Microbial co-occurrence networks as a biomonitoring tool for aquatic environments: a review
Annachiara Codello A B * , Grant C. Hose
A B * , Grant C. Hose  A and Anthony Chariton
A and Anthony Chariton  A
A
A The Environmental (e)DNA and Biomonitoring Lab, School of Natural Sciences, Wallumattagal (North Ryde) Campus, Macquarie University, Darug Nation, NSW 2113, Australia.
B Corresponding author. Email: annachiara.codello@gmail.com
Marine and Freshwater Research - https://doi.org/10.1071/MF22045
Submitted: 16 February 2022 Accepted: 25 February 2022 Published online: 16 November 2022
Journal Compilation © CSIRO 2022 Open Access CC BY-NC-ND
Abstract
Aquatic microbial ecosystems are increasingly under threat from human activities, highlighting the need to for the development and application of biomonitoring tools that can identify anthropogenically induced stress across a wide range of environments. To date, microbial biomonitoring has generally focussed on community composition and univariate endpoints, which do not provide discrete information about how species both interact with each other and as a collective. To address this, co-occurrence networks are being increasingly used to complement traditional community metrics. Co-occurrence network analysis is a quantitative analytical tool that examines the interactions between nodes (e.g. taxa) and their strengths. This information can be integrated and visualised as a network, whose characteristics and topological structures can be quantified. To date, co-occurrence network analysis has rarely been applied to aquatic systems. Here we explore the potential of co-occurrence networks as a biomonitoring tool in aquatic environments, demonstrating its capacity to provide a more comprehensive view of how microbial, notably bacterial, communities may be altered by human activities. We examine the key attributes of networks and providence evidence of how these may change as a response to disturbances while also highlighting some of the challenges associated with making the approach routine.
Introduction
Healthy aquatic ecosystems are pivotal for supporting life on Earth, and of immense social, economic, cultural and environmental value. However, the health of aquatic ecosystems is being put at risk, due to the direct and indirect effects of human activities (Halpern et al. 2008). With the growth of the human population and the expansion of cities globally, pollution, alterations to land use, climate change, overexploitation and invasive species are having an increasingly pronounced detrimental effect on aquatic systems (Friedman et al. 2020). Individually, and in concert, these anthropogenic stressors are placing unprecedented pressure on all aquatic ecosystems, eroding biodiversity and altering ecosystem functions and services at an unparalleled scale and pace (Steffen et al. 2011). Consequently, a deeper understanding of the potential impacts and extent of human activities on aquatic ecosystems is pertinently needed to better inform management actions that are capable of both protecting essential water resources, while also supporting the growth and expansion of the human population.
Freshwater, estuarine and marine ecosystems are complex, heterogeneous, and dynamic environments (Heino et al. 2015). They are host to a great variety of species, with the majority of their biomass, up to 90% (Costello et al. 2010; Snelgrove 2010) being composed of microorganisms. Microorganisms, most notably prokaryotes, provide invaluable functions including carbon and nitrogen fixation and the remineralisation of organic matter, as well as forming the basis of oceanic food webs (Jeffries et al. 2016). The global carbon and nutrient cycles are thus dependent on the activity of these microorganisms (Azam and Malfatti 2007). As such, human induced changes to aquatic microbiota can have secondary effects on productivity, food-web structure and energy flow, and carbon export (Sagova-Mareckova et al. 2021). Moreover, the microbiota of these systems are highly responsive to local and global pressures, because of their high metabolic and growth rates (Allison and Martiny 2008). Consequently, the microbiota can be highly influenced by the input of both organic and inorganic pollutants (Holman et al. 2021; Sagova-Mareckova et al. 2021). For example, it has been found that in an aquatic mesocosm, bacterial community composition, structure and function can be markedly altered by increasing concentrations of copper (Sutcliffe et al. 2019; Codello 2021).
Microbial responses to stress, such as changes diversity, composition and structure can be used to detect disturbances in water ecosystems (Williams et al. 2014). Although the use of aquatic biota as indicators of disturbance and ecological condition is an integral part of water quality and sediment monitoring (Sun et al. 2012), to date, they are very eukaryotic centric, with the routine monitoring of microorganisms, including bacteria, archaea and protists, rarely employed (Bohan et al. 2017; Sagova-Mareckova et al. 2021).
Indicators based on microorganisms can be selected from a broad assortment of taxonomic groups and cover a wide variety of functions and services (Aylagas et al. 2016). Microorganisms are highly sensitive and have an overall population that include both niche specialists and generalist microbes (Bell and Bell 2021). The census of aquatic microorganisms is today possible due to the development of molecular technologies like high throughput sequencing and readily available genetic databases. These advances, which have allowed the construction of effective new approaches such as the culture-free analysis of a microorganism’s genetic material (Shendure and Ji 2008) enable numerous samples of complex assemblages to be sequenced simultaneously.
Analysis of microbial genetic material with metabarcoding and amplicon sequencing techniques is accurate, fast, cost-effective and capable of analysing thousands of environmental samples at the same time, providing a better understanding of microbial communities than previous techniques. These methods can reveal information on the most abundant microbial species in aquatic environments, on rare species, recently introduced invasive species, and can report the presence of organisms that cannot otherwise be sampled or identified with more traditional techniques (Aylagas et al. 2016; Cristescu and Hebert 2018).
To date, the focus has been on composition and univariate endpoints, which does not inform how species interact with each other and as a collective. Most microbial communities are complex systems with intricate interaction patterns showing a collective behavior that cannot be identified by just looking at the single entities or composition (Cimini et al. 2019). Consequently, there is a need to use more advanced analytical methods such as co-occurrence network analysis and their properties to monitor the biological environment and provide a far more complete picture (Tylianakis et al. 2010).
In this review, we demonstrate the potential utility of the microbial community co-occurrence networks and their properties as a biomonitoring tool to assess aquatic health. To date, a number of studies has employed this approach to characterise the system-level responses to environmental change in aquatic systems, including anthropogenic disturbances (Bohan et al. 2017; Cordier et al. 2020; Di Battista et al. 2020). However, it has yet to be used to its full potential because of a lack of empirical data and conflicting views with regards to the ecological implications of any perceived changes in network structure and its associated metrics (Karimi et al. 2017; Derocles et al. 2018; Barroso-Bergada et al. 2020). To address this, firstly, we provide a summary of aquatic microbial networks, specifically co-occurrence networks, and their main structural components, such as nodes and edges. Secondly, we describe network properties, and where possible, what ecological information can be drawn from these networks. Finally, we illustrate the use of co-occurrence network as a biomonitoring tool by providing an overview of existing studies, while articulating the potential advantages and pitfalls associated with applying co-occurrence networks for biomonitoring in aquatic systems.
Co-occurrence networks
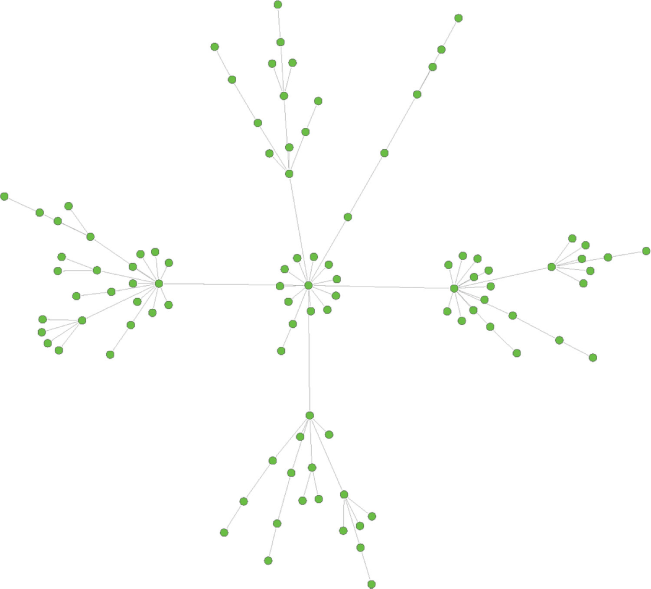
The theory that underpins ecological co-occurrence analysis originates from Jared Diamond’s seminal work on the assembly rules of ecological communities, which predicted that competitive interactions between bird communities in New Guinea would result in non-random co-occurrence patterns (Diamond 1975). The bird population was found to follow a ‘checkerboard’ distribution representing one of the first pieces of evidence that competition can play a key role in determining which species are found within a community. A network is a representation of a community consisting of a multitude of parts, also called nodes, which are connected by a binary interaction that allows exchanges or communications between the parts (Newman 2010). This concept has been used by many fields with myriad applications, ranging from social networks (e.g. Facebook), the World Wide Web to electrical grids. Some of the most common types of biological networks are protein–protein interaction networks, metabolic networks, genetic interaction networks, gene and transcriptional regulatory networks, cell signaling networks, food webs, and networks of interactions between species (co-occurrence) (Newman 2010). Although the biological applications are broad and capture a wide range of systems and processes, they often have a common characteristic, being scale-free networks, also called real-world networks, which inherently contain a relatively low number of nodes that are highly connected to other nodes (Fig. 1) (Bianconi and Barabási 2011).
|
Biological networks are generally compared to a random network, where it is rare to find nodes that have significantly more or fewer connections than the average node (Barabasi and Bonabeau 2003). Most biological complex systems have a similar architecture and similar organisational principles. Consequently, the scale-free structure has important implications, for example, in controlling the spread of viral diseases such as HIV and COVID-19 (Barabasi and Bonabeau 2003; Murali et al. 2011; Pal et al. 2020). Similarly, the interaction between microorganisms have been shown to have scale-free distribution in many freshwater and marine environments (Ruan et al. 2006; Steele et al. 2011; Kara et al. 2013).
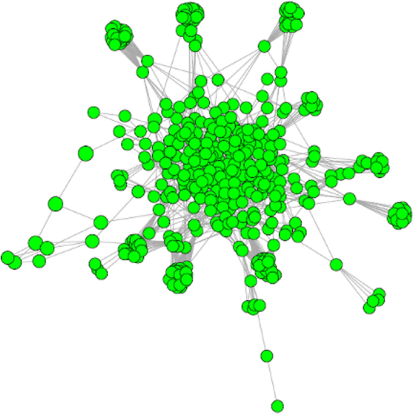
Although there are numerous types of networks, here we focus on co-occurrence networks. Co-occurrence networks are derived from the presence or abundance and interactions of taxa occurrences within a repeatedly sampled unit e.g. multiple samples from a site. Co-occurrence networks are founded on the interactions between taxa derived from significant bipartite relationships. The two fundamental components are nodes and edges. Nodes represent taxa that have a significant interaction with other taxa, and edges represent the co-occurrences bipartite interactions between taxa, allowing for the possibility to represent and study the interactions of the whole microbial community (Fig. 2). A microbial network co-occurrence analysis allows a holistic study of the environment that not only focuses on presence or absence and composition of organisms in the ecosystem but also on how they interact (Layeghifard et al. 2018).
|
A holistic methodology can facilitate our understanding of the complex interactions that occur within biological systems. This is evident in the case of marine bacterioplankton, where ten different ‘ecological species’ of SAR11, phylogenetically similar, have been found due to the unique co-occurrence network relationship between them, other organisms and environmental parameters (Fuhrman and Steele 2008). For a global biomonitoring approach, networks can provide a comprehensive understanding of how ecosystems function across systems and biogeographical regions (Raffaelli 2007; Bohan et al. 2017). Thus, it has been suggested that inclusion of information that examines the interactions between species can greatly benefit our understanding of community ecology, and how communities respond to change (Valiente-Banuet et al. 2015).
An interaction between bacterial species involves three main exchanges: food transfer, information transfer, and gene transfer (Fuhrman 2009). Food transfer in microorganisms creates a food web that is at the centre of the marine ecosystem, allowing the exchange of energy and biochemical compounds (Steele et al. 2011). Information transfer also occurs in microbes since they can receive information cues from the external environment (such as a chemical signal from other microbes) and are able to respond (Westerhoff et al. 2014). Gene transfer can happen both in the form of gamete exchange and lateral gene transfer, e.g. resistance cassettes and antibiotic resistance genes (Gillings et al. 2008; Fuhrman 2009).
Nodes
Nodes represent the first fundamental component of a network. Nodes, are the objects at the start and ends of the connection, and are commonly represented as Operational Taxonomic Units (OTUs), Amplicon Sequence Variants (ASVs), taxa, species, genera, families, classes, phyla or kingdoms, or functional groups (Karimi et al. 2017). The role of the node in relation to the rest of the network allows us to understand the dynamics of the system. ‘Hubs’, ‘bottle necks’ and ‘keystone’ species represent the three main node roles. A ‘hub’ defines a node that has a high level of degree centrality (a high number of connecting neighbouring nodes) (Paine 1995; Delmas et al. 2019), whereas a ‘bottleneck’ is a node that has a fundamental role in connecting other nodes (Peura et al. 2015). Usually, ‘bottlenecks’ are between nodes and get crossed by many pathways inside the network; they have a high betweenness centrality and high transitivity (Peura et al. 2015; Delmas et al. 2019). Common marine and freshwater taxa are typically ‘hub’ and or ‘bottleneck’ species likely due to their ability to adapt to environmental change (Peura et al. 2015; Lin et al. 2019). Species that have a large effect on the community are called ‘keystone’ species, they are less abundant than other organisms in the community, but the other nodes depend on their effect (Power and Scott Mills 1995; Berry and Widder 2014). These keystone species play a critical role in the stability of the system (Peura et al. 2015). Apex predators are an example of a keystone species in macro-ecology, their predatory behaviour assists in the control of the population size of their prey (Berry and Widder 2014). ‘Keystone’ species can be ‘hubs’ but ‘hubs’ are not always ‘keystone’ species. A ‘hub’ species is one that when eliminated can be substituted by another competitor species occupying the same niche with the same or similar function (Berry and Widder 2014; Delmas et al. 2019). However, when a ‘keystone’ species is eliminated, the system undergoes significant redistribution and potential collapse (Valiente-Banuet et al. 2015). It has been hypothesised that in a fragile network, the removal of 25% or less of the highly connected nodes should result in collapse, whereas in a robust and stable network the removal of half of the nodes would not change the network topology (Estrada 2007).
Collectively, rare organisms can influence important biogeochemical processes, thus, in some instances they can also be considered ‘keystone’ species. Although rare taxa may also be important, it should be emphasised that in most cases these taxa do not become nodes as they are only found in a small proportion of the samples used to create the network, and are therefore removed during the initial filtering period, or fail to have robust statistical relationships with node, they must be other taxa (Fuhrman 2009).
Environmental factors can also be incorporated in the network structure as a type of node where the edge exemplifies the effect of the factor on the microbial species node (Li et al. 2018). In seawater samples, a strong archaeal–bacterial association was found due to environmental pressure (Parada and Fuhrman 2017), whereas in thawed ponds and lakes, the importance of environmental factors was highlighted by dissolved organic carbon and conductivity being the most connected nodes (Comte et al. 2016). The inclusion of environmental factors is a powerful characteristic of network analysis that can help to describe ecological niches and discern what factors have the greatest influence over the microbiome (Chaffron et al. 2010; Gao et al. 2019; Mikhailov et al. 2019).
Interactions
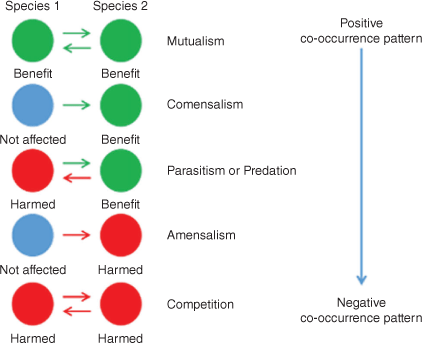
The second fundamental component of networks are edges, which represent interactions between nodes. These can be broadly categorised in accordance with their co-occurrence interactions (see Fig. 3): positive interactions, combination of positive or negative with neutral interactions and negative interactions (Lidicker 1979). Theoretically, positive interactions suggest commensalism and mutualism (Bronstein 2009). Commensalism describes a cooperation between microbes where only one of the two gains an advantage and the second is not affected at all (Lidicker 1979). However, mutualism describes a cooperation where both organisms benefit from the interaction. For example, the bacteria Bacteroides thetaiotaomicron is found in the human distal intestinal microbiome, where it assists in the breakdown of indigestible polysaccharides, making them bioavailable to humans (Bäckhed et al. 2005). In this example, The B. thetaiotaomicron benefits by obtaining its energy from the polysaccharides, whereas the host (human), mutually benefits as the polysaccharides are now converted to digestible form. Mutualism can also be symbiotic, i.e. when both microorganisms are required to survive (Bronstein 2009). Organisms can also have mutual relationships that are syntrophic, that is, when the metabolism of the two organisms is complementary (Barton and Northup 2011). For example, in marine gas–hydrate-rich sediments with high rates of methane-based sulfate reduction, Archaea grow in highly populated conglomerates and are surrounded by sulfate-reducing bacteria, thereby creating a syntrophic consortium that mediates methane oxidation (Boetius et al. 2000).
|
Positive–negative interactions occur when one taxon gains an advantage and the partner is negatively affected, e.g. predation and parasitism (Barton and Northup 2011). In predator–prey interactions, the predator improves its condition by altering the condition of the prey (Lidicker 1979). For example, in marine sediments, Pseudoalteromonas was found to prey on Gram-positive bacteria by secreting pseudoalterin. Pseudoalterin binds to and degrades the glycan strands of Gram-positive bacterial peptidoglycan. This causes the cell to die and release nutrients used for the growth of the predator (Tang et al. 2020). Parasitism represents a host–parasite interaction where the parasite is physically associated with the host and it is in the privileged position to gain an advantage at the expense of the host (Lidicker 1979). In marine environments, abundant viruses can infect cyanobacteria such as Synechococcus sp. Electron microscopy analysis showed that 1–3% of the native cyanobacteria contained viruses close to the end of the lytic cycle. It seems that cyanophages are probably responsible for the death of up to 3% day−1 of Synechococcus in temperate Atlantic waters (Fuhrman 1999). Finally, negative interactions suggest amensalism or competition (Barton and Northup 2011).
Amensalism is characterised by an interaction where one organism is affected by the presence of the other, but the second does not gain any advantage (García et al. 2017). This type of interaction has been observed in the fermentation of residual cheese whey by the Pseudomonas taetrolens and Lactobacillus casei collaboration. During their interaction there was no variation in Lactobacillus, whereas the Pseudomonas community had reduced growth (García et al. 2017). Within the co-occurrence context, competition is an interaction where both nodes are adversely affected, for example, when they directly compete for the same resources (Barton and Northup 2011). An interesting example of bacterial competition was studied between Pseudomonas sp. and Scenedesmus quadricauda, which competed for inorganic phosphorus under laboratory conditions. The dynamics between the two bacteria suggested that, at a steady state with sufficient carbon, Pseudomonas sp. reduced phosphorus below the level required by S. quadricauda, whereas when Pseudomonas sp. did not have enough available carbon S. quadricauda prevailed (Grover 2000).
Although the interpretations of statistically derived interactions observed in co-occurrence networks are driven by ecological theory, in reality, biological relationships between interactions and co-occurrences are often ambiguous (Blanchet et al. 2020). This is particularly true for aquatic microbial communities, where some species are naturally rare or transient, and therefore only interact under specific situations (Blanchet et al. 2020). Furthermore, macro-ecological networks have shown that co-occurrences reflect more niche preferences rather than biotic interactions (Freilich et al. 2018). It is safe to assume that bacteria can reside in the same habitat without interacting so, inference based on network associations, in some instances, may result in erroneous results (Röttjers and Faust 2018). Another issue is that, although the gradient of diversity is continuous, the categories we are describing are discrete. Specifically, it is challenging to discern between parasitism and predation, as well as understanding the difference between mutualism and commensalism, or between amensalism and competition. This is because it is difficult to capture the positive gradient between mutualism and commensalism and the negative gradient between amensalism and competition (Röttjers and Faust 2018). Originally, most networks were inferred from presence–absence data; however, now abundance (count) data provide more information to refine the inferences (Blanchet et al. 2020).
In networks based on metabarcoding data, edges represent the correlation value between OTUs or ASVs. The network edges can be calculated using a wide range of methods from simple correlations that are quick to calculate, to more complex approaches, which require higher computational power. The simplest methods are based on dissimilarities, such as Bray–Curtis and Kullback–Leibler dissimilarity indices, and correlations, such as Pearson and Spearman correlation coefficients (Layeghifard et al. 2017). Multiple regression and probabilistic-based methods, such as Bayesian networks or Markov networks, can also be used to capture more complex interactions (Layeghifard et al. 2017). One of the great challenges, especially when using NGS-derived bacterial data, is addressing biases associated with composition. There are a number of packages specifically designed to minimise the artefacts of such data, including SparCC, MIC, SPIEC-EASI, CoNet and others (Faust and Raes 2012; Layeghifard et al. 2017). An additional important component of network analysis is the capacity to visualise the data. Again, there are numerous approaches, including Cytoscape (Shannon et al. 2003) and iGraph (Csardi and Nepusz 2006), as well other functions within R. The choice of approaches used to produce and analyse a network may influence the overall metrics produced by a network and subsequent interpretation. This is highlighted in Codello (2021), where the same system was examined using SPIEC-EASI and CoNet. Although the findings were not conflicting, they highlighted that the metrics produced from the two approaches varied in their sensitivity to detect stress.
Network structure
In the aquatic environment, environmental factors play a key role in shaping microbial communities (Mikhailov et al. 2019). These include water stratification (Zaccone and Caruso 2019), different levels of light penetration (Parada and Fuhrman 2017), temperature gradient, seasonal turnover (Gilbert et al. 2012; Mikhailov et al. 2019), spatial heterogeneity (Fuhrman 2009), and disturbances present in the water (Liu et al. 2015), which can change the structure or topology of the network. For example, in eutrophic freshwater Lake Taihu, water temperature, chlorophyll a and phytoplankton density were the main factors driving the variation of bacterial community composition and structure between seasons (Zhu et al. 2019). Co-occurrence networks formed larger networks with more nodes in spring and summer than in autumn and winter. Other studies confirmed that warmer seasons support the activity of more bacteria than colder months in water systems (Crump and Hobbie 2005; Gilbert et al. 2012; Kara et al. 2013). However, after the summer network, the autumn network had the largest number of edges, suggesting that the microbial community transition from warmer temperature to colder temperature stimulates the creation of new interactions, be it positive or negative (Zhu et al. 2019). Consequently, the overall structure and topology of networks may provide insight into how a community changes.
Environmental networks are modular but contain a hierarchical structure. At the lowest level are individual nodes. A group of nodes can form small subgraphs called motifs, together motifs create modules and finally modules form a complete network (Mason and Verwoerd 2008). It has been suggested that this hierarchical structure could be the result of co-evolution, natural selection, habitat heterogeneity, the specificity of interaction and phylogenetic relatedness (Deng et al. 2012). For example, Liao et al. (2019) found co-occurrence networks derived from sediments within the tidal zone produced distinctive modular structures that could visually differentiate them from non-tidal and inter-tidal networks. Hence it has been suggested that the structural modularity of a network may also reflect the stability and resilience of the system (Pavlopoulos et al. 2011; Deng et al. 2012; Delmas et al. 2019; Di Battista et al. 2020). In planktonic prokaryotic community profiles from the Taihu Watershed in China, the co-occurrence network was grouped into four major ecological modules where the OTU nodes had a robust co-occurrence correlation (Liu et al. 2020a). The different modules performed various functions, for example, the first module was attributed to have four functions: methanotrophy, aerobic ammonia oxidation, nitrification and ureolysis; the second module had functional groups related to metabolism and human health; and the third module had an association with chlorate reduction, methanol oxidation and methylotrophy. Hereby, clearly demonstrating that in this case, functional groups were linked by ecological associations (Liu et al. 2020a). The analysis of microbial networks can be coupled with specific functions, which allows us to have a general understanding of the microbial phylogenetic distribution, their correlations, and their collective functions (Bohan et al. 2017). The analysis can be integrated with genetic, biochemical, and chemical functions (Bowen et al. 2013). Genes with known functions can profoundly influence the structure of networks, and consequently, in some incidents where the function of a gene is unknown, its role might be revealed by the taxa it interacts with (Galand et al. 2018). For example, if a taxon is highly connected with a group of nitrogen-fixing microorganisms, there is a high probability that it too will be a nitrogen fixer, thus creating a direct link between the taxon’s role in a network and their functional role (Bohan et al. 2017). Hence, determining the network structure of co-occurrence interactions can help define ecological niches and characterise microbial ecotypes.
Network properties
Mathematical approaches are not only used for network graphical representations, but to understand the behaviour of networks, enabling the properties of a network to be summarised using a range of metrics (Proulx et al. 2005). Network properties are features that can provide information on the biological system at the base of the network. There are specific properties to describe and measure network topologies, and can be used to reveal patterns, and inform the structure of the microbial co-occurrence network within aquatic ecosystems (Steele et al. 2011; Röttjers and Faust 2018). The subsequent section provides an overview of some of the key network properties, and how these properties specifically relate to microbial communities, and where possible, in response to disturbance. The basic properties that can describe the whole network and have potential to be applied within a biomonitoring framework include: the number of nodes, number of edges, number of components, diameter or radius, degree distribution, average path length, density, cluster coefficient and centrality measures.
The number of nodes and number of edges measures the total number of nodes and the total number of connections between nodes respectively. In the lake sediment bacterial community during a toxic bloom event caused by untreated sewage, algal bloom and aquaculture, the network structure increased its dimensions. Before the bloom, the network had 100 nodes and 1851 edges, compared to 200 nodes and 3193 edges after the bloom. This expansion highlighted the interference of the toxic bloom on the bacterial community composition (Zhou et al. 2021). It is possible that at first the bacterial community formed a structure that was disrupted by the toxic bloom. The toxic event and the proliferation of the algal bloom increased the microbial complexity with more nodes and increased the number of negative interactions between the bacterial community and the algal bloom (Zhou et al. 2021).
The number of components is the number of modules or groups of nodes that are connected between each other but not to other nodes of the same network, i.e. there are no possible paths between nodes from different components (Newman 2010). In essence, it is a measure of completeness. When there is the fragmentation, former relatively larger networks become dissociated into smaller disconnected networks, resulting in an increase in the number of components (Pavlopoulos et al. 2011). This phenomenon was seen in a river system where the number of components increased downstream compared to upstream because of hydrological changes and metacommunity dynamics (Widder et al. 2014). In this example, it was hypothesised that the elevated physical disturbance related to flow patterns and sedimentary dynamics in the downstream river, caused the loss of keystone taxa, thereby increasing fragmentation in downstream sites (Widder et al. 2014).
The diameter or radius is a measure of the dimension of the network and it is the longest of all the path lengths. It measures how quickly a community can respond to a stimulus (Pavlopoulos et al. 2011). The network constructed with river bacterial communities data changed diameter length according to seasons: spring had the shortest diameter, describing a very efficient and quick to respond community; and autumn had the longest diameter, indicating a slowing down of community collaborations (Lin et al. 2019). These changes suggest that seasonality interferes with the structure of the bacterial network community and with the efficiency to collaborate (Lin et al. 2019).
The average path length (Nij) (Eqn 1) of a pair of nodes is the average number of edges (E) that have to be crossed between two nodes (i, j) to have the shortest distance (dij) (Newman 2010).
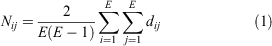
The average path length is another measure of the expediency of the network reaction to a change in the environment or to a change in the node composition (Newman 2010). The length informs how fast two nodes connect and pass information between them, a short path means that the system is ready to react (Mason and Verwoerd 2008). In network experiments from five aquatic time series in dystrophic and eutrophic lakes and open ocean sites, the average path lengths between nodes were shorter in empirical than in random networks (Peura et al. 2015; Liu et al. 2020b). This suggests that in aquatic environments, the average path length maybe comparatively shorter than the whole network dimension (Newman 2010).
Density (D) (Eqn 2) describes how compact or crowded the network is in relation to the number of connections per nodes, and thereby is a measure of complexity (Landi et al. 2018).

where E is the number of edges and n is the number of nodes (Newman 2010).
A sparse network has fewer interactions, lower complexity and needs less energy to be maintained, but a dense network has more interactions, higher complexity and higher energy requirements (Pavlopoulos et al. 2011). Experimental data has shown that biological networks have a lower density than random networks. In the ‘Domino hypothesis’ a less dense network is considered to be more resilient and efficient, with lower costs to maintain the system than a dense network (Leclerc 2008). By contrast, the ‘Insurance hypothesis’ suggests that a denser network has more redundant nodes, which could make the network more resistant to change (Yachi and Loreau 1999). In benthic microbial networks produced from affected offshore oil and gas drilling sites, the density was higher in non-affected sites than those closer to the drilling operations (Laroche et al. 2018). This suggests that there was a reduced complexity in the microbial co-occurrence network when the environment was disturbed by human activities and small spills of oils (Laroche et al. 2018). These two hypotheses highlight the contrary viewpoints when extrapolating the ecological relevance of variations in some network properties.
The cluster coefficient (Cl) (Eqn 3) represents the predisposition to form clusters or modules (Newman 2010). In a graph, a cluster is formed when one or more nodes have a high number of edges E and a low node degree k.

Clusters are common in natural environment networks more than in random networks (Newman 2010). There are network subsets in which species frequently interact between themselves; however, the subset interacts infrequently with species outside of the cluster (Pavlopoulos et al. 2011). A study in agricultural soils, where a single application of alkaline stabilised biosolid (used to destroy pathogens) was applied 10 years prior to sampling, found a 39% increase in the clustering coefficient, in comparison to the control soils (Price et al. 2021). This suggested that the agricultural additive (alkaline-stabilised biosolid) had a strong legacy effect on the soils and on the network structure increasing its complexity.
Centrality measures are properties that can quantitatively describe microbial interactions and identify the most important taxa and offers suggestion on the role or function of the taxa in relation to the rest of the network (Zamkovaya et al. 2021). They are: degree centrality, closeness centrality, betweenness centrality, eigenvector centrality and transitivity (Girvan and Newman 2002). They can be studied as mean values at the level of the full network, or singularly for each node.
The node degree or degree centrality (ki) (Eqn 4) is the number of edges or connections that a node (i) has with other nodes, where n is the total number of nodes (Newman 2010)

A node with high degree has many connections to other nodes, with a lower degree, inferring a sparsely connected node (Pavlopoulos et al. 2011). Owing to the characteristic distribution of scale-free networks, most microbial communities have many nodes with few links and few nodes having a large number of links, also called ‘hubs’, making the network possibly more resistant to random failure, but also vulnerable to organised attacks to the key nodes (Barabasi and Bonabeau 2003). An example of degree centrality in aquatic sediment environment is represented in Fig. 4.
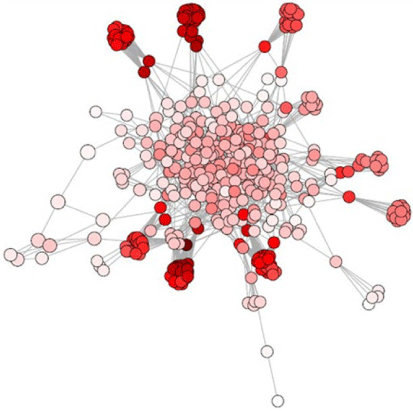
|
Closeness centrality (Ci) (Eqn 5) describes how close a node is to other nodes. It is defined as:

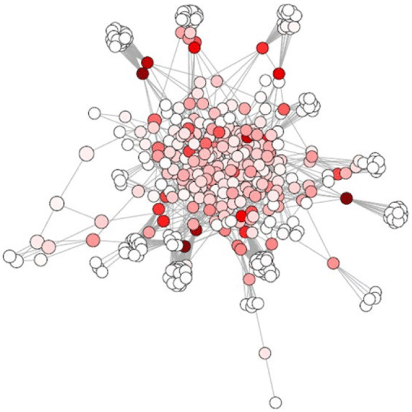
where n is the total number of nodes and dij the shortest distance between i and j (Pavlopoulos et al. 2011). A node has a high closeness when it can communicate quickly with another node (Newman 2010). A key player in a network has high closeness centrality, because it has to quickly interact with many network components (Pavlopoulos et al. 2011). An example of closeness centrality in an aquatic sediment network is illustrated in Fig. 5.

|
Betweenness centrality (xi) (Eqn 6) is the degree of an individual node that lies on a path between other nodes. It is defined as:

where nist is 1 if the node i lies on the shortest path from s to t and 0 if it does not (Pavlopoulos et al. 2011). Nodes with high betweenness connect nodes in the network that otherwise would not be connected (Newman 2010). Between centrality identifies nodes that are intermediates between neighbours and belong to many network pathways (Pavlopoulos et al. 2011). For example, species that act as a bottleneck have high betweenness. An example of betweenness centrality in aquatic sediment environment is represented in Fig. 6.
|
Eigenvector centrality rank (xi) (Eqn 7) measures the importance of the neighbouring connection. In some instances, a node’s importance is increased by the connection to other nodes that are important themselves (Newman 2010). It is defined as:

where xi is the centrality with i nodes and the adjacent matrix Aij (Newman 2010).
Katz centrality and Page rank are also specific types of eigenvector centrality. They are relevant for identifying key populations and a higher order of organisation (Deng et al. 2012). Not all nodes have the same importance, a high eigenvector centrality node does not have to be highly linked, and it can have less connections with more important nodes (Newman 2010).
The transitivity (T) (Eqn 8) is the probability that two nodes are connected directly or indirectly.

where li is the number of links of node i and m is the number of neighbours of i (Newman 2010). For example, hub nodes have high transitivity because of their many connections (Girvan and Newman 2002).
Collectively, these centrality measures are important components of network structure and integrity (Banerjee et al. 2018). For example, in the co-occurrence networks from a large eutrophic Lake Taihu (China), rare taxa, although being low in abundance, showed relatively higher values of degree centrality, closeness centrality and eigenvector centrality than abundant taxa (Zhou et al. 2021). Consequently, rare species can have important roles in the network, supporting the idea that rare species harbour genetic variability with the metabolic potential and capacity to gain a dominant role under specific conditions (Lynch and Neufeld 2015). Also, Zhang et al. (2020) identify rare bacteria as a keystone component of the community network in biochemical processes and community assemblies presenting high degree centrality and low betweenness centrality values. The keystone taxa were Denitratisoma, which played a role in the removal of nitrogen, Anaeromyxobacter, which affected the mobility of metal contaminants, and Candidatus Microthrix, which was involved in the removal of total nitrogen (Zhang et al. 2020).
Biomonitoring
Environmental change is reshaping biodiversity and the provision of ecosystem processes and services globally. The use of metabarcoding and sequencing technologies provides a new generation of biomonitoring tools, which not only capture microbial communities, but also their interactions by network-based approaches (Bohan et al. 2017; Cordier et al. 2020). Co-occurrence networks and their properties can offer new insights into ecosystem degradation and could characterise the system-level responses of environmental change, including pollution, land use, climate change, overexploitation and species invasions (Bohan et al. 2017). An added advantage of using networks is that they can be independent of taxonomy (Makiola et al. 2020), which may be particularly useful for biomonitoring in novel environments where taxonomy of biota is poorly known.
The microbial co-occurrence network is a dynamic system that changes over time and space creating or losing interactions due to abiotic and biotic pressures such as oil spills (Laroche et al. 2018), reductions in the availability of water (Hernandez et al. 2021), and longitudinal changes in rivers (Widder et al. 2014). As described above, specific network properties can indicate different responses in communities and are thus valuable for detecting and potentially revealing causality in a biomonitoring context.
The type of interactions in a network can provide information on the nature of changes in a microbial community (Liu et al. 2020c). Network properties revealed a change in soil microbiome interactions due to the water availability (Hernandez et al. 2021). Specifically, positive associations between taxa dominated the high-stress environments whereas the negative associations dominated in the lower stressed environments (Hernandez et al. 2021).
One of the most obvious changes in network structure is fragmentation. Fragmentation occurs when a single large, well-connected network degenerates to several smaller, weakly connected networks (Delmas et al. 2019). This phenomenon can be caused by anthropogenic contamination and ecological conditions. In rivers, a gradient of contamination had a gradual effect on the microbiome reflected in the disappearance of organisms (Li et al. 2018). These changes were evident in the network structure as loss of connections between microorganisms and reduction of path length. When the average path length of a network is reduced, it has been proposed that the perturbation can more rapidly spread through the network, increasing the suspectability of the network to fragmentation (Laroche et al. 2018).
Fragmentation can also occur in the network constructed from marine biofilm (Lawes et al. 2016). The biofilm has a biological structure where microorganisms stick to each other creating an extracellular matrix, building a multispecies structure to keep beneficial partners close and share functional ability (Little et al. 2008). General microbial survival rules can change when the microbes are involved in a biofilm community and they could also influence the bacterial interaction structure (Marti et al. 2013). In a study on a young marine biofilm co-occurrence network representation, composition and connectivity changed with the availability of nutrients (Lawes et al. 2016). The biofilm with ambient nutrients showed stronger connectivity, whereas the biofilm with enriched nutrients showed a fractured co-occurrence pattern (Lawes et al. 2016). These findings demonstrated a change in how the biofilm used the local resources, from a more collaborative state, where the microbiome used the energetic allocations from within the core microbiome efficiently using local resources; to a fracture network to avoid the unfavourable conditions (exposure to enriched nutrients) and did not exploit local resources to obtain energy (Lawes et al. 2016).
To be able to understand changes in the network topology it is important to use more than one property at a time. For example, Li et al. (2019) and Wu et al. (2019) used the number of nodes, number of edges, density, degree and modularity together to signal a decrease in bacterial community complexity in response to increasing pollution. Wu et al. (2019) showed that the networks of communities at the more polluted sites in the Jinchuan River (China) had the smallest edges to node ratio, cluster coefficient, average degree and density, which highlighted the reduction in complexity relative to the least and moderate polluted sites. Similarly, in Lake Taihu (China) networks from the least nutrient enriched system contained the highest number of nodes and edges. (Li et al. 2019). However, as in the case of Di Battista et al. (2020), Li et al. (2019) these network attributes did not respond in a dose dependent manner, with both studies observing a lower number of nodes and edges in moderately disturbed systems.
Microbial community networks are dynamic structures that can change over a period of time or along a space gradient, so comparing them can provide insight into how environmental conditions and population variation influence the structure of the microbial population (Pellissier et al. 2018). However, there are some major challenges when comparing multiple systems samples due to being a snapshot in the time and space of the community and because of the absence of a delineated start or end of the ecosystem, they are a continuum. The sample size has to be large enough to capture the entire community of interest, otherwise, the network would not represent all possible interactions and would be misleading (Faust and Shorter 1981; Tylianakis and Morris 2017). It is crucial to control for network properties co-variation during the normalisation step, to enable linear regression measurement between network properties and, to study the residual variation (Pellissier et al. 2018). Although comparing networks usually includes only comparing two relations mapped on the same population during the same period, Faust and Shorter (1981) describe a general way to compare networks using network properties when those networks differ widely in size, type of relation, species of the units, and time and space of the observations (Faust and Shorter 1981). For example, Williams et al. (2014) found that co-occurrence relationships occurring in different environments could be ecologically relevant, such as the relationship between Solirubrobacterales and Acidomicrobiales and the related families, Acidimicrobiaceae and Conexibacteraceae, that are found in soils and the human body (Williams et al. 2014).
Network analysis have an important role in risk assessment, as they allow for modelling outcomes of changes to community structure. A microbial community network is robust when its structure remains stable following a disturbance (Bissett et al. 2013). To test the robustness of a network, a node can be removed or altered to affect the network structure (Barnes et al. 2016). Generally, the removal of the most connected species, hubs, causes more secondary extinctions than a random species (Sheykhali et al. 2019). However, when the system returns to the original state after a perturbation the network is considered resilient (Mandakovic et al. 2018). For example, most denitrifiers are resistant to hydrocarbon pollution because they are insensitive to many toxicants and can adapt to various abiotic factors (Powell et al. 2006).
Challenges and pitfalls
Co-occurrence networks have great potential as a tool for biomonitoring of aquatic systems, with network properties being potential metrics of change in the underlying community. However, there is still a lack of examples of how to apply this predictive model experimentally in aquatic ecosystems. Most applications of co-occurrence networks have been field-based, where it is often difficult to separate the effect of contamination from other environmental factors, and many studies have contradictory information (Weiss et al. 2016; Hirano and Kazuhiro 2019). Consequently, there is the need for more experimental studies in which confounding factors can be controlled, and the sensitivity of communities and networks to specific stressors can be tested.
The advent of next-generation sequencing has provided an unprecedented capacity to describe microbial communities, but with the limitation that we can now identify many taxa about which we know little of their ecological roles and functions. This provides a challenge to the interpretation of co-occurrence networks because, although we infer interactions between taxa from the networks, there is little empirical evidence to inform (and confirm) the specific nature of those interactions. Further knowledge of microbial interactions under real-world conditions would enhance our ability to understand the particular network elements.
To date, there are only a few examples of co-occurrence network used to test if they respond to a stressor (Pauvert et al. 2019), but none in aquatic environments. Moreover, sample size affects co-occurrence networks (Kolaczyk et al. 2015), a range between 25 and 300 samples per network has been recommended for obtaining reliable networks (Berry and Widder 2014; Faust et al. 2015). Decreasing costs and increasing analytical power of next generation sequencing means that such levels of replication are now feasible, but this remains a potentially limiting factor because of the time and resources required to observe, collect and process the numerous samples necessary, which is well beyond that used in most current monitoring programs (Bohan et al. 2017).
Although in the last decade there has been an increase in the development of technologies, laboratory procedures and bioinformatics pipelines, the workflow should be optimised to reduce handling steps such as extraction, amplification, or sequencing bias (McGee et al. 2019). Moreover, the quality of taxonomy assigned to current OTUs or ASVs references databases varies greatly, some gene databases are well populated and have robust information, and other gene coverage is still incomplete or has incorrectly assigned sequences. Also, it is important to include unknown taxa, as seen by Zamkovaya et al. (2021) the frequent dominance of unknown taxa as hubs stresses the need for further exploration and functional characterisation of all taxa (Zamkovaya et al. 2021) and capture the full taxonomic diversity or the ecological processes being investigated (Zinger et al. 2019). However, co-occurrence network analysis can avoid these issues with taxonomy enabling the possibility to look at the system as a whole and not focusing only on specific nodes.
Conclusions
Networks and their associated properties are new types of indicators that can evaluate the quality of aquatic ecosystems and are promising environmental biomonitoring tools. Co-occurrence network analysis provides a suite of relevant measures that are frequently used in network studies, albeit in various ways, they all use more than one property at a time to understand changes in the network topology.
Two approaches have been found to be most popular. The first approach described the whole network and reveals patterns allowing comparisons between the entire networks. When looking at the whole network topology, the most used properties are the number of nodes, number of edges and the ratio between positive and negative associations that give an idea of the network dimensions. The number of components is used when comparing networks visually giving a piece of information on the integrity of the network, for example, an increased number of components is typical of fragmentation. However, characteristic path length, cluster coefficient and network density are usually used to describe community cohesion and complexity. The second approach used centrality measures such as degree centrality, betweenness centrality, closeness centrality and transitivity, to look at the role of nodes in relation to the rest of the network and allow for comparison of specific nodes, taxa, roles and functions. Moreover, centrality measures are very informative when studying a network’s robustness while removing nodes to affect the network structure. Although there are some examples of network analysis in aquatic ecosystems, they are all field based, therefore, there is still a need to validate the co-occurrence network analysis approach. Consequently, there is an urgency for more experimental studies that are designed to prove whether co-occurrence networks are indeed sensitive to particular stressors or conditions. Owing to the variety of uses, applications and outcomes obtained from previous studies, there is a further need to develop a common standardised approach with clear indication on the sample size, the number of replicates for the same environment and which program to use.
Ultimately, this field is advancing rapidly and we foresee increasing developments in the technical aspects of network analysis with experimental validation to clarify the statistical results, increase our knowledge of the aquatic microbiome and the use of network analysis as a biomonitoring tool.
Data availability
Data sharing is not applicable as no new data were generated or analysed during this study.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
Annachiara Codello received a PhD scholarship from Macquarie University. This research did not receive any specific funding.
Acknowledgements
We thank Macquarie University for PhD scholarship for Annachiara Codello’s PhD studies.
References
Allison, S. D., and Martiny, J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proceedings of the National Academy of Sciences of the United States of America 105, 11512–11519.Aylagas, E., Borja, Á., Irigoien, X., and Rodríguez-Ezpeleta, N. (2016). Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment. Frontiers in Marine Science 3, 96.
| Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment.Crossref | GoogleScholarGoogle Scholar |
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nature Reviews Microbiology 5, 782–791.
| Microbial structuring of marine ecosystems.Crossref | GoogleScholarGoogle Scholar |
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host–bacterial mutualism in the human intestine. Science 307, 1915–1920.
| Host–bacterial mutualism in the human intestine.Crossref | GoogleScholarGoogle Scholar |
Banerjee, S., Schlaeppi, K., and Van Der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nature Reviews 16, 567–576.
Barabasi, A. L., and Bonabeau, E. (2003). Scale-free network. Scientific American 288, 60–69.
Barnes, M. L., Lynham, J., Kalberg, K., and Leung, P. (2016). Social networks and environmental outcomes. Proceedings of the National Academy of Sciences of the United States of America 113, 6466–6471.
| Social networks and environmental outcomes.Crossref | GoogleScholarGoogle Scholar |
Barroso‐Bergada, D., Pauvert, C., Vallance, J., Delière, L., Bohan, D. A., Buée, M., and Vacher, C. (2020). Microbial networks inferred from environmental DNA data for biomonitoring ecosystem change: strengths and pitfalls. Molecular Ecology Resources 21, 762–780.
| Microbial networks inferred from environmental DNA data for biomonitoring ecosystem change: strengths and pitfalls.Crossref | GoogleScholarGoogle Scholar |
Barton, L. L., and Northup, D. E. (Eds) (2011). ‘Microbial Ecology.’ (Wiley.)
| Crossref |
Bell, T. H., and Bell, T. (2021). Many roads to bacterial generalism. FEMS Microbiology Ecology 97, fiaa240.
| Many roads to bacterial generalism.Crossref | GoogleScholarGoogle Scholar |
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Frontiers in Microbiology 5, 219.
| Deciphering microbial interactions and detecting keystone species with co-occurrence networks.Crossref | GoogleScholarGoogle Scholar |
Bianconi, G., and Barabási, A. L. (2001). Competition and multiscaling in evolving networks. Europhysics Letters 54, 436–442.
Bissett, A., Brown, M. V., Siciliano, S. D., and Thrall, P. H. (2013). Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecology Letters 16, 128–139.
| Microbial community responses to anthropogenically induced environmental change: towards a systems approach.Crossref | GoogleScholarGoogle Scholar |
Blanchet, F. G., Cazelles, K., and Gravel, D. (2020). Co-occurrence is not evidence of ecological interactions. Ecology Letters 23, 1050–1063.
| Co-occurrence is not evidence of ecological interactions.Crossref | GoogleScholarGoogle Scholar |
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B. B., Witte, U., and Pfannkuche, O. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626.
| A marine microbial consortium apparently mediating anaerobic oxidation of methane.Crossref | GoogleScholarGoogle Scholar |
Bohan, D. A., Vacher, C., Tamaddoni-Nezhad, A., Raybould, A., Dumbrell, A. J., and Woodward, G. (2017). Next-generation global biomonitoring: large-scale, automated reconstruction of ecological networks. Trends in Ecology and Evolution 32, 477–487.
| Next-generation global biomonitoring: large-scale, automated reconstruction of ecological networks.Crossref | GoogleScholarGoogle Scholar |
Bowen, J. L., Byrnes, J. E. K., Weisman, D., and Colaneri, C. (2013). Functional gene pyrosequencing and network analysis: an approach to examine the response of denitrifying bacteria to increased nitrogen supply in salt marsh sediments. Frontiers in Microbiology 4, 342.
| Functional gene pyrosequencing and network analysis: an approach to examine the response of denitrifying bacteria to increased nitrogen supply in salt marsh sediments.Crossref | GoogleScholarGoogle Scholar |
Bronstein, J. L. (2009). The evolution of facilitation and mutualism. Journal of Ecology 97, 1160–1170.
| The evolution of facilitation and mutualism.Crossref | GoogleScholarGoogle Scholar |
Chaffron, S., Rehrauer, H., Pernthaler, J., and Von Mering, C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Research 20, 947–959.
| A global network of coexisting microbes from environmental and whole-genome sequence data.Crossref | GoogleScholarGoogle Scholar |
Cimini, G., Squartini, T., Saracco, F., Garlaschelli, D., Gabrielli, A., and Caldarelli, G. (2019). The statistical physics of real-world networks. Nature Reviews Physics 1, 58–71.
| The statistical physics of real-world networks.Crossref | GoogleScholarGoogle Scholar |
Codello, A. (2021). Microbial approaches for monitoring the ecological condition of aquatic environments. PhD thesis, Department of Biological Sciences, Macquarie University, Sydney, NSW, Australia.
Comte, J., Lovejoy, C., Crevecoeur, S., and Vincent, W. F. (2016). Co-occurrence patterns in aquatic bacterial communities across changing permafrost landscapes. Biogeosciences 13, 175–190.
| Co-occurrence patterns in aquatic bacterial communities across changing permafrost landscapes.Crossref | GoogleScholarGoogle Scholar |
Cordier, T., Alonso-Sáez, L., Apothéloz-Perret-Gentil, L., Aylagas, E., Bohan, D. A., Bouchez, A., Chariton, A., Creer, S., Frühe, L., Keck, F., Keeley, N., Laroche, O., Leese, F., Pochon, X., Stoeck, T., Pawlowski, J., and Lanzén, A. (2020). Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Molecular Ecology 30, 2937–2958.
| Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap.Crossref | GoogleScholarGoogle Scholar |
Costello, M. J., Coll, M., Danovaro, R., Halpin, P., Ojaveer, H., and Miloslavich, P. (2010). A census of marine biodiversity knowledge, resources, and future challenges. PLoS One 5, e12110.
| A census of marine biodiversity knowledge, resources, and future challenges.Crossref | GoogleScholarGoogle Scholar |
Cristescu, M. E., and Hebert, P. D. N. (2018). Uses and misuses of environmental DNA in biodiversity science and conservation. Annual Review of Ecology, Evolution, and Systematics 49, 209–230.
| Uses and misuses of environmental DNA in biodiversity science and conservation.Crossref | GoogleScholarGoogle Scholar |
Crump, B. C., and Hobbie, J. E. (2005). Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnology and Oceanography 50, 1718–1729.
| Synchrony and seasonality in bacterioplankton communities of two temperate rivers.Crossref | GoogleScholarGoogle Scholar |
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems 1695, 1–9.
Delmas, E., Besson, M., Brice, M.-H. H., Burkle, L. A., Dalla Riva, G. V., Fortin, M.-J. J., Gravel, D., Guimarães, P. R., Hembry, D. H., Newman, E. A., Olesen, J. M., Pires, M. M., Yeakel, J. D., and Poisot, T. (2019). Analysing ecological networks of species interactions. Biological Reviews 94, 16–36.
| Analysing ecological networks of species interactions.Crossref | GoogleScholarGoogle Scholar |
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13, 113.
| Molecular ecological network analyses.Crossref | GoogleScholarGoogle Scholar |
Derocles, S. A. P., Bohan, D. A., Dumbrell, A. J., Kitson, J. J. N., Massol, F., Pauvert, C., Plantegenest, M., Vacher, C., and Evans, D. M. (2018). Biomonitoring for the 21st century: integrating next-generation sequencing into ecological network analysis. Advances in Ecological Research 58, 1–62.
| Biomonitoring for the 21st century: integrating next-generation sequencing into ecological network analysis.Crossref | GoogleScholarGoogle Scholar |
Diamond, J. M. (1975). Assembly of species communities. In ‘Ecology and Evolution of Communities’. pp. 342–444. (Harvard University Press.)
Di Battista, J. D., Reimer, J. D., Stat, M., Masucci, G. D., Biondi, P., De Brauwer, M., Wilkinson, S. P., Chariton, A. A., and Bunce, M. (2020). Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Scientific Reports 10, 8365.
| Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems.Crossref | GoogleScholarGoogle Scholar |
Estrada, E. (2007). Food webs robustness to biodiversity loss: the roles of connectance, expansibility and degree distribution. Journal of the Theoretical Biology 244, 296–307.
| Food webs robustness to biodiversity loss: the roles of connectance, expansibility and degree distribution.Crossref | GoogleScholarGoogle Scholar |
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nature Reviews Microbiology 10, 538–550.
| Microbial interactions: from networks to models.Crossref | GoogleScholarGoogle Scholar |
Faust, K., and Shorter, J. M. (1981). Comparing networks across space and time, size and species. Mind 90, 61–78.
| Comparing networks across space and time, size and species.Crossref | GoogleScholarGoogle Scholar |
Faust, K., Lima-Mendez, G., Lerat, J. S., Sathirapongsasuti, J. F., Knight, R., Huttenhower, C., Lenaerts, T., and Raes, J. (2015). Cross-biome comparison of microbial association networks. Frontiers in Microbiology 6, 1200.
| Cross-biome comparison of microbial association networks.Crossref | GoogleScholarGoogle Scholar |
Freilich, M. A., Wieters, E., Broitman, B. R., Marquet, P. A., and Navarrete, S. A. (2018). Species co-occurrence networks: can they reveal trophic and non-trophic interactions in ecological communities? Ecology 99, 690–699.
| Species co-occurrence networks: can they reveal trophic and non-trophic interactions in ecological communities?Crossref | GoogleScholarGoogle Scholar |
Friedman, W. R., Halpern, B. S., McLeod, E., Beck, M. W., Duarte, C. M., Kappel, C. V., Levine, A., Sluka, R. D., Adler, S., O’Hara, C. C., Sterling, E. J., Tapia-Lewin, S., Losada, I. J., McClanahan, T. R., Pendleton, L., Spring, M., Toomey, J. P., Weiss, K. R., Possingham, H. P., and Montambault, J. R. (2020). Research priorities for achieving healthy marine ecosystems and human communities in a changing climate. Frontiers in Marine Science 7, 5.
| Research priorities for achieving healthy marine ecosystems and human communities in a changing climate.Crossref | GoogleScholarGoogle Scholar |
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548.
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199.
| Microbial community structure and its functional implications.Crossref | GoogleScholarGoogle Scholar |
Fuhrman, J. A., and Steele, J. A. (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquatic Microbial Ecology 53, 69–81.
| Community structure of marine bacterioplankton: patterns, networks, and relationships to function.Crossref | GoogleScholarGoogle Scholar |
Galand, P. E., Pereira, O., Hochart, C., Auguet, J. C., and Debroas, D. (2018). A strong link between marine microbial community composition and function challenges the idea of functional redundancy. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 12, 2470–2478.
| A strong link between marine microbial community composition and function challenges the idea of functional redundancy.Crossref | GoogleScholarGoogle Scholar |
Gao, H., Lavergne, J. M., Carpenter, C. M. G., Desai, R., Zhang, X., Gray, K., Helbling, D. E., and Wells, G. F. (2019). Exploring co-occurrence patterns between organic micropollutants and bacterial community structure in a mixed-use watershed. Environmental Science: Processes & Impacts 21, 867–880.
| Exploring co-occurrence patterns between organic micropollutants and bacterial community structure in a mixed-use watershed.Crossref | GoogleScholarGoogle Scholar |
García, C., Rendueles, M., and Díaz, M. (2017). Microbial amensalism in Lactobacillus casei and Pseudomonas taetrolens mixed culture. Bioprocess and Biosystems Engineering 40, 1111–1122.
| Microbial amensalism in Lactobacillus casei and Pseudomonas taetrolens mixed culture.Crossref | GoogleScholarGoogle Scholar |
Gilbert, J. A., Steele, J. A., Caporaso, J. G., Steinbrü Ck, L., Reeder, J., Temperton, B., Huse, S., Mchardy, A. C., Knight, R., Joint, I., Somerfield, P., Fuhrman, J. A., and Field, D. (2012). Defining seasonal marine microbial community dynamics. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 6107, 298–308.
| Defining seasonal marine microbial community dynamics.Crossref | GoogleScholarGoogle Scholar |
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., and Stokes, H. W. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. Journal of Bacteriology 190, 5095–5100.
| The evolution of class 1 integrons and the rise of antibiotic resistance.Crossref | GoogleScholarGoogle Scholar |
Girvan, M., and Newman, M. E. J. (2002). Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the United States of America 99, 7821–7826.
| Community structure in social and biological networks.Crossref | GoogleScholarGoogle Scholar |
Grover, J. P. (2000). Resource competition and community structure in aquatic microorganisms: experimental studies of algae and bacteria along a gradient of organic carbon to inorganic phosphorus supply. Journal of Plankton Research 22, 1591–1610.
| Resource competition and community structure in aquatic microorganisms: experimental studies of algae and bacteria along a gradient of organic carbon to inorganic phosphorus supply.Crossref | GoogleScholarGoogle Scholar |
Halpern, B. S., Walbridge, S., Selkoe, K. A., Kappel, C. V., Micheli, F., D’Agrosa, C., Bruno, J. F., Casey, K. S., Ebert, C., Fox, H. E., Fujita, R., Heinemann, D., Lenihan, H. S., Madin, E. M. P. P., Perry, M. T., Selig, E. R., Spalding, M., Steneck, R., Watson, R., D’Agrosa, C., Bruno, J. F., Casey, K. S., Ebert, C., Fox, H. E., Fujita, R., Heinemann, D., Lenihan, H. S., Madin, E. M. P. P., Perry, M. T., Selig, E. R., Spalding, M., Steneck, R., Watson, R., D’Agrosa, C., Bruno, J. F., Casey, K. S., Ebert, C., Fox, H. E., Fujita, R., Heinemann, D., Lenihan, H. S., Madin, E. M. P. P., Perry, M. T., Selig, E. R., Spalding, M., Steneck, R., and Watson, R. (2008). A global map of human impact on marine ecosystems. Science 319, 948–953.
| A global map of human impact on marine ecosystems.Crossref | GoogleScholarGoogle Scholar |
Heino, J., Melo, A. S., Siqueira, T., Soininen, J., Valanko, S., and Bini, L. M. (2015). Metacommunity organisation, spatial extent and dispersal in aquatic systems: patterns, processes and prospects. Freshwater Biology 60, 845–869.
| Metacommunity organisation, spatial extent and dispersal in aquatic systems: patterns, processes and prospects.Crossref | GoogleScholarGoogle Scholar |
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). Environmental stress destabilizes microbial networks. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 15, 1722–1734.
| Environmental stress destabilizes microbial networks.Crossref | GoogleScholarGoogle Scholar |
Hirano, H., and Takemoto, K. (2019). Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics 20, 329.
| Difficulty in inferring microbial community structure based on co-occurrence network approaches.Crossref | GoogleScholarGoogle Scholar |
Holman, L. E., de Bruyn, M., Creer, S., Carvalho, G., Robidart, J., and Rius, M. (2021). Animals, protists and bacteria share marine biogeographic patterns. Nature Ecology & Evolution 5, 738–746.
| Animals, protists and bacteria share marine biogeographic patterns.Crossref | GoogleScholarGoogle Scholar |
Jeffries, T. C., Schmitz Fontes, M. L., Harrison, D. P., Van-Dongen-Vogels, V., Eyre, B. D., Ralph, P. J., and Seymour, J. R. (2016). Bacterioplankton dynamics within a large anthropogenically impacted urban estuary. Frontiers in Microbiology 6, 1438.
| Bacterioplankton dynamics within a large anthropogenically impacted urban estuary.Crossref | GoogleScholarGoogle Scholar |
Kara, E. L., Hanson, P. C., Hu, Y. H., Winslow, L., and McMahon, K. D. (2013). A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 7, 680–684.
| A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA.Crossref | GoogleScholarGoogle Scholar |
Karimi, B., Maron, P. A., Chemidlin-Prevost Boure, N., Bernard, N., Gilbert, D., and Ranjard, L. (2017). Microbial diversity and ecological networks as indicators of environmental quality. Environmental Chemistry Letters 15, 265–281.
| Microbial diversity and ecological networks as indicators of environmental quality.Crossref | GoogleScholarGoogle Scholar |
Krivitsky, P.N., and Kolaczyk, E. D. (2015). On the question of effective sample size in network modeling: an asymptotic inquiry. Statistical Science 30, 184–198.
| On the question of effective sample size in network modeling: an asymptotic inquiry.Crossref | GoogleScholarGoogle Scholar |
Landi, P., Minoarivelo, H. O., Brännström, Å., Hui, C., and Dieckmann, U. (2018). Complexity and stability of ecological networks: a review of the theory. Population Ecology 60, 319–345.
| Complexity and stability of ecological networks: a review of the theory.Crossref | GoogleScholarGoogle Scholar |
Laroche, O., Pochon, X., Tremblay, L. A., Ellis, J. I., Lear, G., and Wood, S. A. (2018). Incorporating molecular-based functional and co-occurrence network properties into benthic marine impact assessments. FEMS Microbiology Ecology 94, fiy167.
| Incorporating molecular-based functional and co-occurrence network properties into benthic marine impact assessments.Crossref | GoogleScholarGoogle Scholar |
Lawes, J. C., Neilan, B. A., Brown, M. V., Clark, G. F., and Johnston, E. L. (2016). Elevated nutrients change bacterial community composition and connectivity: high throughput sequencing of young marine biofilms. Biofouling 32, 57–69.
| Elevated nutrients change bacterial community composition and connectivity: high throughput sequencing of young marine biofilms.Crossref | GoogleScholarGoogle Scholar |
Layeghifard, M., Hwang, D. M., and Guttman, D. S. (2017). Disentangling interactions in the microbiome: a network perspective. Trends in Microbiology 25, 217–228.
| Disentangling interactions in the microbiome: a network perspective.Crossref | GoogleScholarGoogle Scholar |
Layeghifard, M., Hwang, D. M., and Guttman, D. S. (2018). Constructing and analyzing microbiome networks in R. Methods in Molecular Biology 1849, 243–266.
| Constructing and analyzing microbiome networks in R.Crossref | GoogleScholarGoogle Scholar |
Leclerc, R. D. (2008). Survival of the sparsest: robust gene networks are parsimonious. Molecular Systems Biology 4, 213.
| Survival of the sparsest: robust gene networks are parsimonious.Crossref | GoogleScholarGoogle Scholar |
Li, F., Peng, Y., Fang, W., Altermatt, F., Xie, Y., Yang, J., and Zhang, X. (2018). Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environmental Science and Technology 52, 11708–11719.
| Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers.Crossref | GoogleScholarGoogle Scholar |
Li, H.-Y., Wang, H., Wang, H.-T., Xin, P.-Y., Xu, X.-H., Ma, Y., Liu, W.-P., Teng, C.-Y., Jiang, C.-L., Lou, L.-P., Arnold, W., Cralle, L., Zhu, Y.-G., Chu, J.-F., Gilbert, J. A., and Zhang, Z.-J. (2018). The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome 6, 187.
| The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales.Crossref | GoogleScholarGoogle Scholar |
Li, Y., Wu, H., Shen, Y., Wang, C., Wang, P., Zhang, W., Gao, Y., and Niu, L. (2019). Statistical determination of crucial taxa indicative of pollution gradients in sediments of Lake Taihu, China. Environmental Pollution 246, 753–762.
| Statistical determination of crucial taxa indicative of pollution gradients in sediments of Lake Taihu, China.Crossref | GoogleScholarGoogle Scholar |
Liao, H., Yu, K., Duan, Y., Ning, Z., Li, B., He, L., and Liu, C. (2019). Profiling microbial communities in a watershed undergoing intensive anthropogenic activities. Science of The Total Environment 647, 1137–1147.
| Profiling microbial communities in a watershed undergoing intensive anthropogenic activities.Crossref | GoogleScholarGoogle Scholar |
Lidicker, W. Z. (1979). A clarification of interactions in ecological systems. BioScience 29, 475–477.
| A clarification of interactions in ecological systems.Crossref | GoogleScholarGoogle Scholar |
Lin, Y., Zhao, D., Zeng, J., Cao, X., and Jiao, C. (2019). Network analysis reveals seasonal patterns of bacterial community networks in Lake Taihu under aquaculture conditions. Water 11, 1868.
| Network analysis reveals seasonal patterns of bacterial community networks in Lake Taihu under aquaculture conditions.Crossref | GoogleScholarGoogle Scholar |
Little, A. E. F., Robinson, C. J., Peterson, S. B., Raffa, K. F., and Handelsman, J. (2008). Rules of engagement: interspecies interactions that regulate microbial communities. Annual Review of Microbiology 62, 375–401.
| Rules of engagement: interspecies interactions that regulate microbial communities.Crossref | GoogleScholarGoogle Scholar |
Liu, J., Fu, B., Yang, H., Zhao, M., He, B., and Zhang, X.-H. (2015). Phylogenetic shifts of bacterioplankton community composition along the Pearl Estuary: the potential impact of hypoxia and nutrients. Frontiers in Microbiology 6, 64.
| Phylogenetic shifts of bacterioplankton community composition along the Pearl Estuary: the potential impact of hypoxia and nutrients.Crossref | GoogleScholarGoogle Scholar |
Liu, M., Han, X., Tong, J., Zhu, H., and Bai, X. (2020a). Mutual environmental drivers of the community composition, functional attributes and co-occurrence patterns of bacterioplankton in the composite aquatic ecosystem of Taihu watershed in China. FEMS Microbiology Ecology 96, fiaa137.
| Mutual environmental drivers of the community composition, functional attributes and co-occurrence patterns of bacterioplankton in the composite aquatic ecosystem of Taihu watershed in China.Crossref | GoogleScholarGoogle Scholar |
Liu, J., Zhu, S., Liu, X., Yao, P., Ge, T., and Zhang, X. H. (2020b). Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 14, 1463–1478.
| Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship.Crossref | GoogleScholarGoogle Scholar |
Liu, Z., Ma, X., He, N., Zhang, J., Wu, J., and Liu, C. (2020c). Shifts in microbial communities and networks are correlated with the soil ionome in a kiwifruit orchard under different fertilization regimes. Applied Soil Ecology 149, 103517.
| Shifts in microbial communities and networks are correlated with the soil ionome in a kiwifruit orchard under different fertilization regimes.Crossref | GoogleScholarGoogle Scholar |
Lynch, M., and Neufeld, J. (2015). Ecology and exploration of the rare biosphere. Nature Reviews Microbiology 13, 217–229.
| Ecology and exploration of the rare biosphere.Crossref | GoogleScholarGoogle Scholar |
McGee, K. M., Robinson, C. V., and Hajibabaei, M. (2019). Gaps in DNA-based biomonitoring across the globe. Frontiers in Ecology and Evolution 7, 337.
| Gaps in DNA-based biomonitoring across the globe.Crossref | GoogleScholarGoogle Scholar |
Makiola, A., Compson, Z. G., Baird, D. J., Barnes, M. A., Boerlijst, S. P., Bouchez, A., Brennan, G., Bush, A., Canard, E., Cordier, T., Creer, S., Curry, R. A., David, P., Dumbrell, A. J., Gravel, D., Hajibabaei, M., Hayden, B., van der Hoorn, B., Jarne, P., Jones, J. I., Karimi, B., Keck, F., Kelly, M., Knot, I. E., Krol, L., Massol, F., Monk, W. A., Murphy, J., Pawlowski, J., Poisot, T., Porter, T. M., Randall, K. C., Ransome, E., Ravigné, V., Raybould, A., Robin, S., Schrama, M., Schatz, B., Tamaddoni-Nezhad, A., Trimbos, K. B., Vacher, C., Vasselon, V., Wood, S., Woodward, G., and Bohan, D. A. (2020). Key questions for next-generation biomonitoring. Frontiers in Environmental Science 7, 197.
Mandakovic, D., Rojas, C., Maldonado, J., Latorre, M., Travisany, D., Delage, E., Bihouée, A., Jean, G., Díaz, F. P., Fernández-Gómez, B., Cabrera, P., Gaete, A., Latorre, C., Gutiérrez, R. A., Maass, A., Cambiazo, V., Navarrete, S. A., Eveillard, D., and González, M. (2018). Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience. Scientific Reports 8, 5875.
| Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience.Crossref | GoogleScholarGoogle Scholar |
Marti, E., Jofre, J., and Balcazar, J. L. (2013). Prevalence of antibiotic resistance genes and bacterial community composition in river influenced by a wastewater treatment plant. PLoS One 8, e78906.
| Prevalence of antibiotic resistance genes and bacterial community composition in river influenced by a wastewater treatment plant.Crossref | GoogleScholarGoogle Scholar |
Mason, O., and Verwoerd, M. (2008). Graph theory and networks in biology. IET Systems Biology 1, 89–119.
| Graph theory and networks in biology.Crossref | GoogleScholarGoogle Scholar |
Mikhailov, I. S., Bukin, Y. S., Zakharova, Y. R., Usoltseva, M. V., Galachyants, Y. P., Sakirko, M. V., Blinov, V. V., and Likhoshway, Y. V. (2019). Co-occurrence patterns between phytoplankton and bacterioplankton across the pelagic zone of Lake Baikal during spring. Journal of Microbiology 57, 252–262.
| Co-occurrence patterns between phytoplankton and bacterioplankton across the pelagic zone of Lake Baikal during spring.Crossref | GoogleScholarGoogle Scholar |
Murali, T. M., Dyer, M. D., Badger, D., Tyler, B. M., and Katze, M. G. (2011). Network-based prediction and analysis of HIV dependency factors. PLoS Computational Biology 7, e1002164.
| Network-based prediction and analysis of HIV dependency factors.Crossref | GoogleScholarGoogle Scholar |
Newman, M. E. J. (2010). ‘Networks: An Introduction.’ (Oxford University Press: Oxford, UK.)
Paine, R. T. (1995). A conversation on refining the concept of keystone species. Conservation Biology 9, 962–964.
| A conversation on refining the concept of keystone species.Crossref | GoogleScholarGoogle Scholar |
Pal, R., Sekh, A. A., Kar, S., and Prasad, D. K. (2020). Neural network based country wise risk prediction of COVID-19. Applied Sciences 10, 6448.
| Neural network based country wise risk prediction of COVID-19.Crossref | GoogleScholarGoogle Scholar |
Parada, A. E., and Fuhrman, J. A. (2017). Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 11, 2510–2525.
| Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor.Crossref | GoogleScholarGoogle Scholar |
Pauvert, C., Buée, M., Laval, V., Edel-Hermann, V., Fauchery, L., Gautier, A., Lesur, I., Vallance, J., and Vacher, C. (2019). Bioinformatics matters: the accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecology 41, 23–33.
| Bioinformatics matters: the accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline.Crossref | GoogleScholarGoogle Scholar |
Pavlopoulos, G. A., Secrier, M., Moschopoulos, C. N., Soldatos, T. G., Kossida, S., Aerts, J., Schneider, R., and Bagos, P. G. (2011). Using graph theory to analyze biological networks. BioData Mining 4, 10.
| Using graph theory to analyze biological networks.Crossref | GoogleScholarGoogle Scholar |
Pellissier, L., Albouy, C., Bascompte, J., Farwig, N., Graham, C., Loreau, M., Maglianesi, M. A., Melián, C. J., Pitteloud, C., Roslin, T., Rohr, R., Saavedra, S., Thuiller, W., Woodward, G., Zimmermann, N. E., and Gravel, D. (2018). Comparing species interaction networks along environmental gradients. Biological Reviews 93, 785–800.
| Comparing species interaction networks along environmental gradients.Crossref | GoogleScholarGoogle Scholar |
Peura, S., Bertilsson, S., Jones, R. I., and Eiler, A. (2015). Resistant microbial cooccurrence patterns inferred by network topology. Applied and Environmental Microbiology 81, 2090–2097.
| Resistant microbial cooccurrence patterns inferred by network topology.Crossref | GoogleScholarGoogle Scholar |
Powell, S., Ferguson, S., and Snape, I. (2006). Fertilization stimulates anaerobic fuel degradation of Antarctic soils by denitrifying microorganisms. Environmental Science and Technology 40, 2011–2017.
Power, M. E., and Mills, L. S. (1995). The keystone cops meet in Hilo. Trends in Ecology and Evolution 10, 182–184.
| The keystone cops meet in Hilo.Crossref | GoogleScholarGoogle Scholar |
Price, G. W., Langille, M. G. I., and Yurgel, S. N. (2021). Microbial co-occurrence network analysis of soils receiving short- and long-term applications of alkaline treated biosolids. Science of the Total Environment 751, 141687.
| Microbial co-occurrence network analysis of soils receiving short- and long-term applications of alkaline treated biosolids.Crossref | GoogleScholarGoogle Scholar |
Proulx, S. R., Promislow, D. E. L., and Phillips, P. C. (2005). Network thinking in ecology and evolution. Trends in Ecology and Evolution 20, 345–353.
| Network thinking in ecology and evolution.Crossref | GoogleScholarGoogle Scholar |
Raffaelli, D. (2007). Food webs, body size and the curse of the latin binomial. From energy to ecosyst. In ‘Dynamics and Structure of Ecological Systems’. pp. 53–64. (Springer: Dordrecht, Netherlands.)
| Crossref |
Röttjers, L., and Faust, K. (2018). From hairballs to hypotheses – biological insights from microbial environmental factors. Bioinformatics 22, 2532–2538.
| From hairballs to hypotheses – biological insights from microbial environmental factors.Crossref | GoogleScholarGoogle Scholar |
Ruan, Q., Dutta, D., Schwalbach, M. S., Steele, J. A., Fuhrman, J. A., and Sun, F. (2006). Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22, 2532–2538.
| Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors.Crossref | GoogleScholarGoogle Scholar |
Sagova-Mareckova, M., Boenigk, J., Bouchez, A., Cermakova, K., Chonova, T., Cordier, T., Eisendle, U., Elersek, T., Fazi, S., Fleituch, T., Frühe, L., Gajdosova, M., Graupner, N., Haegerbaeumer, A., Kelly, A. M., Kopecky, J., Leese, F., Nõges, P., Orlic, S., Panksep, K., Pawlowski, J., Petrusek, A., Piggott, J. J., Rusch, J. C., Salis, R., Schenk, J., Simek, K., Stovicek, A., Strand, D. A., Vasquez, M. I., Vrålstad, T., Zlatkovic, S., Zupancic, M., and Stoeck, T. (2021). Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring. Water Research 191, 116767.
| Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring.Crossref | GoogleScholarGoogle Scholar |
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504.
| Cytoscape: a software environment for integrated models of biomolecular interaction networks.Crossref | GoogleScholarGoogle Scholar |
Shendure, J., and Ji, H. (2008). Next-generation DNA sequencing. Nature Biotechnology 26, 1135–1145.
Sheykhali, S., Fernández-Gracia, J., Traveset, A., and Eguíluz, V. M. (2019). Extinction-induced community reorganization in bipartite networks. Applied Network Science 4, 23.
| Extinction-induced community reorganization in bipartite networks.Crossref | GoogleScholarGoogle Scholar |
Snelgrove, P. V. R. (2010). ‘Discoveries of the Census of Marine Life: Making Ocean Life Count, Discoveries of the Census of Marine Life.’ (Cambridge University Press.)
| Crossref |
Steele, J. A., Countway, P. D., Xia, L., Vigil, P. D., Beman, J. M., Kim, D. Y., Chow, C. E. T., Sachdeva, R., Jones, A. C., Schwalbach, M. S., Rose, J. M., Hewson, I., Patel, A., Sun, F., Caron, D. A., and Fuhrman, J. A. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 5, 1414–1425.
| Marine bacterial, archaeal and protistan association networks reveal ecological linkages.Crossref | GoogleScholarGoogle Scholar |
Steffen, W., Persson, Å., Deutsch, L., Zalasiewicz, J., Williams, M., Richardson, K., Crumley, C., Crutzen, P., Folke, C., Gordon, L., Molina, M., Ramanathan, V., Rockström, J., Scheffer, M., Schellnhuber, H. J., and Svedin, U. (2011). The anthropocene: from global change to planetary stewardship. Ambio 40, 739–761.
| The anthropocene: from global change to planetary stewardship.Crossref | GoogleScholarGoogle Scholar |
Sun, M., Dafforn, K., Brown, M., and Johnston, E. (2012). Bacterial communities are sensitive indicators of contaminant stress. Marine Pollution Bulletin 64, 1029–1038.
| Bacterial communities are sensitive indicators of contaminant stress.Crossref | GoogleScholarGoogle Scholar |
Sutcliffe, B., Hose, G. C., Harford, A. J., Midgley, D. J., Greenfield, P., Paulsen, I. T., and Chariton, A. (2019). Microbial communities are sensitive indicators for freshwater sediment copper contamination. Environmental Pollution 247, 1028–1038.
| Microbial communities are sensitive indicators for freshwater sediment copper contamination.Crossref | GoogleScholarGoogle Scholar |
Tang, B.-L., Yang, J., Chen, X.-L., Wang, P., Zhao, H.-L., Su, H.-N., Li, C.-Y., Yu, Y., Zhong, S., Wang, L., Lidbury, I., Ding, H., Wang, M., McMinn, A., Zhang, X.-Y., Chen, U., and Zhang, Y.-Z. (2020). A predator–prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nature Communications 11, 285.
| A predator–prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.Crossref | GoogleScholarGoogle Scholar |
Tylianakis, J. M., and Morris, R. J. (2017). Ecological networks across environmental gradients. Annual Review of Ecology, Evolution, and Systematics 48, 25–48.
| Ecological networks across environmental gradients.Crossref | GoogleScholarGoogle Scholar |
Tylianakis, J. M., Laliberté, E., Nielsen, A., and Bascompte, J. (2010). Conservation of species interaction networks. Biological Conservation 143, 2270–2279.
| Conservation of species interaction networks.Crossref | GoogleScholarGoogle Scholar |
Valiente-Banuet, A., Aizen, M. A., Alcántara, J. M., Arroyo, J., Cocucci, A., Galetti, M., García, M. B., García, D., Gómez, J. M., Jordano, P., Medel, R., Navarro, L., Obeso, J. R., Oviedo, R., Ramírez, N., Rey, P. J., Traveset, A., Verdú, M., and Zamora, R. (2015). Beyond species loss: the extinction of ecological interactions in a changing world. Functional Ecology 29, 299–307.
Weiss, S., Van Treuren, W., Lozupone, C., Faust, K., Friedman, J., Deng, Y., Xia, L. C., Xu, Z. Z., Ursell, L., Alm, E. J., Birmingham, A., Cram, J. A., Fuhrman, J. A., Raes, J., Sun, F., Zaccone, R., and Caruso, G. (2019). Microbial enzymes in the Mediterranean Sea: relationship with climate changes. AIMS Microbiology 5, 251–271.
| Microbial enzymes in the Mediterranean Sea: relationship with climate changes.Crossref | GoogleScholarGoogle Scholar |
Westerhoff, H. V., Brooks, A. N., Simeonidis, E., García-Contreras, R., He, F., Boogerd, F. C., Jackson, V. J., Goncharuk, V., and Kolodkin, A. (2014). Macromolecular networks and intelligence in microorganisms. Frontiers in Microbiology 5, 379.
| Macromolecular networks and intelligence in microorganisms.Crossref | GoogleScholarGoogle Scholar |
Widder, S., Besemer, K., Singer, G. A., Ceola, S., Bertuzzo, E., Quince, C., Sloan, W. T., Rinaldo, A., and Battin, T. J. (2014). Fluvial network organization imprints on microbial co-occurrence networks. Proceedings of the National Academy of Sciences of the United States of America 111, 12799–12804.
| Fluvial network organization imprints on microbial co-occurrence networks.Crossref | GoogleScholarGoogle Scholar |
Williams, R. J., Howe, A., and Hofmockel, K. S. (2014). Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Frontiers in Microbiology 5, 358.
| Demonstrating microbial co-occurrence pattern analyses within and between ecosystems.Crossref | GoogleScholarGoogle Scholar |
Wolfram, S. (1999). ‘The MATHEMATICA book, version 4.’ (Cambridge University Press.)
Wu, H., Li, Y., Zhang, W., Wang, C., Wang, P., Niu, L., Du, J., and Gao, Y. (2019). Bacterial community composition and function shift with the aggravation of water quality in a heavily polluted river. Journal of Environmental Management 237, 433–441.
| Bacterial community composition and function shift with the aggravation of water quality in a heavily polluted river.Crossref | GoogleScholarGoogle Scholar |
Yachi, S., and Loreau, M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proceedings of the National Academy of Sciences of the United States of America 96, 1463–1468.
| Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis.Crossref | GoogleScholarGoogle Scholar |
Zamkovaya, T., Foster, J. S., de Crécy-Lagard, V., and Conesa, A. (2021). A network approach to elucidate and prioritize microbial dark matter in microbial communities. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 15, 228–244.
| A network approach to elucidate and prioritize microbial dark matter in microbial communities.Crossref | GoogleScholarGoogle Scholar |
Zhang, L., Tu, D., Li, X., Lu, W., and Li, J. (2020). Impact of long-term industrial contamination on the bacterial communities in urban river sediments. BMC Microbiology 20, 254.
| Impact of long-term industrial contamination on the bacterial communities in urban river sediments.Crossref | GoogleScholarGoogle Scholar |
Zhou, C., Miao, T., Jiang, L., Zhang, H., Zhang, Y., and Zhang, X. (2021). Conditions that promote the formation of black bloom in aquatic microcosms and its effects on sediment bacteria. Science of The Total Environment 751, 141869.
| Conditions that promote the formation of black bloom in aquatic microcosms and its effects on sediment bacteria.Crossref | GoogleScholarGoogle Scholar |
Zhu, C., Zhang, J., Zohaib, M., Mahboob, N. S., Al-Ghanim, K. A., Khan, I. A., Lu, Z., and Chen, T. (2019). Seasonal succession and spatial distribution of bacterial community structure in a eutrophic freshwater lake, Lake Taihu. Science of The Total Environment 669, 29–40.
| Seasonal succession and spatial distribution of bacterial community structure in a eutrophic freshwater lake, Lake Taihu.Crossref | GoogleScholarGoogle Scholar |
Zinger, L., Bonin, A., Alsos, I., Bálint, M., Bik, H., Boyer, F., Chariton, A., Creer, S., Coissac, E., Deagle, B., De Barba, M., Dickie, I., Dumbrell, A., Ficetola, G., Fierer, N., Fumagalli, L., Gilbert, T., Jarman, S., Jumpponen, A., Kauserud, H., Orlando, L., Pansu, J., Pawlowski, J., Tedersoo, L., Thomsen, P., Willerslev, E., and Taberlet, P. (2019). DNA metabarcoding – need for robust experimental designs to draw sound ecological conclusions. Molecular Ecology 28, 1857–1862.
| DNA metabarcoding – need for robust experimental designs to draw sound ecological conclusions.Crossref | GoogleScholarGoogle Scholar |


