Evidence for convergent evolution among phylogenetically distant rare species of Tetratheca (Elaeocarpaceae, formerly Tremandraceae) from Western Australia
Ryonen Butcher A B E , Margaret Byrne C and Darren M. Crayn DA Western Australian Herbarium, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, WA 6983, Australia.
B School of Plant Biology (Botany MO90), The University of Western Australia, 35 Stirling Highway, Crawley, WA 6009, Australia.
C Science Division, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, WA 6983, Australia.
D National Herbarium of New South Wales, Botanic Gardens Trust, Mrs Macquaries Road, Sydney, NSW 2000, Australia.
E Corresponding author. Email: Ryonen.Butcher@dec.wa.gov.au
Australian Systematic Botany 20(2) 126-138 https://doi.org/10.1071/SB06017
Submitted: 1 June 2006 Accepted: 22 January 2007 Published: 26 April 2007
Abstract
Morphological and molecular investigations of taxon relationships among rare species of Tetratheca Sm. occurring near Koolyanobbing, Western Australia, have confirmed the distinctness of T. aphylla F.Muell., T. harperi F.Muell. and T. paynterae Alford and identified three new rare taxa from collections affiliated with T. aphylla and T. paynterae. The recognition of these taxa at specific and sub-specific ranks is based on their different degrees of morphological and molecular divergence, combined with geographic disjunction. Cladistic analysis of nrDNA internal transcribed spacer and cpDNA trnL-trnF sequences from a range of Tetratheca species from Western Australia and the eastern states indicates that T. aphylla, T. harperi and T. paynterae belong to three separate evolutionary lineages and that the endemism displayed among these taxa to small, disjunct ranges within the same geographic area, is a result of in situ speciation due to historical fragmentation. These results exemplify the extremely high conservation value of the Yilgarn banded ironstone ranges. The superficial similarity among the study taxa in having a ‘leafless’ habit can be seen to be adaptive convergence in response to the marginal and semi-arid environments in which they occur, and this character is highly homoplastic within the genus.
Introduction
Tetratheca Sm. is the largest of the three genera (the others are Tremandra R.Br.ex D.C. and Platytheca Steetz) previously comprising the small, endemic Australian family Tremandraceae (Watson and Dallwitz 1992). Recent molecular studies (e.g. Savolainen et al. 2000; Soltis et al. 2000; Bradford and Barnes 2001; Crayn et al. 2006) have indicated that Tremandraceae is nested within Elaeocarpaceae (Oxalidales) and the results of studies of floral morphology, anatomy and histology (Matthews and Endress 2002) are consistent with this relationship. Therefore Tremandraceae has been included in Elaeocarpaceae in recent classifications (Stevens 2001; The Angiosperm Phylogeny Group 2003; Coode 2004).
Tremandra and Platytheca each contain two species restricted to south-west Western Australia. Tetratheca is widespread across the southern half of the continent and currently contains 42 described species, 23 of which are restricted to Western Australia (Thompson 1976; Alford 1995; Western Australian Herbarium 1998; Butcher and Sage 2005). In both eastern and western Australia there are a few widespread species of Tetratheca and numerous restricted taxa, many of these appearing to be relicts of once far more widespread species (Thompson 1976; Brown et al. 1998). As a result of the large number of additional collections that have been made since the last revision of Tetratheca (Thompson 1976), it is evident that the number of species in this genus is much higher than is currently recorded, and that many of these new taxa are geographically restricted and may require conservation listing.
The last revision of Tetratheca (Thompson 1976) resulted in the description of a large number of new species and the conjectural identification of various species pairs, or small groups, containing morphologically similar taxa. In examining the Western Australian species, Thompson (1976) recognised the following associations: T. aphylla F.Muell. and T. paucifolia Joy Thomps.; T. harperi F.Muell. and T. halmaturina J.M.Black; and T. affinis Endl., T. efoliata F.Muell. and T. retrorsa Joy Thomps. Of interest is that T. halmaturina is restricted to Kangaroo Island, off the South Australian coast, and it is hypothesised that this island has been colonised with Tetratheca from both Western Australia and the eastern states in the past (Thompson 1976). In the key to species, Thompson (1976, p. 154) regarded all these taxa as having ‘…a generally leafless aspect, although some leaves may be present’, and included T. nuda Lindl., T. virgata Steetz and T. remota Joy Thomps. in this ‘leafless’ grouping. However, T. retrorsa, T. virgata and T. remota are also recognised as being ‘leafy’ (Thompson 1976, p. 154–55) and it is acknowledged that this ‘leafless’ group is an artificial, but convenient, construct, although close relationships probably exist between species within it.
As part of her investigation into the conservation and taxonomic status of Western Australian Tetratheca, Alford (1995) described two new, geographically restricted ‘leafless’ species; T. chapmanii Alford and T. paynterae Alford, the initial collections of which were thought to represent range extensions for T. aphylla. These species are similar to T. aphylla in having terete, shortly tuberculate stems and minute, narrowly triangular, early deciduous leaves, but can be distinguished by a suite of morphological characters (Alford 1995). Although Alford (1995) placed these two taxa, along with T. halmaturina, in the ‘Tetratheca aphylla group’ (Alford 1995, p. 144) and suggested that a close evolutionary relationship exists between these four species, this was not substantiated by any cladistic analyses and the significance of the majority of morphological characters for phylogeny reconstruction is not known.
In assigning taxa to species pairs or groups, Thompson (1976) placed considerable emphasis on ovule number per loculus as an indicator of relatedness between similar taxa. In the associations outlined above, Thompson (1976) records T. aphylla, T. paucifolia, T. harperi and T. halmaturina as possessing one ovule per loculus, T. efoliata and T. retrorsa as possessing two ovules per loculus and T. affinis as being highly distinctive in usually possessing four or five ovules per loculus. Similarly, Alford (1995) suggests that the shared possession of two ovules per loculus by T. chapmanii and T. paynterae indicates that these two taxa may be closely related relictual species. However, the close resemblance between these taxa and T. aphylla also suggests possible convergent evolution in response to habitat, as all three species grow on steep slopes and in rock crevices of exposed, banded ironstone or sandstone massifs in semi-arid areas (Alford 1995; Brown et al. 1998), with T. chapmanii geographically isolated in the Carnarvon Range of the Murchison region and T. aphylla and T. paynterae restricted to single hills or small ranges in the Coolgardie region (Thackway and Cresswell 1995; Brown et al. 1998).
Interest in the ‘leafless’ species of Tetratheca has been renewed recently because of an expansion in mining activity in the Koolyanobbing area of Western Australia, where T. aphylla, T. paynterae and T. harperi are recorded as geographically restricted, rare species. According to Brown et al. (1998), T. aphylla occurs over an area of c. 10 km in the Helena and Aurora Range, whereas T. paynterae is restricted to an area of c. 2 km on an unnamed range north of Windarling Peak, known colloquially as the ‘Windarling Range’. T. harperi is known only from collections at Mt Jackson and Muddarning Hill (Brown et al. 1998), where it occupies nearly identical habitat to T. aphylla and T. paynterae. As a result of their highly restricted distributions, T. aphylla, T. paynterae and T. harperi are currently gazetted as Declared Rare Flora (Atkins 2006) with T. aphylla and T. harperi listed as Vulnerable and T. paynterae as Critically Endangered (IUCN 2001). Recognised threats to these species include mineral exploration, mining activity and increased pastoral stocking (Brown et al. 1998).
Recent flora surveys have located a new population of Tetratheca in the Die Hardy Range, c. 10 km NNW of ‘Windarling Range’, which is morphologically similar to T. paynterae. As with other regional ranges (e.g. Helena and Aurora Range ‘Windarling Range’, Mt Jackson, Koolyanobbing Range) the ironstone vegetation of the Die Hardy Range has been classified under Beard’s (1972) Bungalbin System (Gibson et al. 1997) and the distinctness of plants from this new population from T. paynterae, which is currently threatened by mining activities, has been queried. In addition there is some question in relation to the taxonomic status of collections identified as T. aphylla from geographically disjunct locations near Newdegate, c. 300 km south of the Helena and Aurora Range, and from near Eneabba, in Western Australia’s Geraldton Sandplain region (Thackway and Cresswell 1995). This study aimed to examine the morphological variation in these new collections relative to the existing collections of T. paynterae, T. aphylla and other ‘leafless’ species at PERTH to resolve their taxonomic status.
Molecular systematics can provide additional, morphologically independent evidence of species relationships and DNA sequencing studies are now commonplace for the investigation of organismal relationships at the generic and specific ranks. Determination of consistent differences in DNA sequences is a complementary tool to morphological analysis for assessing taxon distinctness. The most frequently sequenced regions of the plant genome for the investigation of species-level relationships are the internal transcribed spacer (ITS) of nuclear rDNA (see Baldwin et al. 1995 for a review; Bena et al. 1998a, 1998b) and the trnL-trnF spacer in the chloroplast genome (see Sang et al. 1997 for a review; Bayer et al. 2000), and an analysis of these regions was undertaken to investigate relationships within the T. aphylla–T. paynterae–T. harperi group of ‘leafless’ species from the Koolyanobbing area and the taxonomic status of the newly identified, unclassified collections affiliated with these species. A molecular systematic study of these taxa can also be used to investigate the phylogenetic signal of putatively significant morphological characters, such as ovule number and the leafless condition, as well as elucidate the biogeographical relationships between these rare ironstone endemics and their affiliates. To increase the robustness of any phylogenies hypothesised within this group, DNA sequences from these taxa need to be placed in broader analyses, and to this end sequences from an additional seven Western Australian species and five eastern states species, as well as the outgroup taxa, Tremandra diffusa D.C. and Platytheca galioides Steetz, were obtained and included in analyses.
Methods
Morphology
Herbarium material from PERTH was examined for all Western Australian ‘leafless’ species, sensu Thompson (1976) and Alford (1995), including type collections of T. paynterae, T. chapmanii and T. paucifolia. In addition to herbarium specimens, fresh flowering material was examined for T. aphylla and T. paynterae as well as the unclassified populations from the Die Hardy Range and from Eneabba. Fresh sterile material was examined for T. harperi and the unclassified population from Newdegate.
Measurements of leaves and flowers were made from reconstituted fragments and spirit-preserved fresh material, and observations and measurements of stem characters were based on herbarium specimens. Morphological characters that were assessed in detail concentrated on those considered to have high taxonomic value by Thompson (1976) and included the vestiture of the stem, stem thickness in the flowering region, leaf size, form and pubescence, pedicel length and vestiture, receptacle width and shape, calyx segment length, shape and vestiture, petal number, shape and coloration, the length, shape, colour, vestiture and fusion of the various parts of the stamens, vestiture of the ovary and style, and ovule number per loculus.
Thompson (1976) notes that there is infraspecific morphological variation in flower colour, with white flowered individuals occurring in many taxa, and in the density and distribution of hairs on the various parts of the plant. Particular note is made of the importance of combinations of hair types in species determination, with the glabrous condition in an individual viewed as less significant than the absence of a specific type of hair when multiple types occur in combination. Note is also made of anomalous flowers in which there is variation from the usual number of ovules per locule, aberrant locule number, ‘double’ flowers or split styles. Knowledge of these variants highlights the importance of combinations of characters in accurate species discrimination and the need to examine as many specimens as possible from across a taxon’s range before formalising taxonomic rank.
Molecular
Deoxyribonucleic acid sequence data from the trnL-trnF region of cpDNA and the ITS region of nrDNA were generated for seven individuals of T. aphylla, four each of T. paynterae and T. harperi, three of T. (Newdegate), and two each of T. (Die Hardy Range) and T. (Eneabba) at the Western Australian Herbarium (PERTH). One individual of one species each of Tremandra and Platytheca and a range of other Tetratheca species chosen to represent the geographic and taxonomic diversity of the genus were sequenced at the Royal Botanic Gardens, Sydney (NSW). Molecular protocols were similar between the institutions. The protocols detailed below are for PERTH, with those in square brackets for NSW where they differed from PERTH. Collection and voucher information including GenBank accessions for sequences are presented in Table 1.
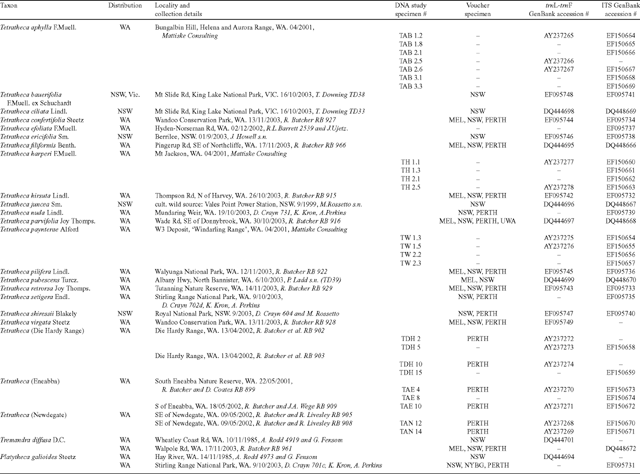
|
Deoxyribonucleic acid extractions were made from a variety of tissues including fresh mature leaves and the bases of deciduous leaves, bracts, young buds and stem scrapings, as well as young buds and bracts collected and stored in liquid nitrogen in the field and from whole stem material dehydrated in silica gel. Extractions were performed with a DNeasy Plant Mini Prep Kit (Qiagen Inc, Valencia, CA, USA) according to the manufacturer’s specifications, with 0.01–0.10 g of starting material yielding generally less than 25 ng of DNA per μL. Deoxyribonucleic acid extracted from stem scrapings was more degraded than that obtained from buds or leaf/bract material. Extractions from T. (Newdegate) and T. (Eneabba) plants were made utilising a 20% higher volume of Buffer AP1 and Buffer AP2 than in the manufacturer’s instructions (after J. Bradford, pers. comm.) with apparently good results, but amplification difficulties in these two taxa suggested that additional compounds in the fresh buds were interfering with PCR reactions.
Amplification of the ITS region was carried out in a reaction mix of 5 μL 5× PCR buffer (Invitrogen, Carlsbad, CA, USA) [2.5 μL 10× buffer; Bioline, Luckenwalde, Germany], 2 [1.5–3.0] mm MgCl2, 200 μM each dNTP, 200 μM each primer, 1 unit Gibco (Invitrogen) Taq polymerase [0.5 units BIOTAQ; Bioline, Luckenwalde, Germany], 25–50 ng DNA and dH2O to a total of 25 μL, with the following cycle: 95°C for 5 min [94°C for 3 min], followed by 30 cycles of 95°C for 1 min [94°C for 30 s], 56°C for 1 min [52°C or 55°C for 30 s] and 72°C for 1 min, followed by 7 min at 72°C [5 min at 72°C]. For the taxa sequenced at PERTH, two primer pairs were used: ITSLeu1 and ITS4 (Mast 1998, modified from White et al. 1990), and P3L (5′-TTGAATGGTCCGGTGAAGTGTTCGG-3′) and P2R (5′-CTTTTCCTCCGCTTATTGATA-3′) (designed for Proteaceae by P. Weston). ITSLeu1 and ITS4 provided clean sequence reads in T. harperi but P3L and P2R produced the best sequence results for T. paynterae and T. aphylla (Butcher et al. 2001) and were used to amplify and sequence the T. (Die Hardy Range), T. (Eneabba) and T. (Newdegate) individuals. For taxa sequenced at NSW, the primers GN1 (Scott and Playford 1996) and ITS4 (White et al. 1990) were used.
Amplification mixtures for the trnL–trnF region differed from those for ITS in containing 3 [1.5–3.0] mm MgCl2, 160 [200] μM each dNTP, 50 [25–50] ng DNA. Amplification and sequencing utilised the primers trn-c and trn-f (Taberlet et al. 1991).
Amplification products were purified with the HighPure PCR Purification Kit (Roche Applied Science, Basel, Switzerland) or QIAQuick PCR Purification Kit (Qiagen, Hilden, Germany) [JetQuick columns (Genomed, Bad Oeynhausen, Germany)] according to the manufacturer’s specifications. Sequence reactions were performed by the Big Dye Terminator (BDT) method and 10 [15] μL reaction volumes. ITS mixtures contained 4 [1] μL BDT, [3 μL CSA buffer], 40–100 ng [60–100 ng] DNA and 2 [3] pmol primer. trnL-trnF mixtures contained 2 [1] μL BDT, [3 μL CSA buffer], 40–100 ng [60–100 ng] DNA and 1.6 pmol [3] primer. Reaction mixtures were subjected to 96°C for 4 min, followed by 25 cycles of 95°C for 30 s [96°C for 10 s], 43°C for 15 s [50°C for 5 s] and 60°C for 4 min [plus a final 1 min incubation at 60°C]. Fragments were ethanol-purified and electrophoresed with an ABI Prism 373 slab-gel sequencer [ABI Prism 3730 capillary sequencer (Applied Biosystems, Foster City, CA, USA)]. Chromatograms were checked and manually corrected where polymorphisms were observed with SeqEd v 1.0.3 (Kececioglu and Myers 1992), and pair-wise, multiple sequence alignments for all datasets were performed with ClustalW (Thompson et al. 1994) using the default settings. The Clustal alignments were further improved manually in BioEdit 5.0.9 (Hall 1999).
Following correction and alignment the ITS and trnL-trnF sequence matrices were analysed separately, and combined, with PAUP* (v. 4.0b10, Swofford 2003). The combined dataset contained only those 25 taxa for which both ITS and trnL-trnF data were available. Variable sites (where two or more different bases were evident at the same position in the chromatogram) were coded as uncertainties (rather than polymorphisms) and gaps (representing indels) were treated as missing data. Unambiguous, potentially informative indels were binary coded and added to the data matrices (Simmons and Ochoterena 2000). Fitch parsimony (characters treated as unweighted and unordered, Fitch 1971) analyses were performed by heuristic search with 1000 random addition sequence replicates saving a maximum of 100 trees per replicate, with tree bisection-reconnection branch swapping. Clade support was evaluated by bootstrap (Felsenstein 1985) with 2000 replicates of heuristic search, saving a maximum of 1000 trees per replicate. Trees were rooted using Platytheca galioides and Tremandra diffusa as outgroups. Broader analyses indicate that these taxa are the nearest relatives of Tetratheca (Crayn et al. 2006).
A model-based estimate of the phylogeny was obtained by Bayesian analysis using the Markov Chain Monte Carlo (MCMC) implemented in Mr Bayes 3.1.1 (Huelsenbeck and Ronquist 2001). Two runs each consisting of four Markov chains were started simultaneously from random trees. The most general likelihood model was used: number of substitution types set to six (nst = 6), and among-site rate variation modelled by the gamma distribution with four rate categories and a proportion of invariable sites (rates = invgamma, ngammacat = 4). All other priors for the analysis were set flat (Dirichlet priors). One million generations were performed for each run sampling a tree every 100 generations. Trees generated before stationarity being reached (the burn-in) were discarded before the posterior probability of each node being determined using the ‘sumt’ command. The indel characters were not used in the Bayesian analyses.
Results
Morphological variation
The rare species of the Koolyanobbing region (T. aphylla, T. harperi and T. paynterae) are easily distinguished from one another based on a large number of morphological characters and these differences are outlined in detail in Thompson (1976) and in Alford (1995).
Analysis of plants of Tetratheca (Die Hardy Range) highlighted their morphological similarity to T. paynterae and the two populations shared important diagnostic characters such as glabrous stems with rounded tubercules, pedicels of similar length which were curved at the base and expanded abruptly into the receptacle at their apex, petals which were dark pink with a yellow spot at the base, similarly proportioned calyx segments and stamens, two ovules per locule, and a strong, musky floral scent. Plants of T. (Die Hardy Range) could be distinguished from typical T. paynterae in both fresh and herbarium material, however, by their sparser pubescence across all parts, the shape of their receptacle, the colour of the anther tubes, anther filaments and style apex, and the pubescence of the ovary and style. In the field, plants of T. (Die Hardy Range) were also noted to be larger and more intricately branched with a sprawling habit, such that they hung down cliff walls from rock fissures, compared with typical plants of T. paynterae which usually had shorter, erect stems. A morphometric investigation of 34 plants of T. paynterae and 35 plants of T. (Die Hardy Range) which encompassed their entire distributions at their disjunct localities, found that these two populations were distinct based on a suite of consistent, small differences in morphology (Butcher et al. 2002).
The unclassified Tetratheca collections from Eneabba and Newdegate were morphologically similar to T. aphylla collections from Bungalbin Hill in their possession of short, acute stem tubercules, short, hispid pedicels, hispid calyx segments and one ovule per locule. Plants of T. (Newdegate) were distinguishable from T. aphylla only by small differences in stem characters, the curvature of the anthers and the length and thickness of the anther filaments, but differed in the width of the fruits. By comparison, plants of T. (Eneabba) possessed several morphological differences from existing T. aphylla collections in both vegetative and floral characters including leaf morphology, the length of the calyx segments and petals, the form of the glandular hairs on the pedicels, calyx segments and ovary, and in the relative lengths, curvature and vestiture of the staminal parts.
Molecular analyses
ITS
Amplification of the ITS region in T. aphylla, T. harperi, T. paynterae, T. (Die Hardy Range), T. (Eneabba) and T. (Newdegate) produced a PCR product of 651–663 bp in length. Variation between individuals within these taxa was negligible and occurred mainly as single nucleotide substitutions, usually at a base position in which intraindividual polymorphisms were identified. At these nucleotide positions, chromatograms revealed peaks of approximately equal height, representing the co-occurrence of two or more different bases. Within each of the taxa sampled, the nucleotide position and base composition of these multiple peaks was consistent between individuals, but there was variation in the relative dominance of each base at that site.
The ITS sequence matrix contained 774 nucleotide characters plus two indel characters (776 altogether). Alignment of a section of the ITS1 region close to 5.8S (comprising 75 positions) was problematical and this region was excluded from analysis. The analysed data therefore comprised 701 characters, 280 of which were variable and 126 potentially informative.
Most of the informative characters were in the ITS1 and ITS2 regions. A single base transversion substitution common to T. paynterae and T. (Die Hardy Range) was observed in the highly conserved 5.8S gene and differentiated these from all other taxa.
The level of ITS sequence divergence varied within and among the Koolyanobbing taxa sampled. For example, pair-wise distances between individuals of T. paynterae and T. aphylla were high and ranged from 0.05837 to 0.07099 (mean character differences), with total character differences ranging from 38 to 46, whereas pair-wise distances between T. paynterae and T. (Die Hardy Range) were very low and ranged from 0 to 0.00306 (mean character differences) with total character differences ranging from 0 to 2 among the individuals sampled. Similarly, pair-wise distances between T. aphylla and T. (Newdegate) were also low with mean character differences ranging from 0.00303 to 0.00760 and total character differences ranging from 2 to 5 among the individuals sampled.
Parsimony analysis of the ITS data yielded 96 400 shortest trees (L = 487 steps, CI excluding uninformative characters = 0.621, RI = 0.737). Despite the large number of optimal trees, the strict consensus tree is well resolved and many clades obtain bootstrap support (Fig. 1). Bayesian analysis indicated high posterior probabilities (based on 19 002 trees – the first 500 trees of each run were discarded as the burn-in) for many clades. The individuals of T. (Eneabba) were strongly monophyletic (1.0/87%), as were those of T. harperi (1.0/100%). Other entities were part of strongly supported groups but were not resolved as monophyletic: individuals of T. paynterae and T. (Die Hardy Range) (1.0/100%), and individuals of T. aphylla and T. (Newdegate) (1.0/100%). Other well-supported relationships include Tetratheca (0.89/99%), T. filiformis Benth. as sister to the remaining Tetratheca (0.92/91%), T. bauerifolia F.Muell. ex Schuchardt, T. juncea Sm. and T. shiressii Blakely (1.0/96%), and T. hirsuta Lindl. and T. pubescens Turcz. (0.96/81%).
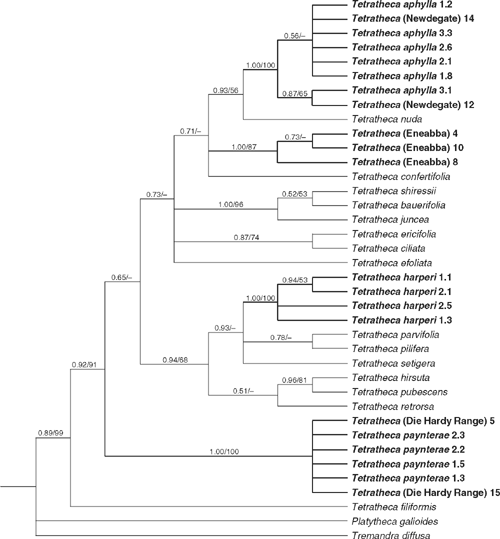
|
trnL-trnF
Amplification of the trnL-trnF region in T. aphylla, T. harperi, T. paynterae, T. (Die Hardy Range), T. (Eneabba) and T. (Newdegate) produced a fragment between 724–875 bp length. Alignment of the trnL-trnF sequence indicated that the trnL-trnF sequences of T. aphylla, T. harperi and T. paynterae were clearly distinct with T. paynterae differentiated from T. aphylla by nine substitutions and three indels and T. harperi differentiated from T. aphylla by two substitutions and two indels. Comparatively, the sequences of T. paynterae and T. (Die Hardy Range) were very similar, differing at only three base positions including a two bp indel at position 336–337 and a transversion substitution at position 792 within a shared indel region. The sequences of T. aphylla and T. (Newdegate) varied at only one base position, with T. (Newdegate) having a unique transversion substitution at position 859. T. (Eneabba) was differentiated from the other taxa primarily by a unique 87 bp indel as well as an autapomorphic transversion substitution at position two. The alignment comprised 998 nucleotide characters plus eight indel characters (1006 altogether). Of these, 91 were variable and 47 potentially informative.
Parsimony analysis of trnL-trnF data found 1189 shortest trees (L = 105 steps, CI excl. uninf. = 0.831, RI = 0.926) distributed among 10 islands. Posterior probabilities were based on 19 802 trees, the first 100 of each run being discarded as the burn-in. There is strong support for a clade comprising T. paynterae and T. (Die Hardy Range) (1.0/100%), and strong to moderate support for T. harperi (1.0/87%) and T. (Eneabba) (0.93/80%). An eastern Australian clade comprising T. ericifolia Sm., T. shiressii, T. juncea and T. bauerifolia obtained moderate support (0.94/83%). Within this clade, parsimony resolved T. juncea as sister to the other three taxa, whereas Bayesian analysis resolved a T. juncea plus T. bauerifolia clade. Both arrangements received very weak support, however. Weak support was obtained for the monophyly of T. (Newdegate) (0.93/62%) and there was a tendency for individuals of T. paynterae to form a group (<0.5/56%). These groups are incongruent with the ITS tree but again the differences obtain low support. (Fig. 2).
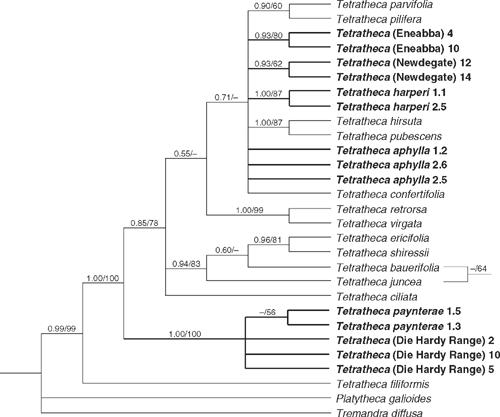
|
Combined ITS – trnL-trnF
An incongruence length difference test (Farris et al. 1994) indicated significantly different phylogenetic signals between the ITS and trnL-trnF data (100 replicates, P = 0.01). However, because interpretation of test results is somewhat unclear and false significance or insignificance can be attributed under certain circumstances (Dolphin et al. 2000; Barker and Lutzoni 2002; Yoder et al. 2001; Ramirez 2006), and because the ITS and trnL-trnF tree topologies were mostly congruent and observed conflicts were very weakly supported, analysis of the combined data was undertaken. Only the 25 taxa for which data from both regions was available were included, which meant that the representation of most taxa was reduced to one or two individuals. Therefore, the results of the combined analyses are not directly comparable to those from the ITS and trnL-trnF analyses in some respects. Parsimony analysis of the combined data yielded 12 shortest trees (L = 507 steps, CI excl. uninf. = 0.699, RI = 0.756). Posterior probabilities were based on 19 802 trees, the first 100 of each run being discarded as the burn-in. The monophyly of Tetratheca and the placement of T. filiformis as sister to the rest are strongly supported (both 1.0/100%). Despite their geographical proximity, the Koolyanobbing area taxa are not closely related, falling into three groups widely separated on the phylogeny (Fig. 3). The individuals of T. (Die Hardy Range) and T. paynterae form a robust clade (1.0/100%) but the data do not support monophyly of T. paynterae with respect to T. (Die Hardy Range), however. This clade is resolved with confidence as sister (1.0/100%) to a robust clade (1.0/80%) comprising all other Tetratheca except T. filiformis. T. harperi is strongly monophyletic (1.0/100%) and is nested within a well supported clade (1.0/81%) containing a range of other WA species, namely T. hirsuta, T. parvifolia Joy Thomps., T. pilifera Lindl., T. pubescens and T. retrorsa. A clade comprising samples of T. aphylla, T. (Eneabba) and T. (Newdegate) is resolved (0.91/67%) with T. confertifolia Steetz as its sister. Within this clade T. (Newdegate) is monophyletic (0.95/70%) within a robust clade (1.0/100%) that also includes the two T. aphylla samples. This clade is sister (0.91/67%) to a well supported (1.0/100%) T. (Eneabba) clade.
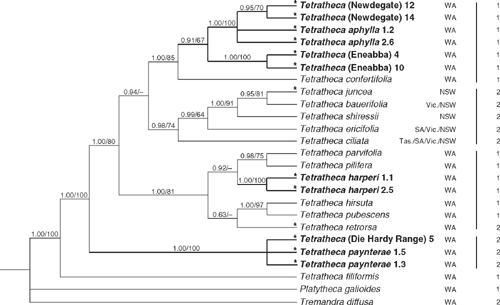
|
Monophyly of the eastern Australian species (T. bauerifolia, T. ciliata Lindl., T. ericifolia, T. juncea, T. shiressii) is moderately well supported (0.98/74%) and there is some evidence from the Bayesian analysis that this clade is sister (0.94/<50%) to the T. aphylla clade.
The ‘leafless’ condition appears polyphyletic on the phylogeny with taxa regarded as ‘leafless’ by Thompson (1976) occurring in each of the main clades (Fig. 3).
Discussion
Taxonomic and systematic conclusions
There is a clear correlation between morphological variation and DNA sequence divergence among the rare Tetratheca species from the Koolyanobbing area and their affiliates. Analysis of ITS and trnL-trnF sequence data corroborates the morphological evidence that T. harperi, T. aphylla and T. paynterae are distinctly different species, and thus permits their use as a baseline for comparison in assessing issues of taxonomic distinctness and rank among the unclassified populations.
Tetratheca (Eneabba) and T. (Newdegate) share several morphological characters with T. aphylla, including shortly acute-tuberculate stems, short, hispid pedicels, hispid calyx segments and one ovule per locule, and these taxa co-occur in weakly to moderately well-supported clades (Figs 1–3) on the basis of their nuclear and chloroplast DNA sequences. While T. (Eneabba) is clearly distinct from T. aphylla in both its morphological and molecular characters, T. (Newdegate) is extremely similar in both, such that their relationship is unresolved in ITS analysis and only weakly resolved based on trnL-trnF sequences as a result of a single, unambiguous, nucleotide substitution. The sister relationship between T. aphylla individual 3.1 and T. (Newdegate) individual 12 (Fig. 1) is probably because of long branch attraction resulting from poor sequence reads for each of these samples and the correspondingly high number of missing characters for these in the data matrix. On the basis of the degree of morphological and molecular divergence observed between T. aphylla, T. paynterae and T. harperi, it is concluded that T. (Eneabba) warrants recognition as a distinct species, whereas T. (Newdegate) should be recognised at sub-specific rank relative to T. aphylla. These taxa are described as new in this issue (Butcher 2007). T. paucifolia has been hypothesised to be the sister species to T. aphylla (Thompson 1976) and has close morphological affinities with T. (Eneabba), with which it shares its geographic distribution. These three species appear to form a close group and the inclusion of T. paucifolia in future cladistic analyses of Tetratheca is needed in order to test this.
Tetratheca (Die Hardy Range) was found to be distinct from T. paynterae in both minor morphological and molecular characters, and the taxa are evidently very closely related. As was seen between T. aphylla and T. (Newdegate), cladistic analysis of ITS sequence data did not resolve the relationships between the T. paynterae and T. (Die Hardy Range) individuals, with the inclusion of trnL-trnF sequences nesting the Die Hardy Range plants within the T. paynterae clade because of their shared possession of a single base substitution and a two bp indel. A preliminary investigation of genetic diversity within and between T. paynterae and T. (Die Hardy Range) populations with amplified fragment length polymorphism identified marked genetic differentiation between plants from these two localities relative to the degree of variation within each population, with 33.3% of total markers scored for T. (Die Hardy Range) showing fixed or highly significant genetic differences (S. Krauss, pers. comm.). Assessment of both morphological (Butcher et al. 2002) and molecular (S. Krauss, pers. comm.) variation within and between T. (Die Hardy Range) and T. paynterae populations, in conjunction with a detailed investigation of morphological variation throughout the Western Australian species of Tetratheca, and accounts of infraspecific plasticity in eastern states taxa (Thompson 1976), indicates that T. (Die Hardy Range) is a distinct taxon from T. paynterae that should be recognised at sub-specific rank. This taxon is described as new in this issue (Butcher 2007).
The combined use of nrDNA and cpDNA datasets for the study group affords greater resolution of relationships among these taxa than either dataset alone, with the more recent evolutionary divergence of T. aphylla and T. (Newdegate), and T. paynterae and T. (Die Hardy Range) reflected in the similarity of their DNA sequences. It is interesting that there is a similarly low level of morphological and molecular divergence between these subspecies pairs given the differences in geographic distance between them, with T. paynterae and T. (Die Hardy Range) occupying adjacent ranges separated by c. 10 km, and T. aphylla and T. (Newdegate) separated by c. 300 km. The high level of morphological and molecular divergence seen between T. aphylla, T. paynterae and T. harperi is significant given their close geographic proximity and similarity of ecological niches in the Koolyanobbing area, and they are members of distinctly different and well-supported clades, some containing species from well outside the Koolyanobbing area. It is evident that these species are not closely related despite their superficial similarity and it can be hypothesised that they are relictual members of different evolutionary lineages that have speciated in situ on each of these small, banded ironstone ranges over millions of years. The extremely high conservation value of these Yilgarn ranges, with their large numbers of endemic and conservation listed species (Gibson et al. 1997; Mattiske Consulting 2001; ecologia Environmental Consultants 2002; Western Australian Herbarium 1998) cannot be emphasised enough.
The importance of ovule number for distinguishing between species and for assessing relationships has been emphasised by Thompson (1976, p. 141) and this has been partly supported in this study. Although the taxonomic utility of ovule number per locule is reasonably sound for the differentiation of similar taxa, such as T. aphylla and T. paynterae, and the number of ovules per locule is generally consistent within each lineage (Fig. 3), observations of recently collected material at PERTH have found more variation than has been previously recorded within some species. For example, some T. aphylla collections (PERTH 06352103; PERTH 06352049) have two ovules per locule, with either one or two of these developing into seed and the remainder aborted, and a collection of T. (Die Hardy Range) (PERTH 06958923) had no ovules in one locule and only one in the other. Additionally, several collections of T. affinis were found to vary from usual in possessing only one, two or three ovules per locule, rather than the typical four or five. This variability represents an interesting condition in a genus in which ovule number is considered consistent for each taxon (Thompson 1976, p. 151).
Alford’s (1995) placement of T. paynterae in the ‘T. aphylla group’, based on their overall similarity in habit is not supported as a natural grouping by these molecular data, which indicate that the ‘leafless’ condition is highly homoplastic. Taxa regarded as ‘leafless’ in Thompson’s (1976) key to species are polyphyletic (Fig. 3) and this condition has evidently arisen multiple times within the genus, presumably in response to xeric habitat. Similar patterns of adaptive radiation and convergent evolution in ‘leaflessness’ can be seen across the Western Australian flora in multiple genera from multiple families [e.g. Exocarpos (Exocarpaceae), Lechenaultia and Dampiera (Goodeniaceae), and Comesperma (Polygalaceae)], as well as in numerous species within a genus [e.g. Daviesia (Fabaceae)], with the ‘leafless’ habit commonly seen in species occurring in drier, hotter, frequently more inland, conditions than their leafy congeners (Carlquist 1974). Adaptations to drier habitat, to minimise transpiration, include microphyllous and deciduous leaves, both of which are seen in the Koolyanobbing area species, as well as ericoid leaves and inrolled leaf margins, which are common in many wheat belt and semi-arid Tetratheca species from Western Australia (e.g. T. confertifolia, T. retrorsa and T. efoliata).
Biogeography
Although far from exhaustive, the Tetratheca phylogeny presented here (Fig. 3) shows support for the recognition of both refugial and relictual species among the Western Australian taxa. For example, in this analysis, T. filiformis is sister to the rest of the genus and is restricted to winter wet, refugial areas in the far south-west. A Western Australian origin for the genus has been hypothesised (Thompson 1976) and it is apparent that Tetratheca underwent an early transcontinental radiation, and following the Eocene separation of the eastern and western populations, there was substantial diversification of lineages in the western region, some of which have undergone additional historical fragmentation events. The status of the Koolyanobbing area rare species as relictual taxa is supported based on their position in different clades and their high degree of local endemism on disjunct, ancient geological formations in close geographic proximity. Among these species and their affiliates it can be seen that T. aphylla and T. (Eneabba) are members of a common lineage that has undergone historical fragmentation, and that the disjunction between T. aphylla and T. (Newdegate) is a possible result of fragmentation and range contraction, with the taxa diversified into very different habitats. By comparison, the lineage comprising T. paynterae and T. (Die Hardy Range) seems to have arisen earlier in the evolutionary history of Tetratheca, with the current patterns of distribution and diversification between the subspecies resulting from either a more recent fragmentation or dispersal event, followed by reproductive isolation.
The combination of ancient relictual species and more recently diverged taxa is consistent with the biogeographical history of the region. Speciation processes across the topographic and edaphic mosaics that characterise the Transitional Rainfall Province and South Coastal Province of Western Australia are hypothesised to have been driven, in part, by late Tertiary–Quaternary climatic oscillations and resultant episodes of population fragmentation and genetic isolation (Hopper 1979; Coates et al. 2003; Hopper and Gioia 2004). This has led to both old relictual taxa and relatively ancient fragmented population systems within some species complexes (e.g. Byrne et al. 2001; Coates et al. 2003). For these taxa, a secondary consequence of the climatic and habitat instability that forged much of the diversity of the South-west Australian Floristic Region (Hopper and Gioia 2004) has been extinction throughout much of their range, such that they now persist in geographically restricted and fragmented or disjunct populations (Coates et al. 2003). The relationships identified here in the Koolyanobbing Tetratheca show similar combinations of relict species diverged over long time frames and more recently evolved taxa.
The disjunct ironstone ranges of the Koolyanobbing area are, in effect, islands (Fig. 4) and as such are subject to many of the processes recognised in classical island biogeography, including the difficulty of long distance dispersal, post-establishment isolation and adaptation into, and exploitation of, ecological opportunities in vacant niches (Whittaker 1998). Unlike the majority of oceanic islands, which are relatively recently derived, these Goldfield ironstone ranges are ancient geological relicts in the semi-arid landscape, and are more similar to old continental islands on which long-scale isolation has led to the evolution of a highly endemic flora with obvious relictual elements. The high levels of endemism and complex vegetation patterning seen in the Helena and Aurora Range, Jackson Range, ‘Windarling Range’ and the Die Hardy Range are in contrast with nearby areas such as the Highclere Hills, which are characterised by a more subdued topography of Archaean granites weathered into undulating plains and broad valleys, and across which no species have been recorded as endemic (Gibson and Lyons 2001). Although historically isolated, the current patterns of endemism displayed among these Koolyanobbing area Tetratheca species seem to be a combination of habitat niche specificity and low dispersal ability.
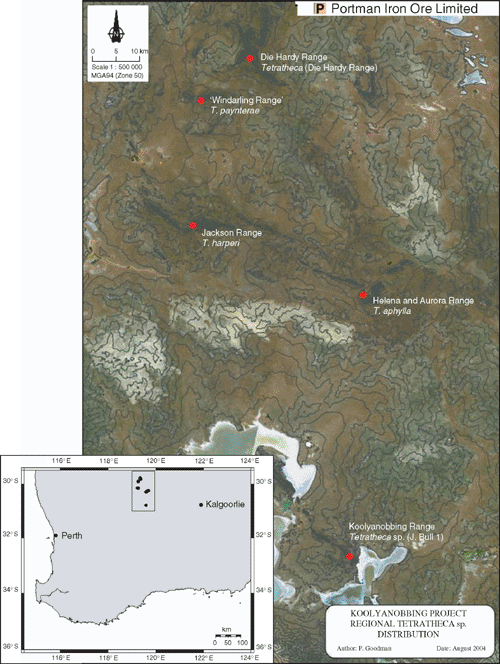
|
The exposed rock faces and scree slopes occupied by these Tetratheca are inhospitable, but there are few other taxa that share this niche and therefore little competition for establishment and success. Survey for plants of T. paynterae has determined that while the subspecies occur on ranges c. 10 km apart, surveys have not located plants of Tetratheca on nearby Mt Geraldine, or the Yokradine Hills, which lie adjacent and more-or-less parallel to the Die Hardy Range (Fig. 4). Field investigation has determined that the habitat preferred by these regionally endemic Tetratheca species is not available on Mt Geraldine, which comprises relatively uniform slopes and an even covering of shrubland and scrub, and that small areas of potentially suitable habitat exist on the western side of the Yokradine Hills. The correlation between low dispersal ability and endemism has been well documented for island plants (Whittaker 1998) and in the case of these Tetratheca, it would appear that the prominent elaiosome on the seeds ensures that ants transport the seeds into rock fissures, and that this usually occurs in close proximity to the parent plant.
Within the Die Hardy Range, the Tetratheca subpopulations are located on the exposed cliff faces comprising the eastern face of the main ridge and the western and south-eastern faces of smaller ridges, separated from the main ridge by moderately broad, shallow valleys. The subpopulations are likely to have been established on these inward facing cliffs through short-distance abiotic or biotic means. Although there was no morphological variation between plants from all three subpopulations (Butcher et al. 2002), a preliminary study of genetic diversity conducted in 2002 noted a low level of differentiation between two of the subpopulations (S. Krauss, pers. comm.) and it is possible that the topographic position of these subpopulations may act as a barrier to gene flow. As such, the existence of potentially suitable but vacant habitat on the opposite side of the Yokradine Hills to the population of Tetratheca in the Die Hardy Ranges supports the hypothesis that seed dispersal is the limiting factor to recruitment.
Acknowledgements
RB thanks Jerome Bull for his enthusiasm in the field, Matthew Williams for his assistance with morphometric analyses, Jenny Chappill (dec.), Marco Duretto, Neil Gibson, Terry Macfarlane and Juliet Wege for taxonomic discussions, Bronwyn Macdonald and Paul Rymer for laboratory assistance, Siegy Krauss for preliminary genetic results, Erica Alecs, Daniel Sievers and Bob Dixon for field assistance and Dot and Ron Budge, and Joy and Herb (dec.) Budge for their hospitality during field work. The Department of Clinical Immunology at Royal Perth Hospital and the DNA Analysis Facility at University of New South Wales performed the sequence electrophoresis. Funding from Portman Iron Ore Pty Ltd (to MB), the Australian Biological Resources Study (to DMC) and the Hermon Slade Foundation (to DMC) is acknowledged. Piers Goodman (Portman Iron Ore Pty Ltd) is thanked for the provision of the Koolyanobbing region Tetratheca distribution map and for facilitating financial support for the publication of colour images.
Alford JJ
(1995) Two species of Tetratheca (Tremandraceae), from the Coolgardie and Austin Botanical Districts, Western Australia. Nuytsia 10, 143–149.
[Verified 30 January 2007]
Yoder AD,
Irwin JA, Payseur BA
(2001) Failure of the ILD to determine data combinability for slow loris phylogeny. Systematic Biology 50, 408–424.
| Crossref | GoogleScholarGoogle Scholar | PubMed |



