A molecular journey in conservation genetics
Margaret Byrne
Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Locked Bag 104, Bentley Delivery Centre, WA 6983, Australia. Email: margaret.byrne@dbca.wa.gov.au
Pacific Conservation Biology 24(3) 235-243 https://doi.org/10.1071/PC18025
Submitted: 19 February 2018 Accepted: 4 June 2018 Published: 5 July 2018
Journal compilation © CSIRO 2018 Open Access CC BY-NC-ND
Abstract
Genetics, and more recently genomics, has become an integral part of conservation science. From the early days of DNA fingerprinting through development of hybridisation based and polymerase chain reaction based markers, to applications of genomics, genetics has provided many insights to improve management of plants, animals and their ecosystems. I share my journey of discovery in genetics and genomics, and their application in conservation of plants through understanding evolutionary history, population genetics of rare and threatened species, molecular taxonomy, fragmentation and the role of pollen dispersal, restoration in a risk management context, and adaptation to climate change.
Additional keywords: biodiversity conservation, DNA, genetics, genomics, plants.
Introduction
You may have seen a television program that aired in mid-2017 called Code of a Killer (I recommend watching it if you didn’t). It told the story of Sir Alec Jeffreys, the brilliant British scientist who identified minisatellites that became the basis of DNA fingerprinting and revolutionised forensics. In a nutshell, the story goes like this. Alec Jeffreys was a scientist at the University of Leicester and was researching mammalian and human genetics when, in 1984, he found a core sequence in tandem repeated DNA in the human myoglobin gene that could be used to assay a large number of loci that demonstrated the relationships between individuals (Jeffreys et al. 1985a, 1985b). He was contacted to determine the identity of a young boy in an immigrant debate, and showed that the boy was indeed the son of the purported mother and he was allowed to stay in the United Kingdom. At the same time, the police in Leicester were investigating the 1983 murder of a teenage girl with few clues, and the senior officer heard about Jeffreys’ work in the immigration case and sought him out to see whether he could assist with screening all the men in the district to identify the murderer. In short, the police set up an extensive screening program and were able to identify the man who had committed that murder, as well as that of another young girl during the time of the investigation in 1986. This unlikely teaming up of the scientist and the police officer established DNA fingerprinting as a tool that revolutionised forensics, individual identification and paternity analysis, and has been celebrated as a paradigm shift in evolutionary and ecological genetics (Chambers et al. 2014).
So what has this to do with me and with conservation? In 1986, I undertook Honours at The University of Western Australia and my supervisor, Associate Professor Sid James, showed me the two papers of Alec Jeffreys (Jeffreys et al. 1985a, 1985b) describing minisatellites and their application in identification, and quite simply said ‘I want to do this in plants’. So, rather naively, we set out to develop molecular DNA techniques with the aid of the book ‘Molecular cloning: a laboratory manual’ by Maniatis et al. (1982) that was a mainstay of molecular biology at the time because of its comprehensive description of the emerging application of gene cloning technologies. When I think now of that approach I would not have allowed a student to take on a project like that considering it too risky, but it was a time of major technical developments in molecular biology and we were not so concerned with measurable outputs (papers), but rather advancing capability. So I set about learning molecular techniques according to Maniatis, and I wrote to Alec Jeffreys who very kindly provided an aliquot of the probes for the minisatellites, because this used restriction fragment-length polymorphism (RFLP) technology at that time. My supervisor, Sid James, would only allow me to test the probes on his own family, so we assayed DNA samples from Sid and Marie, his wife, and his four children, Phillip, Stephen, Denise and Marcelle, and thankfully showed that they were all related as parents and children as expected. Obviously, this in itself was not ground-breaking, but it enabled me to show that we had established the methodology (DNA extraction, restriction enzyme digestion, gel electrophoresis, Southern blotting, radioactive labelling of the probes, probe hybridisation, autoradiography) and had it working as it should, which was no mean feat at that time, before the internet and Google. Feeling encouraged, I went on to do a PhD with Sid and, with some assistance from the Biochemistry Department, developed genomic libraries to identify probes and undertake a molecular genetic diversity study of the granite rock dwelling plant, Isotoma petraea, Sid’s favourite plant in which he discovered complex hybridity and that he had worked on for so many years, and Macrozamia reidlei, an ancient cycad (Byrne et al. 1997c). I remain deeply indebted to Sid for his belief in me, sharing his intrepid pursuit of ideas, challenging me to think ‘outside the box’, exposing me to research as a partnership, and for his and Marie’s friendship. I couldn’t have asked for a better supervisor, or a better introduction to research. As Tony Brown, an eminent geneticist at CSIRO, once said to me ‘You’ve come from an excellent stable and you’ve been taught to think’.
Revolutions in technology
The laboratory scenes in the television series, Code of a Killer, had been accurately recreated for the time, and the sight of the Hybaid oven, the autoradiographs, light boxes, radiation signs all took me back to the time when I did this RFLP work, and painstakingly inserted Southern blots into the glass tubes and added just the right amount of radioactively labelled solution and sealed the tubes to ensure they didn’t dry out as they rotated around the oven for 24 h (I later made improvements by putting them in a box and mixing on a rotator). It made me think of how far we have come in the journey of molecular developments in conservation genetics as we now readily produce complete genome sequences or generate tens of thousands of single nucleotide polymorphism (SNP) loci, something inconceivable 30 years ago. Prior to RFLPs, isozymes had been the workhorse for genetics (Tanksley and Orton 1983) and had shown how genetic analysis could provide great insights into the diversity and mating systems of plants and animals. The advent of RFLPs took us beyond the limited number of isozyme loci and opened up the potential for larger numbers of loci in the nuclear, chloroplast and mitochondrial genomes. As I finished my PhD, the next revolution was occurring with the first papers describing application of polymerase chain reaction (PCR; Erlich et al. 1991) in random amplification of polymorphic DNA (RAPD; Williams et al. 1990; Hadrys et al. 1992). While RAPDs were dominant markers (i.e. the locus band was either absent or present and the heterozygote could not be discriminated from the dominant homozygote), which reduced their effectiveness in comparison to co-dominant markers, such as isozymes and RFLPs, the method produced many more loci and could be used on any species without development of specific probes. It didn’t take long for the next advance to occur and amplified fragment length polymorphism (AFLP; Vos et al. 1995) became a favoured tool because it combined the reliability of the RFLP technique with the power of the PCR technique. While AFLP was also a dominant marker, it was more reliable than RAPDs and could be applied to any species, although species with large genomes, such as pines, required longer primers to provide greater stringency and reduce the number of bands produced. Interestingly, AFLPs didn’t get taken up by animal geneticists as readily as they did by plant geneticists, possibly because animal geneticist had access to mitochondrial sequencing that provided a means of assaying variation (Moritz and Brown 1986). About the time that the applications of AFLPs became recognised, microsatellites came on the scene. While others took up AFLPs, because they required no up front development in different species, I went straight onto developing and using microsatellites, developing the first microsatellites in an Australian plant, Eucalyptus nitens (Byrne et al. 1996). Although microsatellites required development of primers for particular species, or close congeners, they were codominant and so provided the analytical power that was missing with RAPDs and AFLPs. This was where the skills learned in making genomic libraries to develop RFLP probes came to the fore again, in constructing genomic libraries to develop microsatellite primers (these were the early days well before the times when you simply sent DNA to a commercial company to develop primers through 454 sequencing!). I was lucky during these times to be working at CSIRO Forestry and Forest Products in Canberra, in a team led by Gavin Moran, pursuing innovative avenues for application of molecular techniques to genetic mapping in forest trees, in my case, eucalypts (Byrne et al. 1995). As is often the case, advances in technologies are made in agricultural or forestry environments where commercial drivers are present, and at that time marker assisted selection was a very strong driver for development of molecular methods allowing discrimination and tracking of individuals (Byrne et al. 1997a, 1997b). Of course, the power and applicability of microsatellites also unearthed many secrets as the use of molecular techniques throughout the world showed major errors in breeding programs and plant production programs. Microsatellites became the standard marker for molecular ecology for many years and enabled studies on population structure and diversity, mating system and paternity analysis, clonality, hybridisation, and molecular taxonomy at the species-population interface.
More recently, the genomics revolution has enabled us to readily sequence complete genomes. While iconic species, such as the tammar wallaby (Renfree et al. 2011) and Eucalyptus (Myburg et al. 2014), were the focus of early painstakingly slow genome construction projects using Sanger sequencing, genomes can now be sequenced for any species at will. Next year will be the 20th anniversary of the initiation of the International Eucalypt Genome Consortium, where we agreed to progress construction of the eucalypt genome and, subsequently, obtained a grant of US$10 million (a large sum in those days) from the Joint Genome Initiative of the US Department of Energy. Once again, the development of methodologies, such as DArTseq (Sansaloni et al. 2011), ddRAD (Peterson et al. 2012) and Genotyping by Sequencing (Elshire et al. 2011), occurred in model organisms or agricultural and production systems, but with the prevalence of far superior means of communication, they were much more readily taken up in the conservation arena. Of course, as wet laboratory methods develop, there is a corresponding need for analytical tools and pipelines for handling the very large amounts of data generated by these genomic projects.
Genomics is providing greater power to undertake what have become ‘classical’ avenues of investigation in conservation genetics, such as population diversity, structure, inbreeding and hybridisation, but is also providing us with the capacity to investigate areas that we couldn’t explore with previous markers, such as local adaptation, inbreeding depression and outbreeding depression (Allendorf et al. 2010). We are still only at the beginning of understanding how molecular variation intersects with environmental conditions, conferring capacity for persistence and adaptation to environmental conditions (Hoffmann et al. 2015). Genomic developments have also spawned the assays of micro amounts of DNA in pooled environmental samples, such as soil (Bissett et al. 2016), and environmental DNA (eDNA) methods are becoming integral in a wide range of areas, including conservation (Thomsen and Willerslev 2015), biosecurity and forensics, as well as in monitoring and tracking cryptic species. Most recently, new advances in CRISPR-Cas9 gene editing technologies have provided the promise of new approaches to control invasive pests that are a fundamental threat to Australian mammals (Moro et al. 2018).
Conservation genetics and genomics
Genetics and genomics have become essential aspects of conservation science as we seek to conserve and manage the three components of biodiversity: ecosystems, species and genes. Genetic studies on their own can provide information to inform conservation management of species; however, I have found that the greatest gains arise when genetics is combined with other disciplines in interdisciplinary studies. In my experience, single studies may influence a particular action, but it is the synthesis of a range of information that is most informative for decision making and setting policy (Lynch et al. 2015). Over the years I have combined work on evolutionary history, population genetics of rare and threatened species, molecular taxonomy, fragmentation and the role of pollen dispersal, restoration in a risk management context and adaptation to climate change to provide a comprehensive conceptual basis for understanding the conservation and management of Australian plants. There is too much to fully elaborate on here, but I would like to mention what I consider to be some highlights.
My early foray into conservation genetics was focussed on understanding population diversity and structure in eucalypts (Byrne et al. 1998), and in particular using cpDNA to look at phylogeographic patterns to infer evolutionary history (Byrne and Moran 1994). Looking back into evolutionary history captured my interest and I have continued to work on this throughout my career. Together with colleagues and students, I gained new insights and have driven a paradigm shift in concepts about evolutionary history and refugia, in particular showing that plants in south-western Australia have responded to historical climate change through persistence in localised refugia (Byrne and Macdonald 2000; Byrne et al. 2002, 2003; Byrne and Hines 2004; Wheeler and Byrne 2006; Byrne 2007, 2008; Byrne and Hopper 2008; Keppel et al. 2012; Tapper et al. 2014a, 2014b), and not through broad-scale contraction and expansion as has been the classical paradigm based on temperate northern hemisphere responses (Hewitt 2004). I was particularly thrilled to lead an Environmental Futures Network project that brought together a range of colleagues and produced interdisciplinary syntheses of the evolutionary history of the three Australian biomes, showing the complexity of the evolutionary history of the Australian biota (Byrne et al. 2008b, 2011a; Bowman et al. 2010).
A key feature of work on rare and threatened plants has been demonstrating that many restricted species are not genetically depauperate, as predicted from classical genetic theory, and have adaptations to persist in small populations (Millar et al. 2010, 2013; Clarke et al. 2012; Millar and Byrne 2013; Coates et al. 2015). This has important implications for the way we manage our rare species, demonstrating that all populations are important, no matter how small, and follows on from my PhD supervisor’s insights into the complexity of a flora adapted to persistence in an ancient landscape (James 2000). Use of molecular markers enabled me to investigate other features of plant populations, particularly clonality that informs more accurate census sizes in small populations (Millar et al. 2010; Binks et al. 2015a), and hybridisation to resolve conservation status of putative hybrid species (Walker et al. 2009, 2018). Working with taxonomists to bring molecular tools to bear on resolution of species complexes has been enlightening, and together we have resolved entities in Eucalyptus, Melaleuca, Acacia, Atriplex and Pityrodia (Byrne 1999, 2004; Broadhurst et al. 2004; Millar et al. 2011; Shepherd et al. 2013, 2015; Binks et al. 2015b).
Another highlight has been using paternity analysis to understand mating patterns and demonstrate the role of extensive pollen dispersal, even in fragmented landscapes (Byrne et al. 2007, 2008a; Millar et al. 2008, 2012, 2014; Sampson and Byrne 2008; Llorens et al. 2012; Sampson et al. 2014) where extensive pollen dispersal was not anticipated on the basis of classical expectations of leptokurtic dispersal curves. Much of this work on pollen dispersal and mating systems has been conducted in conjunction with ecological studies, thus providing a more integrated understanding of population processes that inform effective restoration of fragmented landscapes. In the context of revegetation, it enabled a more integrated approach to managing environmental risk by incorporating the potential for genetic risk from planting non-local vegetation (Byrne et al. 2009, 2011b; Byrne and Stone 2011).
The prospect of being able to understand climate change adaptation was a driver for delving into genomics. Funding from the National Climate Change Adaptation Facility enabled me to team up with like-minded colleagues to undertake the first investigation of signatures of adaptive variation in Australian plants, in conjunction with variation in ecophysiological traits (Steane et al. 2014, 2017a, 2017b; McLean et al. 2014), and use this information to develop adaptation strategies for restoration practice (Prober et al. 2015). I have followed on with this approach to determine adaptation along riparian systems to inform revegetation and, in a key forest tree, to inform forest management. Our investigation of genomics in Eucalyptus salubris, a widespread species through the Great Western Woodlands, showed an unexpected separation of two cryptic lineages that were differentiated on both genetic and specific leaf area traits (Steane et al. 2015), and we’re now using genomics in molecular taxonomy at the species–population interface with work on species in Seringia.
The personal journey
For me, it has been a fantastic journey of discovery and development of innovations, as I have travelled from the early days of DNA markers to the current applications of genomics. I have been fortunate, indeed, to be part of development of plant conservation genetics in Australia and worldwide. I was always interested in science and got hooked on genetics in secondary school; I think it was the neatness and simplicity of Mendelian genetics and Punnett squares that captivated me. Later on, I remember the fascination of looking at the inheritance of microsatellite and RFLP bands in a three generation pedigree of Eucalyptus nitens and seeing the segregation of alleles across 118 second generation progeny (Fig. 1). My initial plans (as far as I had any) were to enter academia; after all, that was really the only world I knew at that stage, and as I had initially thought I would follow in my parents’ footsteps and be a secondary teacher before being drawn into research, it seemed a natural progression to be a university academic. However, fate (or whatever) intervened and, at the end of my PhD, I was offered a position at CSIRO in Canberra with Gavin Moran. I had just had my second child (born the day my PhD was submitted), but we packed up and moved, and I became fully exposed to the world of applied science, and that opportunity to make a difference empowered me. Gavin was a population geneticist, so he encouraged me to work with him in population genetics, in addition to the explicit application in forestry breeding programs that was my core work, and so my interest in conservation was borne. Five years later, this unusual coupling of interests made me the most suitable applicant for a job at the Department of Conservation and Land Management in Perth, where the position was required to provide genetic expertise for forest management and development of species for agroforestry, as well as plant conservation programs with David Coates who was then using isozymes. This was the start of an exciting journey in conservation genetics, with my first task being to establish a molecular genetics laboratory in the State conservation agency. It was daunting; even though application of molecular techniques was becoming routine in agriculture and forestry, it was still in its infancy in conservation in Australia, and we had lots of false starts and learning from mistakes. However, the opportunities were endless and it was at a time when funding was adequate for the task.
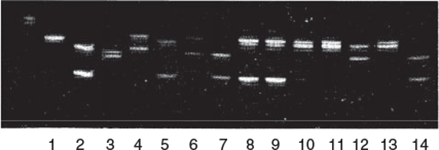
|
As I grew in my knowledge and understanding of conservation and how science supports management, I was provided with many opportunities to broaden my horizons and move into the conservation policy and strategy. Management was not something I had given much thought until, in 2000, I was asked to act in the position of Director of Science for 3 months, and found I liked the new challenge of providing direction to a great team of scientists. Over the next 10 years, I sought out opportunities to further my knowledge and understanding, and gain experience in the policy and management world. This opportunity to combine policy and science is incredibly rewarding and, over the past 7 years, I have had the privilege of leading a conservation science team providing excellent science to support conservation and ecosystem management (Fig. 2). There have been challenges through those years with departmental changes and the amalgamation of science and conservation. Although I thought I had a good understanding of conservation and management, being a scientist embedded in a conservation agency, taking over responsibility for the conservation program of the department opened my eyes to the complexity of the policy–management interface, and I came to see the role of science in a new light. This came into sharp focus for me in November 2015 when extensive lightning-sparked fires on the southern coast destroyed 90% of the habitat of our most threatened bird, the western ground parrot, and the most threatened mammal, Gilbert’s potoroo. We held a workshop to garner input from a range of experts, and, as a scientist, it was clear to me that a range of actions was needed; however, as a manager, with responsibility for 625 threatened plants and animals and limited resources, the decisions weighed on my mind. More recently, the amalgamation of science teams from a conservation agency and three conservation orientated natural attractions is providing new challenges and opportunities to make a difference in applied science to support conservation. Life is never dull!
I have been privileged to have the support of three great Director Generals who have supported me throughout my leadership journey; they all have different styles of leadership and I have learnt different aspects from each of them. And, along the journey, I have also had the fantastic support of my Assistant Directors, Dr Fran Stanley who also has a PhD but went directly into the management sphere, and Dr Stephen van Leeuwen, a Noongar man who has shown the way for Indigenous engagement in science.
Learnings along the way
In 2014 and 2015, I was invited to give presentations at women in leadership conferences and have reflected on the key features of my journey and what advice I would give to young women coming through the system. I offer some thoughts here on the basis of my own experience.
The token woman
In my early days in the State conservation agency, as the most senior woman in the Science Division and one of the most senior women in the department, I was frequently ‘invited’ to serve on selection panels and other fora because they needed a woman. I was sometimes asked why I agreed to so many of these requests to be what some considered to be a ‘token woman’; however, I found that they were fantastic opportunities to get to know staff in the department that I wouldn’t normally associate with, and also to get to know the work of a large and complex department. My message is: don’t take offence, use these situations as opportunities and make the most of them, as you never know what they will lead to. In 2000, I came to the attention of the then new Director General through being on a selection panel with him, and, a few months later, he provided me with the opportunity to act as Director, even though there were three senior managers who would have expected to be given that opportunity. If I hadn’t been on that selection panel, I may not have made the transition into management and leadership.
Role models
I can’t say I had any prominent female role models, and, for all of my career, I have been in male dominated work environments. Although there were senior females in CSIRO, such as Liz Dennis and Adrienne Clark, I didn’t know them myself; nonetheless, I knew that doing what you wanted was possible, and it didn’t really occur to me that I couldn’t build a career in science. I do very much acknowledge the male colleagues who saw skills in me that I hadn’t necessarily seen myself and provided opportunities to explore those skills, such as a Wally Cox, the Director General who gave me the opportunity to act as Director, even though I was only a mid-career officer; Glen Kile, Chief of CSIRO Forestry and Forest Products, who gave me an early opportunity to contribute to organisational change when he asked me to be a staff representative in the change process for amalgamation of two CSIRO divisions; Neil Burrows, Director of Science who provided me the opportunities to develop leadership skills and ‘sponsored’ me through the process; Bob Hill, Dean of Science at University of Adelaide, who invited me to work with the ARC funded Environmental Futures Network that opened up opportunities for national collaboration; John McGrath who gave me my first opportunity to manage a group of staff, even though I was located in another building and knew little of the practicalities of forest management; and, more recently, Jim Sharp, the Director General who gave me the opportunity to broaden my horizons into policy and take responsibility for delivery of conservation in the department.
Carpe diem
To be successful at anything requires motivation and resilience. It doesn’t mean that there won’t be difficulties, because there will be difficult times, and times when you need to pick yourself up and keep going regardless. I have had many challenges and times when I have felt completely deflated, but I have kept my eye on the goal and just kept at it or found ways around the problem. In most cases, the ones that stand out are not the technical challenges, which usually just require a logical evidence based approach, but rather the people challenges because none of us work in isolation; more and more we work in teams and collaborations, and these require a special set of so called ‘softer skills’ that we as women are endowed with. Use your intuition, people skills and capacity to build rapport to build and engage in fantastic teams and you will reap the benefits. I have had the privilege of working with so many great colleagues over the years, and still have a great team of young, enthusiastic plant and animal genetics staff that achieve so much more together than we could ever do separately. Although there is a very strong streak of competitiveness in science, as we compete for things like funds, positions and ideas, collaborative approaches allow everyone to gain and, collectively, we deliver better outcomes. In my experience, there are two key elements that make successful collaborations: enthusiastic engagement and mutual respect for everyone’s contributions.
Self-doubt
We all have it (at least as women we do), but it does not need to be a limiting factor; it can be empowering, because it keeps you grounded and realistic. When I was first asked to act as Director in 2000, and, again in 2013 when I was asked to take on the responsibility for conservation as well as science, I did what most women do, sold myself short and expressed all my perceived limitations. However, I then pulled myself up, agreed to take it on and give it my best shot, and I found it was nowhere near as daunting as I had thought.
Change
As they say, change is the new constant. We will always be in situations of change, whether that is in developing new methodologies or concepts, or in organisational change, and my recommendation is to embrace it. There are three particular aspects of change management that I have taken from leadership courses and that I have found particularly useful in effectively navigating these, often stressful, situations. The phrase ‘change is threatening when done to us but exciting when done by us’ sums up a critical component of change in terms of engagement; in change situations, get involved, as it’s only when we participate and engage that we can influence the future. The phrase ‘honour the past and embrace the future’ resonates with me because it acknowledges that we don’t need to leave the past behind, but rather, we carry it forward with us as we build the new future. The third strategy is the ‘four doors’ concept. In any change situation, there are four doors. One represents what we couldn’t do before and we still can’t, another represents what we could do before and still can; both of these present little change but are often overlooked and not acknowledged. Then there are the other two doors, one representing the things we could do before but can’t now and these are the things we need to let go of, and the fourth door represents the things we couldn’t do before and now can; these are the things to embrace. Whether you are leading or responding to change, operating from these positions will make you a change agent and a valued member of the team.
Eyes on the goal
Stay focused on what you are seeking to achieve in the context of self-awareness. There will be challenges wherever you are and whatever you do, and your response often depends on your level of self-awareness and emotional intelligence. In general, the days of overt discrimination are gone and I can thankfully say that I have experienced direct discrimination on only two occasions. But, as a competent woman in a male dominated environment, I have seen and personally experienced the double standards, bullying and power games that you read about in any ‘women in management’ book, and I know how challenging it is to stay collegiate and focused on outcomes when faced with this divisive behaviour. I realise that there is still much unconscious bias in our scientific workplaces at all levels, and we will only move beyond it by understanding it together. Mark Lonsdale, when Chief of CSIRO Ecosystem Science a few years ago, ran some unconscious bias workshops, and I was privileged to have some discussions with several of my long-standing male colleagues in CSIRO following those workshops when they graciously shared a new appreciation of their own unconscious biases and challenged me on mine. My message is to stay focused on the outcome you’re seeking. I’ve learnt that it is OK to internally hold our often negative and justifiable response to a situation for a short while (suppressing it is not healthy) and use it to build our own self-awareness, but then to gently let it go and keep working towards the outcome we want.
Concluding remarks
I’ve had a great career where I have enjoyed going to work every day. I’ve learnt heaps, explored lots, been puzzled much, met great people, had a multitude of opportunities, contributed to key initiatives and been well challenged. A younger female colleague recently said that I make it seem easy. It has not been easy, but it has been very rewarding and hugely satisfying. I hope young women today starting careers in conservation science will persist through the crunch points where women drop out of the system, and find a satisfying career where they too can make a difference.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgements
I am indebted to all the colleagues, post-doctoral fellows and students who I have had the pleasure of working with. I particularly thank Bronwyn Macdonald who has shared the journey with me as my technical officer for 20 years and whose friendship I value deeply.
References
Allendorf, F. W., Hohenlohe, P. A., and Luikart, G. (2010). Genomics and the future of conservation genetics. Nature Reviews. Genetics 11, 697–709.| Genomics and the future of conservation genetics.Crossref | GoogleScholarGoogle Scholar |
Binks, R. M., Millar, M. A., and Byrne, M. (2015a). Contrasting patterns of clonality and fine-scale genetic structure in two rare sedges with differing geographic distributions. Heredity 115, 235–242.
| Contrasting patterns of clonality and fine-scale genetic structure in two rare sedges with differing geographic distributions.Crossref | GoogleScholarGoogle Scholar |
Binks, R. M., O’Brien, M., Macdonald, B., Maslin, B., and Byrne, M. (2015b). Genetic entities and hybridization within the Acacia microbotrya species complex in Western Australia. Tree Genetics & Genomes 11, 65.
| Genetic entities and hybridization within the Acacia microbotrya species complex in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Bissett, A., Fitzgerald, A., Meintjes, T., Mele, P. M., Reith, F., Dennis, P. G., Breed, M. F., Brown, B., Brown, M. V., Brugger, J., Byrne, M., Caddy-Retalic, S., Carmody, B., Coates, D. J., Correa, C. C., Ferrari, B. C., Gupta, V. V. S. R., Hamonts, K., Haslem, A., Hugenholtz, P., Karan, M., Koval, J., Lowe, A. J., Macdonald, S., McGrath, L., Martin, D., Morgan, M., North, K. I., Pendal, E., Paungfoo-Lonhienne, C., Philips, L., Pirzl, R., Powell, J. R., Ragan, M. A., Schmidt, S., Seymour, N., Snape, I., Stephen, J. R., Stevens, M., Tinning, M., Williams, K., Yeoh, Y. K., Zammit, C. M., and Young, A. (2016). Introducing BASE: the biomes of Australian soil environments soil microbial diversity database. GigaScience 5, 1–11.
| Introducing BASE: the biomes of Australian soil environments soil microbial diversity database.Crossref | GoogleScholarGoogle Scholar |
Bowman, D. M. J. S., Brown, G. K., Braby, M. F., Brown, J. R., Cook, L. G., Crisp, M. D., Ford, F., Haberle, S., Hughes, J., Isagi1, Y., Joseph, L., McBride, J., Nelson, G., and Ladiges, P. Y. (2010). Biogeography of the Australian monsoon tropics. Journal of Biogeography 37, 201–216.
| Biogeography of the Australian monsoon tropics.Crossref | GoogleScholarGoogle Scholar |
Broadhurst, L., Byrne, M., Craven, L., and Lepschi, B. (2004). Genetic congruence with new species boundaries in the Melaleuca uncinata complex (Myrtaceae). Australian Journal of Botany 52, 729–737.
| Genetic congruence with new species boundaries in the Melaleuca uncinata complex (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Byrne, M. (1999). High genetic identities between three oil mallee taxa, Eucalyptus kochii subsp. kochii, subsp. plenissima and E. horistes, based on nuclear RFLP analysis. Heredity 82, 205–211.
| High genetic identities between three oil mallee taxa, Eucalyptus kochii subsp. kochii, subsp. plenissima and E. horistes, based on nuclear RFLP analysis.Crossref | GoogleScholarGoogle Scholar |
Byrne, M. (2004). Recognition of Eucalyptus quaerenda (Myrtaceae) at specific rank. Nuytsia 15, 321–323.
Byrne, M. (2007). Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora. Australian Journal of Botany 55, 316–325.
| Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora.Crossref | GoogleScholarGoogle Scholar |
Byrne, M. (2008). Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quaternary Science Reviews 27, 2576–2585.
| Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., and Hines, B. (2004). Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae). Australian Journal of Botany 52, 459–470.
| Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae).Crossref | GoogleScholarGoogle Scholar |
Byrne, M., and Hopper, S. D. (2008). Granite outcrops as ancient islands in old landscapes: evidence from the phylogeography and population genetics of Eucalyptus caesia in Western Australia. Biological Journal of the Linnean Society. Linnean Society of London 93, 177–188.
| Granite outcrops as ancient islands in old landscapes: evidence from the phylogeography and population genetics of Eucalyptus caesia in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., and Macdonald, B. (2000). Phylogeography and conservation of three oil mallee taxa Eucalyptus kochii ssp. kochii, ssp. plenissima and E. horistes. Australian Journal of Botany 48, 305–312.
| Phylogeography and conservation of three oil mallee taxa Eucalyptus kochii ssp. kochii, ssp. plenissima and E. horistes.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., and Moran, G. F. (1994). Population divergence in the chloroplast genome of Eucalyptus nitens. Heredity 73, 18–28.
| Population divergence in the chloroplast genome of Eucalyptus nitens.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., and Stone, L. M. (2011). The need for ‘duty of care’ when introducing new plants for agriculture. Current Opinion in Environmental Sustainability 3, 50–54.
| The need for ‘duty of care’ when introducing new plants for agriculture.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Murrell, J. C., Allen, B., and Moran, G. F. (1995). An integrated genetic linkage map for Eucalyptus nitens. Theoretical and Applied Genetics 91, 869–875.
Byrne, M., Marquez-Garcia, I., Uren, T., and Moran, G. F. (1996). Conservation and genetic diversity of microsatellites in the genus Eucalyptus. Australian Journal of Botany 44, 331–341.
| Conservation and genetic diversity of microsatellites in the genus Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Murrell, J. C., Owen, J. V., Kriedemann, P., Williams, E. R., and Moran, G. (1997a). Identification and mode of action of quantitative trait loci affecting seedling height and leaf area in Eucalyptus nitens. Theoretical and Applied Genetics 94, 674–681.
| Identification and mode of action of quantitative trait loci affecting seedling height and leaf area in Eucalyptus nitens.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Murrell, J. C., Owen, J. V., Williams, E. R., and Moran, G. (1997b). Identification of quantitative trait loci influencing frost tolerance in Eucalyptus nitens. Theoretical and Applied Genetics 95, 975–979.
| Identification of quantitative trait loci influencing frost tolerance in Eucalyptus nitens.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Waycott, M., Hobbs, A. A., and James, S. H. (1997c). Variation in ribosomal DNA within and between populations of Isotoma petraea and Macrozamia riedlei. Heredity 79, 578–583.
| Variation in ribosomal DNA within and between populations of Isotoma petraea and Macrozamia riedlei.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Parrish, T., and Moran, G. F. (1998). RFLP diversity in the nuclear genome of Eucalyptus nitens. Heredity 81, 225–233.
| RFLP diversity in the nuclear genome of Eucalyptus nitens.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Macdonald, B., and Coates, D. (2002). Phylogeographic patterns within the Acacia acuminata (Myrtaceae) complex in Western Australia. Journal of Evolutionary Biology 15, 576–587.
| Phylogeographic patterns within the Acacia acuminata (Myrtaceae) complex in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Macdonald, B., and Brand, J. (2003). Phylogeography and divergence in the chloroplast genome of Western Australian sandalwood (Santalum spicatum). Heredity 91, 389–395.
| Phylogeography and divergence in the chloroplast genome of Western Australian sandalwood (Santalum spicatum).Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Elliott, C. P., Yates, C., and Coates, D. J. (2007). Extensive pollen dispersal in a bird-pollinated shrub, Calothamnus quadrifidus, in a fragmented landscape. Molecular Ecology 16, 1303–1314.
| Extensive pollen dispersal in a bird-pollinated shrub, Calothamnus quadrifidus, in a fragmented landscape.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Elliott, C., Yates, C., and Coates, D. (2008a). Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conservation Genetics 9, 97–105.
| Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Yeates, D. K., Joseph, L., Kearney, M., Bowler, J., Williams, M. A. J., Cooper, S., Donnellan, S. C., Keogh, J. S., Leys, R., Melville, J., Murphy, D. J., Porch, N., and Wyrwoll, K.-H. (2008b). Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Molecular Ecology 17, 4398–4417.
| Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Stone, L., and Millar, M. (2009). Environmental risk in agroforestry. In ‘Agroforestry for Natural Resource Management’. (Eds I. Nuberg, B. George, and J. Reid.) pp. 107–126. (CSIRO Publishing: Melbourne.)
Byrne, M., Steane, D. A., Joseph, L., Yeates, D. K., Jordan, G. J., Crayn, D., Aplin, K., Cantrill, D. J., Cook, L. G., Crisp, M. D., Keogh, J. S., Melville, J., Moritz, C., Porch, N., Sniderman, J. M. K., Sunnucks, P., and Weston, P. (2011a). Decline of a biome: contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38, 1635–1656.
| Decline of a biome: contraction, fragmentation, extinction and invasion of the Australian mesic zone biota.Crossref | GoogleScholarGoogle Scholar |
Byrne, M., Stone, L., and Millar, M. A. (2011b). Assessing genetic risk in revegetation. Journal of Applied Ecology 48, 1365–1373.
| Assessing genetic risk in revegetation.Crossref | GoogleScholarGoogle Scholar |
Chambers, G., Curtis, C., Millar, C., Huynen, L., and Lambert, D. (2014). DNA fingerprinting in zoology: past, present and future. Investigative Genetics 5, 3.
| DNA fingerprinting in zoology: past, present and future.Crossref | GoogleScholarGoogle Scholar |
Clarke, L. J., Jardine, D., Byrne, M., Shepherd, K., and Lowe, A. J. (2012). Significant population genetic structure detected for a new and highly restricted species of Atriplex (Chenopodiaceae) from Western Australia, and implications for conservation management. Australian Journal of Botany 60, 32–41.
| Significant population genetic structure detected for a new and highly restricted species of Atriplex (Chenopodiaceae) from Western Australia, and implications for conservation management.Crossref | GoogleScholarGoogle Scholar |
Coates, D. J., McArthur, S. L., and Byrne, M. (2015). Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae). Biological Conservation 192, 353–360.
| Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae).Crossref | GoogleScholarGoogle Scholar |
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., and Mitchell, S. E. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379.
| A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species.Crossref | GoogleScholarGoogle Scholar |
Erlich, H. A., Gelfland, D. H., and Sninsky, J. J. (1991). Recent advances in the polymerase chain reaction. Science 252, 1643–1651.
| Recent advances in the polymerase chain reaction.Crossref | GoogleScholarGoogle Scholar |
Hadrys, H., Balick, M., and Schierwater, B. (1992). Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Molecular Ecology 1, 55–63.
| Applications of random amplified polymorphic DNA (RAPD) in molecular ecology.Crossref | GoogleScholarGoogle Scholar |
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359, 183–195.
| Genetic consequences of climatic oscillations in the Quaternary.Crossref | GoogleScholarGoogle Scholar |
Hoffmann, A., Griffin, P., Dillon, S., Catullo, R., Rane, R., Byrne, M., Jordan, R., Oakeshott, J., Weeks, A., Joseph, L., Lockhart, P., Borevitz, J., and Sgrò, C. (2015). A framework for incorporating evolutionary genomics into biodiversity conservation and management. Climate Change Responses 2, 1–23.
| A framework for incorporating evolutionary genomics into biodiversity conservation and management.Crossref | GoogleScholarGoogle Scholar |
James, S. H. (2000). Genetic systems in the south-west flora: implications for conservation strategies for Australian plant species. Australian Journal of Botany 48, 341–347.
| Genetic systems in the south-west flora: implications for conservation strategies for Australian plant species.Crossref | GoogleScholarGoogle Scholar |
Jeffreys, A., Wilson, V., and Thein, S. L. (1985a). Hypervariable ‘minisatellite’ regions in human DNA. Nature 314, 67–73.
| Hypervariable ‘minisatellite’ regions in human DNA.Crossref | GoogleScholarGoogle Scholar |
Jeffreys, A. J., Wilson, V., and Thein, S. L. (1985b). Individual-specific ‘fingerprints’ of human DNA. Nature 316, 76–79.
| Individual-specific ‘fingerprints’ of human DNA.Crossref | GoogleScholarGoogle Scholar |
Keppel, G., Van Niel, K. P., Wardell-Johnson, G. W., Yates, C. J., Byrne, M., Mucina, L., Schut, A. G. T., Hopper, S. D., and Franklin, S. E. (2012). Refugia: identifying and understanding safe havens for biodiversity from climate change. Global Ecology and Biogeography 21, 393–404.
| Refugia: identifying and understanding safe havens for biodiversity from climate change.Crossref | GoogleScholarGoogle Scholar |
Llorens, T. M., Byrne, M., Yates, C. J., Nistelberger, H. M., and Coates, D. J. (2012). Evaluating the influence of different aspects of habitat fragmentation on mating patterns and pollen dispersal in the bird pollinated Banksia sphaerocarpa var. caesia. Molecular Ecology 21, 314–328.
| Evaluating the influence of different aspects of habitat fragmentation on mating patterns and pollen dispersal in the bird pollinated Banksia sphaerocarpa var. caesia.Crossref | GoogleScholarGoogle Scholar |
Lynch, A. J. J., Thackway, R., Specht, A., Beggs, P. J., Brisbane, S., Burns, E., Byrne, M., Capon, S. J., Casanova, M., Clarke, P. A., Davies, J. M., Dovers, S., Dwyer, R., Ens, E., Fisher, D. O., Flanigan, M., Garnier, E., Guru, S., Kilminster, K., Locke, J., Mac Nally, R., McMahon, K. M., Mitchell, P. J., Pierson, J., Rodgers, E. M., Russell-Smith, J., Udy, J., and Waycott, M. (2015). Transdisciplinary synthesis for ecosystem science, policy and management: the Australian experience. The Science of the Total Environment 534, 173–184.
| Transdisciplinary synthesis for ecosystem science, policy and management: the Australian experience.Crossref | GoogleScholarGoogle Scholar |
Maniatis, T., Ftitsch, E. F., and Sambrook, J. (1982). ‘Molecular Cloning: a Laboratory Manual.’ 1st edn. (Cold Spring Harbor Press: Cold Spring Harbor, NY.)
McLean, E. H., Steane, D. A., Prober, S. M., Stock, W. D., Potts, B. M., Vaillancourt, R. E., and Byrne, M. (2014). Plasticity of functional traits varies clinally along a rainfall gradient in Eucalyptus tricarpa. Plant, Cell & Environment 37, 1440–1451.
| Plasticity of functional traits varies clinally along a rainfall gradient in Eucalyptus tricarpa.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., and Byrne, M. (2013). Cryptic divergent lineages of Pultenaea pauciflora M.B.Scott (Fabaceae, Mirbelieae) exhibit different patterns of evolutionary history. Biological Journal of the Linnaean Society 108, 871–881.
| Cryptic divergent lineages of Pultenaea pauciflora M.B.Scott (Fabaceae, Mirbelieae) exhibit different patterns of evolutionary history.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Byrne, M., Nuberg, I., and Sedgley, M. (2008). High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites. Tree Genetics & Genomes 4, 367–377.
| High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Byrne, M., and Coates, D. (2010). The maintenance of disparate levels of clonality, genetic diversity and genetic differentiation in disjunct subspecies of the rare Banksia ionthocarpa. Molecular Ecology 19, 4217–4227.
| The maintenance of disparate levels of clonality, genetic diversity and genetic differentiation in disjunct subspecies of the rare Banksia ionthocarpa.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Byrne, M., and O’Sullivan, W. O. (2011). Defining entities in the Acacia saligna (Fabaceae) species complex using a population genetics approach. Australian Journal of Botany 59, 137–148.
| Defining entities in the Acacia saligna (Fabaceae) species complex using a population genetics approach.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Byrne, M., Nuberg, I., and Sedgley, M. (2012). High levels of genetic contamination in remnant populations of Acacia saligna from a genetically divergent planted stand. Restoration Ecology 20, 260–267.
| High levels of genetic contamination in remnant populations of Acacia saligna from a genetically divergent planted stand.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Coates, D. J., and Byrne, M. (2013). Genetic connectivity and diversity in inselberg populations of Acacia woodmaniorum, a rare endemic plant of the Yilgarn Craton banded iron formations. Heredity 111, 437–444.
| Genetic connectivity and diversity in inselberg populations of Acacia woodmaniorum, a rare endemic plant of the Yilgarn Craton banded iron formations.Crossref | GoogleScholarGoogle Scholar |
Millar, M. A., Coates, D. J., and Byrne, M. (2014). Extensive long-distance pollen dispersal and highly outcrossed mating in historically small and disjunct populations of Acacia woodmaniorum (Fabaceae), a rare banded iron formation endemic. Annals of Botany 114, 961–971.
| Extensive long-distance pollen dispersal and highly outcrossed mating in historically small and disjunct populations of Acacia woodmaniorum (Fabaceae), a rare banded iron formation endemic.Crossref | GoogleScholarGoogle Scholar |
Moritz, C., and Brown, W. M. (1986). Tandem duplication of D-loop and ribosomal RNA sequences in lizard mitochondrial DNA. Science 233, 1425–1427.
| Tandem duplication of D-loop and ribosomal RNA sequences in lizard mitochondrial DNA.Crossref | GoogleScholarGoogle Scholar |
Moro, D., Byrne, M., Kennedy, M., Campbell, S., and Tizard, M. (2018). Identifying knowledge gaps for gene drive research to control invasive animal species: the next CRISPR step. Global Ecology and Conservation 13, e00363.
| Identifying knowledge gaps for gene drive research to control invasive animal species: the next CRISPR step.Crossref | GoogleScholarGoogle Scholar |
Myburg, A. A., Grattapaglia, D., Tuskan, G. A., Hellsten, U., Hayes, R. D., Grimwood, J., Jenkins, J., Lindquist, E., Tice, H., Bauer, D., Goodstein, D., Dubchak, I., Poliakov, A., Mizrachi, E., Kullan, A. R. K., Hussey, S. G., Pinard, D., van der Merwe, K., Singh, P., van Jaarsveld, I., Silva-Junior, O. B., Togawa, R., Pappas, M. R., Faria, D. A., Sansaloni, C. P., Petroli, C. D., Yang, X., Ranjan, P., Tschaplinski, T., Ye, C.-Y., Li, T., Sterck, L., Vanneste, K., Murat, F., Soler, M., San Clemente, H., Saidi, N., Cassan-Wang, H., Dunand, C., Hefer, C. A., Bornberg-Bauer, E., Kersting, A. R., Vining, K., Amarashighe, V., Ranik, M., Naithani, S., Elser, J., Boyd, A. E., Liston, A., Spatafora, J. W., Dharmwardhana, P., Raja, R., Sullivan, C., Romanel, E., Alves-Ferreira, M., Külheim, C., Foley, W., Carocha, V., Paiva, J., Kudrna, D., Brommonschenkel, S. H., Pasquali, G., Byrne, M., Rigault, P., Tibbits, J., Spokevicius, A., Jones, R. C., Steane, D. A., Vaillancourt, R. E., Potts, B. M., Joubert, F., Barry, K., Pappas, G. J., Strauss, S. H., Jaiswal, P., Grima-Pettenati, J., Salse, J., Van de Peer, Y., Rokhsar, D., and Schmutz, J. (2014). The genome of Eucalyptus grandis. Nature 510, 356–362.
| The genome of Eucalyptus grandis.Crossref | GoogleScholarGoogle Scholar |
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S., and Hoekstra, H. E. (2012). Double Digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7, e37135.
| Double Digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species.Crossref | GoogleScholarGoogle Scholar |
Prober, S. M., Byrne, M., McLean, E. H., Steane, D. A., Potts, B. M., Vaillancourt, R. E., and Stock, W. D. (2015). Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration. Frontiers in Ecology and Evolution 3, 65.
| Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration.Crossref | GoogleScholarGoogle Scholar |
Renfree, M. B., Papenfuss, A. T., Deakin, J. E., Lindsay, J., Heider, T., Belov, K., Rens, W., Waters, P. D., Pharo, E. A., Shaw, G., Wong, E. S. W., Lefèvre, C. M., Nicholas, K. R., Kuroki, Y., Wakefield, M. J., Zenger, K. R., Wang, C., Ferguson-Smith, M., Nicholas, F. W., Hickford, D., Yu, H., Short, K. R., Siddle, H. V., Frankenberg, S. R., Chew, K. Y., Menzies, B. R., Stringer, J. M., Suzuki, S., Hore, T. A., Delbridge, M. L., Patel, H. R., Mohammadi, A., Schneider, N. Y., Hu, Y., O’Hara, W., Al Nadaf, S., Wu, C., Feng, Z-P., Cocks, B. G., Wang, J., Flicek, P., Searle, S. M. J., Fairley, S., Beal, K., Herrero, J., Carone, D. M., Suzuki, Y., Sugano, S., Toyoda, A., Sakaki, Y., Kondo, S., Nishida, Y., Tatsumoto, S., Mandiou, I., Hsu, A., McColl, K. A., Lansdell, B., Weinstock, G., Kuczek, E., McGrath, A., Wilson, P., Men, A., Hazar-Rethinam, M., Hall, A., Davis, J., Wood, D., Williams, S., Sundaravadanam, Y., Muzny, D. M., Jhangiani, S. N., Lewis, L. R., Morgan, M. B., Okwuonu, G. O., Ruiz, S. J., Santibanez, J., Nazareth, L., Cree, A., Fowler, G., Kovar, C. L., Dinh, H. H., Joshi, V., Jing, C., Lara, F., Thornton, R., Chen, L., Deng, J., Liu, Y., Shen, J. Y., Song, X.-Z., Edson, J., Troon, C., Thomas, D., Stephens, A., Yapa, L., Levchenko, T., Gibbs, R. A., Cooper, D. W., Speed, T. P., Fujiyama, A., Graves, J. A. M., O’Neill, R. J., Pask, A. J., Forrest, S. M., and Worley, K. C. (2011). Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biology 12, R81.
| Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development.Crossref | GoogleScholarGoogle Scholar |
Sampson, J. F., and Byrne, M. (2008). Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Molecular Ecology 17, 2769–2781.
| Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba.Crossref | GoogleScholarGoogle Scholar |
Sampson, J. F., Byrne, M., Yates, C. J., Gibson, N., Thavornkanlapachai, R., Stankowski, S., Macdonald, B., and Bennett, I. (2014). Contemporary pollen-mediated gene immigration reflects the historical isolation of a rare shrub in a fragmented landscape. Heredity 112, 172–181.
| Contemporary pollen-mediated gene immigration reflects the historical isolation of a rare shrub in a fragmented landscape.Crossref | GoogleScholarGoogle Scholar |
Sansaloni, C. P., Petroli, C. D., Jaccoud, D., Carling, J., Detering, F., Grattapaglia, D., and Kilian, A. (2011). Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proceedings 5, P54.
| Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus.Crossref | GoogleScholarGoogle Scholar |
Shepherd, K. A., Perkins, A., Collins, J., Byrne, M., and Thiele, K. R. (2013). Morphological and molecular evidence supports the recognition of a new subspecies of the critically endangered Pityrodia scabra (Lamiaceae). Australian Systematic Botany 26, 1–12.
| Morphological and molecular evidence supports the recognition of a new subspecies of the critically endangered Pityrodia scabra (Lamiaceae).Crossref | GoogleScholarGoogle Scholar |
Shepherd, K. A., Thiele, K. R., Sampson, J., Coates, D., and Byrne, M. (2015). A rare, new species of Atriplex (Chenopodiaceae) comprising two genetically distinct but morphologically cryptic populations in arid Western Australia: implications for taxonomy and conservation. Australian Systematic Botany 28, 234–245.
| A rare, new species of Atriplex (Chenopodiaceae) comprising two genetically distinct but morphologically cryptic populations in arid Western Australia: implications for taxonomy and conservation.Crossref | GoogleScholarGoogle Scholar |
Steane, D. A., Potts, B. M., McLean, E., Prober, S. M., Stock, W. D., Vaillancourt, R. E., and Byrne, M. (2014). Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology 23, 2500–2513.
| Genome-wide scans detect adaptation to aridity in a widespread forest tree species.Crossref | GoogleScholarGoogle Scholar |
Steane, D. A., Potts, B. M., McLean, E., Collins, L., Prober, S. M., Stock, W. D., Vaillancourt, R. E., and Byrne, M. (2015). Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt. Tree Genetics & Genomes 11, 33.
| Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt.Crossref | GoogleScholarGoogle Scholar |
Steane, D. A., McLean, E., Potts, B. M., Prober, S. M., Stock, W. D., Stylianou, V. M., Vaillancourt, R. E., and Byrne, M. (2017a). Evidence for adaptation and acclimation of a widespread eucalypt of semi-arid Australia. Biological Journal of the Linnean Society. Linnean Society of London 121, 484–500.
| Evidence for adaptation and acclimation of a widespread eucalypt of semi-arid Australia.Crossref | GoogleScholarGoogle Scholar |
Steane, D. A., Potts, B. M., McLean, E., Collins, L., Holland, B. R., Prober, S. M., Stock, W. D., Vaillancourt, R. E., and Byrne, M. (2017b). Genome scans across three eucalypts indicate that adaptation is a genome wide phenomenon. Genome Biology and Evolution 9, 253–265.
| Genome scans across three eucalypts indicate that adaptation is a genome wide phenomenon.Crossref | GoogleScholarGoogle Scholar |
Tanksley, S. D., and Orton, T. J. (1983). ‘Isozymes in Plant Genetics and Breeding Part B.’ (Elsevier Science Publishers: Amsterdam.)
Tapper, S.-L., Byrne, M., Yates, C. J., Keppel, G., Hopper, S. D., Van Niel, K., Schut, A. G. T., Mucina, L., and Wardell-Johnson, G. W. (2014a). Isolated with persistence or dynamically connected?: genetic patterns in a common granite outcrop endemic. Diversity & Distributions 20, 987–1001.
| Isolated with persistence or dynamically connected?: genetic patterns in a common granite outcrop endemic.Crossref | GoogleScholarGoogle Scholar |
Tapper, S.-L., Byrne, M., Yates, C. J., Keppel, G., Hopper, S. D., Van Niel, K., Schut, A. G. T., Mucina, L., and Wardell-Johnson, G. W. (2014b). Prolonged isolation and persistence of a common endemic on granite outcrops in both mesic and semi-arid environments in south-western Australia. Journal of Biogeography 41, 2032–2044.
| Prolonged isolation and persistence of a common endemic on granite outcrops in both mesic and semi-arid environments in south-western Australia.Crossref | GoogleScholarGoogle Scholar |
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183, 4–18.
| Environmental DNA: an emerging tool in conservation for monitoring past and present biodiversity.Crossref | GoogleScholarGoogle Scholar |
Vos, P., Hogers, R., Bleeker, M., Reijans, T., van de Lee, M., Hornes, A., Frijters, A., Jerina, P., Peleman, J., Kuiper, M., and Zabeau, M. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23, 4407–4414.
| AFLP: a new technique for DNA fingerprinting.Crossref | GoogleScholarGoogle Scholar |
Walker, E., Byrne, M., Macdonald, B., Nicolle, D., and McComb, J. (2009). Clonality and hybrid origin of the rare Eucalyptus bennettiae (Myrtaceae) in Western Australia. Australian Journal of Botany 57, 180–188.
| Clonality and hybrid origin of the rare Eucalyptus bennettiae (Myrtaceae) in Western Australia.Crossref | GoogleScholarGoogle Scholar |
Walker, E., McComb, J., and Byrne, M. (2018). Genetic and morphological evidence supports the hybrid status of Adenanthos cunninghamii (now Adenanthos × cunninghamii). South African Journal of Botany , .
| Genetic and morphological evidence supports the hybrid status of Adenanthos cunninghamii (now Adenanthos × cunninghamii).Crossref | GoogleScholarGoogle Scholar |
Wheeler, M., and Byrne, M. (2006). Congruence between phylogeographic patterns in cpDNA variation in Eucalyptus marginata (Myrtaceae) and geomorphology of the Darling Plateau. Australian Journal of Botany 54, 17–26.
| Congruence between phylogeographic patterns in cpDNA variation in Eucalyptus marginata (Myrtaceae) and geomorphology of the Darling Plateau.Crossref | GoogleScholarGoogle Scholar |
Williams, J. G., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18, 6531–6535.
| DNA polymorphisms amplified by arbitrary primers are useful as genetic markers.Crossref | GoogleScholarGoogle Scholar |



