently assessed only through genotyping.
Principal component analysis biplot based on 17 morphological traits from Table 1 summarising morphological variability across ‘pure’ (asterisk) and hybrid (open circle) seedlings of Eucalyptus cryptica (as defined by NewHybrids analysis) sourced from the Clarke, Logie, Saltwater and Shoplands sites.
Taxonomic treatment
The section of the online key to Eucalyptus in New South Wales (PlantNET 2023), originally written by Ken Hill, is here amended to incorporate E. cryptica T.C.Wilson, S.Rutherf. & S.M.Douglas. Eucalyptus cryptica keys out with other New South Wales members of E. subgenus Symphyomyrtus section Latoangulatae series Annulares on the basis of the following characters: (i) bark rough, persistent over entire trunk, (ii) not an ironbark, and (iii) not a stringybark. However, it is then separated from its closest congeners (E. notabilis, E. resinifera and E. scias) by the valves of its fruit being shallowly enclosed to weakly exserted with the rim of the disc (v. strongly exserted). It, like E. microcorys F.Muell. and the more closely related E. botryoides and E. robusta Sm. (both currently in E. subgenus Symphyomyrtus section Latoangulatae), is distinguished from other Eucalyptus species by the adult leaves being strongly discolourous and closely penniveined. It can then be distinguished from E. botryoides by its fruit being pedicellate (v. fruit sessile). Below are modifications to the key that distinguish E. cryptica from E. microcorys and E. robusta.
| 76 | Habit a single-stemmed, small to medium-sized tree or mallee; buds ampulliform, fusiform, obpyriform or obconical, operculum usually beaked, operculum scar present; fruit cylindrical, hemispherical, cupular or obconical, >5 mm in diameter....76a Habit a medium-sized to tall single-stemmed tree; buds clavate, operculum never beaked, operculum scar absent; fruit obconical to obpyriform, <6 mm in diameter...E. microcorys |
| 76a | Bark fissured, eximious (i.e. with thin flakes on ridges); buds ampulliform or obpyriform or obconical, 6–15 mm long; fruit urceolate, valves free...E. cryptica Bark furrowed, persistent; buds fusiform, >15 mm long; fruit cylindrical, valves usually joined across orifice...E. robusta |
Eucalyptus cryptica T.C.Wilson, S.Rutherf. & S.M.Douglas, sp. nov.
Type: Australia: New South Wales: Central Coast: Sydney: Annangrove, 14 Oct. 2019, T.C.Wilson 830, S.Rutherford & J.Yap (holo: NSW 1058373! iso: BRI!, CANB!, MEL!).
Eucalyptus sp. Cattai (Gregson s.n., 28 August 1954) NSW Herbarium: Royal Botanic Gardens and Domain Trust, PlantNET database record (see https://plantnet.rbgsyd.nsw.gov.au).
Morphologically most similar to E. notabilis. Both species have strongly discolourous and narrowly to broadly ovate adult leaves with a glossy abaxial surface, buds with obtusely conical or hemispherical opercula bearing a scar, and a hemispherical or obconical fruit with a level to slightly raised disc. It differs from E. notabilis by its fibrous and somewhat eximious (i.e. with thin flakes on ridges) bark (v. fibrous but not eximious) and by having fruit with valves reaching only slightly past the rim of the disc (v. valves strongly exserted).
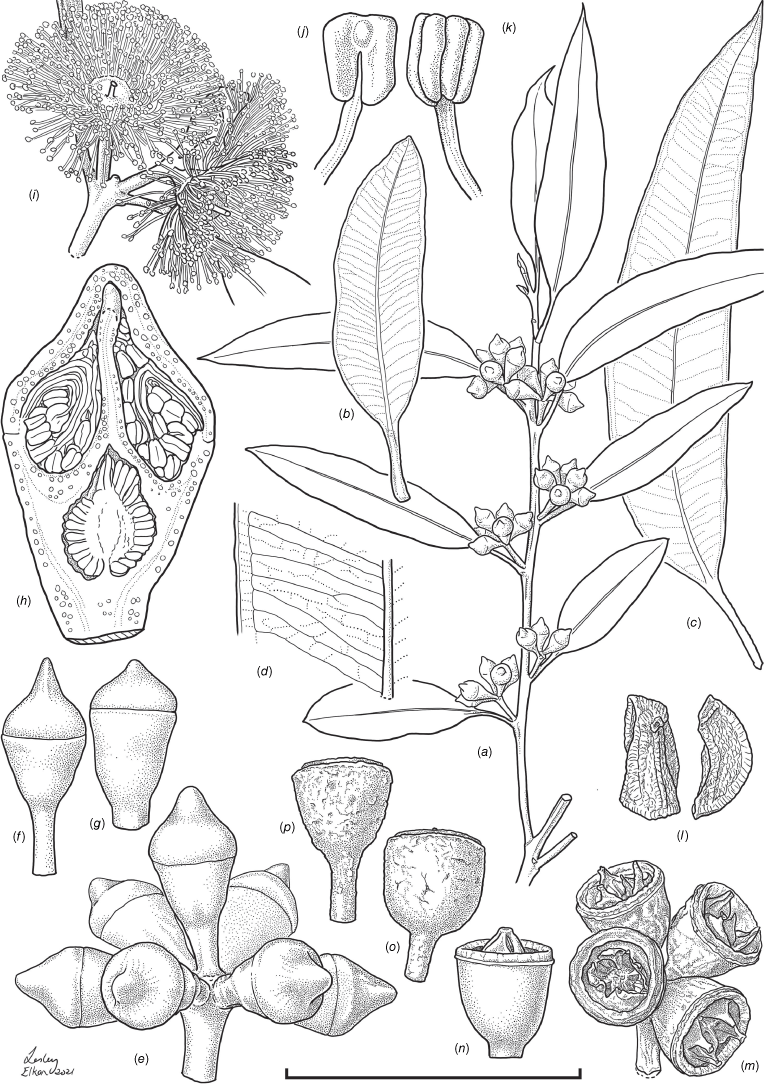
Erect, multi-stemmed mallee or tree with a single but crooked stem, 5(–10) m tall, lignotuber present. Bark fissured, somewhat fibrous and eximious, greyish-brown on exposed areas, reddish-brown underneath. Juvenile Stem bright green to red; Adult Branchlets quadrangular, glabrous, pale or bright green to red. Cotyledon petiole (0.1–)1–1. 8(–3.9) mm long, (0.1–)0.3–0.55(–0.7) mm wide; cotyledon lamina surface matte, (0.9–)1.8–3.8(–9.5) mm long, (0.5–)1.6–3.8(–5.2) mm wide, reniform, apex shallowly and broadly emarginate, base truncate to shortly attenuate. Juvenile leaves opposite for 2 or 3(8) nodes prior to transition to alternate phyllotaxy, petiole (usually present) (0.12–)0.9–4.3(–21.6) mm long, (0.2–)0.3–1(–2) mm wide; lamina narrow ovate to ovate or nearly oblong, discolourous, bright to dark yellowish-green, margin entire, glabrous, (1–)5–36(–60) mm long, (0.6–)2–12(–61) mm wide, 0.1–1 mm thick, apex acute, base equal or unequal sided and cuneate to shortly tapering. Adult Leaves petiolate, alternate, flat to partially pendulous, glabrous; petiole green to reddish- or yellowish-green, 5–17(–22) mm long, 1–2 mm wide; lamina discolourous, margin entire with recurved ridge, adaxial surface dark green and glossy, abaxial surface glaucous and dull, (32–)40–170 mm long, 10–27(–40) mm wide, narrow-ovate to broad-ovate, elliptic or sometimes weakly falcate, apex acute, rarely obtuse or uncinate, base cuneate to attenuate and sometimes oblique, veins sparsely to moderately reticulate, secondary veins 45–90°, intramarginal vein visible, 0.5–3.2 mm from margin; oil glands mainly occurring between veins (island-type), up to 0.15 mm wide, 1–7(–10) glands mm−2. Inflorescence axillary, unbranched, held erect, 7-flowered (occasionally fewer by abortion); peduncles rounded to angular, 4–13(–33) mm long, 2–5 mm wide; pedicels thickened, flattened and angular, 0.3–4 mm long, 0.5–2.1 mm wide. Flower Buds glabrous, smooth, shallowly ribbed when dry, 6–15 mm long, outline ampulliform, obpyriform or obconical, outer operculum scar present, hypanthium:operculum length ratio 1–1.9; hypanthium 3.5–9.5 mm long, 3–6 mm wide, occasionally ribbed; operculum 2–7 mm long, 4–8 mm wide, apex compressed and acute to obtuse or sometimes beaked. Stamens white or yellowish-white at anthesis, all fertile; filaments irregularly inflexed in bud, 2.5–6.5 mm long, 0.1–0.2 mm wide; anthers 0.4–0.8 mm long, 0.3–0.6 mm wide, oblong, dorsifixed, versatile, dehiscing by parallel slits. Gynoecium: style 2.5–5 mm long, 0.5–0.7 mm wide, ovules 0.3–0.7 mm long, 0.2–0.3 mm wide. Fruit 4.9–9 mm long, 5–8 mm wide, hemispherical, cupular or obconical (excluding disc and valves), pedicel 0.7–2.6 mm long; disc 0.4–1 mm wide, level to slightly ascending; valves 3 or 4(5), free, erect, enclosed, often with apices exserted up to 2 mm past disc. Seeds yellowish-orange to orange–brown, cuboid or crescent- or wedge-shaped in outline, 0.7–1.7 mm long, 0.2–0.5 mm wide, hilum terminal. (Fig. 5, 6, 7.)
Eucalyptus cryptica. (a) Branchlet with inflorescences; (b) leaf, adaxial surface; (c) leaf, adaxial surface; (d) close-up of leaf adaxial surface; (e) inflorescence in bud; (f) bud, profile view; (g) bud, profile view; (h) bud, longitudinal section; (i) flowers at anthesis; (j) anther, abaxial view; (k) anther, adaxial view; (l). seed, front and side views; (m) infructescence showing oblique view of dried fruit with shallowly exserted valves dehisced; (n) profile view of fresh fruit with shallowly exserted valves not dehisced; (o) profile view of dried cup-shaped fruit with valves hidden from view; (p) profile view of dried wineglass-shaped fruit with valves hidden from view. Scale bar: (a) 80 mm; (b, c) 80 mm; (d–g, m–p) 20 mm; (h) 8 mm; (i) ~30 mm; (j–l) 2.4 mm. Voucher: T.C.Wilson 830 et al. (holotype, NSW 1058373). Illustrations: L. Elkan.
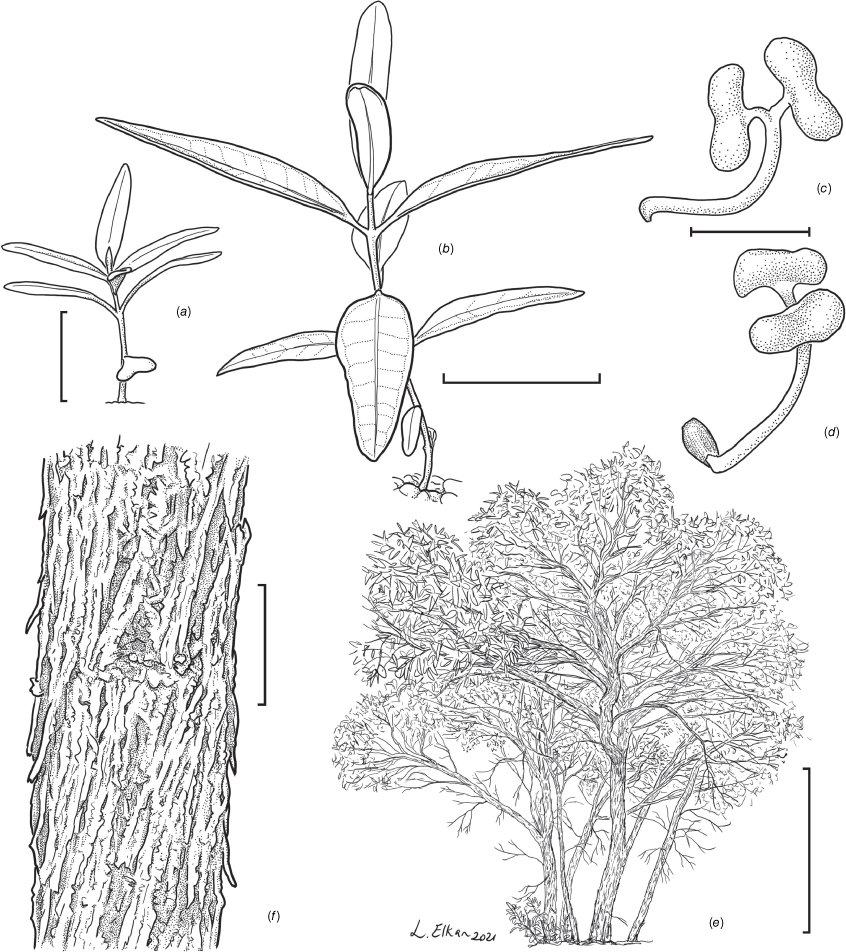
Eucalyptus cryptica. (a) Habit of seedling grown from K.E.Willis 37 (NSW 1043128); (b) habit of seedling, grown from K.E.Willis 34 (NSW 1047892); (c) seedling with cotyledons, grown from K.E.Willis 34 (tissue sample unknown); (d) seedling with cotyledons, grown from K.E.Willis 34 (tissue sample unknown); (e) mallee habit of adult; (f) detail of bark. Scale bar: (a) 20 mm; (b) 20 mm; (c, d) 2 mm; (e) 1 m; (f) 100 mm. Illustrations: (L. Elkan).
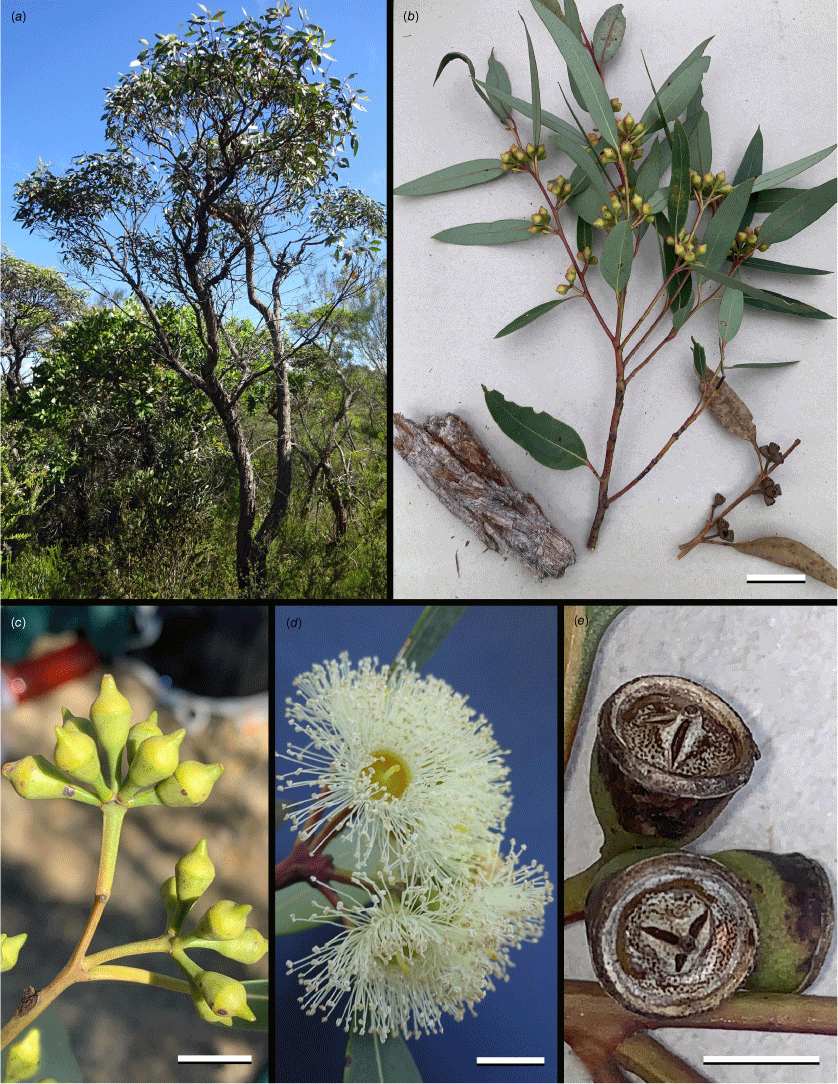
Images of Eucalyptus cryptica. (a, d) Type locality, without associated voucher; (b, e) T.C.Wilson 830; (c) T.C.Wilson 829. (a) Mallee tree habit in heath; (b) sample of bark, branchlet with unopened inflorescences and senesced branchlet with fruit; (c) inflorescence; (d) flowers at anthesis; (e) oblique view of infructescence showing undehisced fruit. Scale bar: (b) 30 mm; (c) 10 mm; (d, e) 5 mm. Photographs: E. Lee (a, d) and T. C. Wilson (b, c, e).
Approximately 700–800 individuals are restricted to 14 known sites in water catchments within or in close proximity of Cattai Creek of north-western Sydney, New South Wales, Australia, spanning the suburbs of Kellyville, Maraylya and Glenorie (Threatened Species Scientific Committee 2018). Our searches for the species in seemingly identical habitat in adjoining water catchments and local government areas have not detected any individuals.
Plateaux and gentle slopes with a western aspect, in shallow or poorly drained soils derived from upper Hawkesbury Group sandstone and overlying Mittagong Formation, sometimes also with laterite. Associated soil landscapes include Lucas Heights and Faulconbridge (Bannerman et al. 2010; Bannerman and Hazelton 2011). The species occurs in heathland or low, open woodland mixed with a dense understorey of shrubs, sometimes associated with sandstone pavements or outcrops.
Flowering has been recorded between October and January. Anecdotal observations suggest that insects are pollinators, although further study of reproductive biology is warranted.
The number of individuals is estimated between 700 and 800 individuals. No occurrences are known from formal conservation estate, some occur on unreserved Crown (State-owned) land, but most are present on freehold rural–residential and non-arable rural land and associated Council-managed roadsides. The species is listed as Critically Endangered in a conservation advice by the Threatened Species Scientific Committee (2018), which indicates that the area where E. cryptica occurs is highly fragmented as a result of urbanisation and is threatened further by an intensification of urban expansion. Genetic swamping from unidentified eucalypt species was also detected by Rutherford et al. (2022) and is a natural threat observed in rare and restricted eucalypts (Larcombe et al. 2014; Rutherford et al. 2019).
From the latinised Greek ‘crypticus’ (i.e. hidden), the epithet refers to the recent near obscurity of this species. Despite herbarium specimens being collected since 1967 and the populations being situated in the largest city in Australia, it has remained unclear to science whether it is a species or a collection of populations of hybrid origin.
In addition to Eucalyptus sp. Cattai (Gregson s.n. 28 Aug 1954), E. cryptica is also cited informally as ‘Eucalyptus sp. Cattai (NSW218983)’ and ‘E. notabilis – resinifera subsp. resinifera’ in the Flora of NSW (PlantNET 2023).
Similarities and differences between E. cryptica and the species of E. subgenus Symphyomyrtus section Latoangulatae series Annulares (Nicolle 2019) it closely resembles are summarised in Table 3. Eucalyptus cryptica is sympatric with the more widely distributed E. squamosa H.Deane & Maiden, a similar-looking mallee that is placed in E. subgenus Symphyomyrtus section Bisectae series Squamosae Chippend. It can be readily distinguished from E. squamosa by its longitudinally fissured bark (v. scaly or tessellated), discolourous leaves (v. concolourous), single axillary inflorescence (v. inflorescence paired), buds generally having a smooth surface (v. often verrucose) and fruit with valves reaching only slightly past the rim of the disc (v. valves distinctly exserted).
| Character | E. cryptica | E. notabilis | E. resinifera | E. scias | |
|---|---|---|---|---|---|
| Habit | Often a multi-stemmed tree (mallee), <6 (–8) m | Single-stemmed tree often >6 m (rarely mallee) | Single-stemmed tree often >6 m | Single-stemmed tree often >6 m | |
| Bark | Longitudinally fissured, fibrous with thin flakes on ridges (eximious) | Longitudinally fissured, fibrous | Longitudinally fissured, fibrous | Longitudinally furrowed, fibrous | |
| Buds | Surface smooth, calyptra compressed | Surface smooth or sometimes ribbed, calyptra compressed | Surface smooth, calyptra tapered | Surface smooth or sometimes verrucose or ribbed, calyptra tapered | |
| Fruit | <9 mm in diameter | <11 mm in diameter | <11 mm in diameter | >10 mm in diameter | |
| Valves | 3 or 4, enclosed with tips sometimes reaching past disc | 3 or 4, exserted | 3 or 4, exserted | 3–6, exserted | |
| Disc | Slightly ascending | Level to slightly ascending | Slightly ascending, level, slightly descending | Level to ascending |
AUSTRALIA: NEW SOUTH WALES (specific localities removed according to endangered listing): 20 Sep. 2018, K.E.Willis 31, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051607); 14 Oct. 2019, T.C.Wilson 829, S.Rutherford & J.Yap (NSW 1058372, NSW 1059242, NSW 1059243, NSW 1059244, NSW 1059246); 31 Oct. 1964, A.Rodd s.n. (NSW 318985); 20 Sep. 2018, K.E.Willis 25, G.Errington, J.Wait & E.Lee (NSW 1057506); 20 Sep. 2018, K.E.Willis 37, G.Errington, J.Wait & E.Lee (NSW 1051639); 20 Sep. 2018, K.E.Willis 38a, G.Errington, J.Wait & E.Lee (NSW 1051640); 20 Sep. 2018, K.E.Willis 38b, G.Errington, J.Wait & E.Lee (NSW 1051641); 20 Nov. 2018, T.C.Wilson & E.Lee s.n. (NSW 1050034); 14 Oct. 2019, T.C.Wilson 831, S.Rutherford & J.Yap (NSW 1058374, NSW 1059222); 21 Feb. 2019, E.Lee & N.Izquiedo s.n. (NSW 1029038); 13 May 2001, M.I.H.Brooker 13208 (NSW 923591); 13 May 2001, M.I.H.Brooker 13209 (NSW 923587); 13 May 2001, M.I.H.Brooker 13211 (NSW 923377); 20 Sep. 2018, K.E.Willis 32a, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051609); 20 Sep. 2018, K.E.Willis 32b, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051611); 20 Sep. 2018, K.E.Willis 33, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051613); 20 Sep. 2018, K.E.Willis 34, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051614); 20 Sep. 2018, K.E.Willis 35, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051615); 20 Sep. 2018, K.E.Willis 36, G.Errington, J.Wait, E.Lee & E.Roper (NSW 1051616); 5 July 2019, T.C.Wilson, S.Rutherford, G.Errington, E.Roper, Z.Aliberti & S.Hunt s.n. (NSW 1028368, NSW 1048012, NSW 1048007); 14 Feb. 2011, M.I.H.Brooker 13207 (NSW 923482); 20 Nov. 2018, T.C.Wilson, E.Lee & P.Barry s.n. (NSW 1050035); 23 Sep. 2010, Klaphake 1b & R.Haq (NSW 874083); 2 Nov. 2018, T.C.Wilson, S.Rutherford, G.Errington & E.Lee (NSW 1048003); 23 Sep. 2010, Klaphake 6b & R.Haq (NSW 874088, NSW 2299635); 5 Mar. 2019, T.C.Wilson & S.Rutherford s.n. (NSW 1042424); 28 Aug. 1954, E.J.Gregson s.n. (NSW 318983, NSW 2299633); 15 Sep. 2015, M.Stables s.n. (NSW 986406).
Data availability
The nuclear genomic data set is available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.f7m0cfz2h.
Acknowledgements
The authors acknowledge the Traditional Custodians of the land on which the study of this species was completed and pay respects to Elders past and present. This paper was enhanced by the excellent botanical illustrations prepared by Lesley Elkan (Royal Botanic Gardens & Domain Trust). The study was also enhanced by assistance with seed germination from Graeme Errington (Australian Plant Bank, Mount Annan). Multiple contributions regarding collections and the knowledge of distribution, ecology and morphology have been provided by Van Klaphake, Peter Ridgeway (Greater Western Sydney Local Land Services), Andrew Orme (Royal Botanic Gardens & Domain Trust) and the late Teresa James (consultant botanist). We thank Erin Roper (then with The Hills Shire Council, Sydney), Stephen Wright (then with Deerubbin Local Aboriginal Land Council) and Patricia Barry and Kevin Barry for assistance with accessing sites; Peter Hannam, whose article in The Sydney Morning Herald precipitated the discovery of previously unknown populations; Peter Jobson and Peter Wilson for revisions and assistance with preparing the paper; and the Horticultural team at the Australian Botanic Gardens, Mount Annan.
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW, Zinner D (2013) Hybridization and speciation. Journal of Evolutionary Biology 26, 229-246.
| Crossref | Google Scholar |
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160, 1217-1229.
| Crossref | Google Scholar |
Bell S, Nicolle D (2020) Glen Gallic Mallee (Eucalyptus dealbata subsp. aperticola, Myrtaceae), a new taxon from the sandstone escarpment of the Hunter Valley, New South Wales. Telopea 23, 141-150.
| Crossref | Google Scholar |
Bragg JG, Yap JYS, Wilson T, Lee E, Rossetto M (2021) Conserving the genetic diversity of condemned populations: optimizing collections and translocation. Evolutionary Applications 14, 1225-1238.
| Crossref | Google Scholar |
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Hér. (Myrtaceae). Australian Systematic Botany 13, 79-148.
| Crossref | Google Scholar |
Collins TL, Andrew RL, Bruhl JJ (2019) Morphological, phytochemical and molecular analyses define species limits in Eucalyptus magnificata (Myrtaceae) and lead to the discovery of a new rare species. Australian Systematic Botany 32, 12-28.
| Crossref | Google Scholar |
Draper D, Laguna E, Marques I (2021) Demystifying negative connotations of hybridization for less biased conservation policies. Frontiers in Ecology and Evolution 9, 637100.
| Crossref | Google Scholar |
Field DL, Ayre DJ, Whelan RJ, Young AG (2009) Molecular and morphological evidence of natural interspecific hybridization between the uncommon Eucalyptus aggregata and the widespread E. rubida and E. viminalis. Conservation Genetics 10, 881-896.
| Crossref | Google Scholar |
Flores-Rentería L, Rymer PD, Riegler M (2017) Unpacking boxes: integration of molecular, morphological and ecological approaches reveals extensive patterns of reticulate evolution in box eucalypts. Molecular Phylogenetics and Evolution 108, 70-87.
| Crossref | Google Scholar |
Goulet BE, Roda F, Hopkins R (2017) Hybridization in plants: old ideas, new techniques. Plant Physiology 173, 65-78.
| Crossref | Google Scholar |
Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA (2012) Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genetics & Genomes 8, 463-508.
| Crossref | Google Scholar |
Griffin AR, Burgess IP, Wolf L (1988) Patterns of natural and manipulated hybridisation in the genus Eucalyptus L’Herit.: a review. Australian Journal of Botany 36, 41-66.
| Crossref | Google Scholar |
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23, 254-267.
| Crossref | Google Scholar |
Huson DH, Kloepper T, Bryant D (2008) SplitsTree 4.0. Computation of phylogenetic trees and networks. Bioinformatics 14, 68-73.
| Google Scholar |
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403-1405.
| Crossref | Google Scholar |
Jones RC, Nicolle D, Steane DA, Vaillancourt RE, Potts BM (2016) High density, genome-wide markers and intra-specific replication yield an unprecedented phylogenetic reconstruction of a globally significant, speciose lineage of Eucalyptus. Molecular Phylogenetics and Evolution 105, 63-85.
| Crossref | Google Scholar |
Larcombe MJ, Barbour RC, Vaillancourt RE, Potts BM (2014) Assessing the risk of exotic gene flow from Eucalyptus globulus plantations to native E. ovata forests. Forest Ecology and Management 312, 193-202.
| Crossref | Google Scholar |
Le S, Nock C, Henson M, Shepherd M (2009) Genetic differentiation among and within three red mahoganies (series Annulares), Eucalyptus pellita, E. resinifera and E. scias (Myrtaceae). Australian Systematic Botany 22, 332-343.
| Crossref | Google Scholar |
Levin DA, Francisco-Ortega J, Jansen RK (1996) Hybridization and the extinction of rare plant species. Conservation Biology 10, 10-16.
| Crossref | Google Scholar |
Mallet J (2005) Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20, 229-237.
| Crossref | Google Scholar |
Nicolle D (2019) Classification of the eucalypts (Angophora, Corymbia and Eucalyptus) Version 4. (Published by the author) Available at http://www.dn.com.au/Classification-Of-The-Eucalypts.pdf
Nicolle D, Jones R (2018) A revised classification for the predominantly eastern Australian Eucalyptus subgenus Symphyomyrtus sections Maidenaria, Exsertaria, Latoangulatae and related smaller sections (Myrtaceae). Telopea 21, 129-145.
| Crossref | Google Scholar |
PlantNET (2023) Genus Eucalyptus. In ‘New South Wales Flora Online: The NSW Plant Information Network System’. (Royal Botanic Gardens and Domain Trust, Sydney, NSW, Australia) Available at https://plantnet.rbgsyd.nsw.gov.au/cgi-bin/NSWfl.pl?page=nswfl&lvl=gn&name=Eucalyptus [Verified 28 August 2023]
Rieseberg LH (1997) Hybrid origins of plant species. Annual Review of Ecology and Systematics 28, 359-389.
| Crossref | Google Scholar |
Rossetto M, Yap JYS, Lemmon J, Bain D, Bragg J, Hogbin P, Gallagher R, Rutherford S, Summerell B, Wilson TC (2021) A conservation genomics workflow to guide practical management actions. Global Ecology and Conservation 26, e01492.
| Crossref | Google Scholar |
Rutherford S, Rossetto M, Bragg JG, McPherson H, Benson D, Bonser SP, Wilson PG (2018) Speciation in the presence of gene flow: population genomics of closely related and diverging Eucalyptus species. Heredity 121, 126-141.
| Crossref | Google Scholar |
Rutherford S, van der Merwe M, Wilson PG, Kooyman RM, Rossetto M (2019) Managing the risk of genetic swamping of a rare and restricted tree. Conservation Genetics 20, 1113-1131.
| Crossref | Google Scholar |
Rutherford S, Wilson TC, Yap JYS, Lee E, Errington G, Rossetto M (2022) Evolutionary processes in an undescribed eucalypt: implications for the translocation of a critically endangered species. Annals of Botany 130, 491-508.
| Crossref | Google Scholar |
Steane DA, Nicolle D, Sansaloni CP, Petroli CD, Carling J, Kilian A, Myburg AA, Grattapaglia D, Vaillancourt RE (2011) Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Molecular Phylogenetics and Evolution 59, 206-224.
| Crossref | Google Scholar |
Threatened Species Scientific Committee (2018) ‘Conservation Advice Eucalyptus sp. Cattai (Gregson s.n., 28 Aug 1954).’ (Department of the Environment and Energy: Canberra, ACT, Australia) Available at https://www.environment.gov.au/biodiversity/threatened/species/pubs/89499-conservation-advice-11052018.pdf
Wilson TC, Rossetto M, Bain D, Yap J-YS, Wilson PD, Stimpson ML, Weston PH, Croft L (2022) A turn in species conservation for hairpin banksias: demonstration of oversplitting leads to better management of diversity. American Journal of Botany 109, 1652-1671.
| Crossref | Google Scholar |


