Using integrative taxonomy to distinguish cryptic halfbeak species and interpret distribution patterns, fisheries landings, and speciation
Indiana J. Riley A B * , Joseph D. DiBattista C , John Stewart D , Hayden T. Schilling
A B * , Joseph D. DiBattista C , John Stewart D , Hayden T. Schilling  A B and Iain M. Suthers
A B and Iain M. Suthers  A B
A B
A Centre for Marine Science and Innovation, School of Biological, Earth and Environmental Sciences, University of New South Wales, Sydney, NSW 2052, Australia.
B Sydney Institute of Marine Science, Chowder Bay Road, Mosman, NSW 2088, Australia.
C Australian Museum Research Institute, Australian Museum, Sydney, NSW 2010, Australia.
D NSW Department of Primary Industries, Sydney Institute of Marine Science, Chowder Bay Road, Mosman, NSW 2088, Australia.
Marine and Freshwater Research 74(2) 125-143 https://doi.org/10.1071/MF22048
Submitted: 7 February 2022 Accepted: 6 December 2022 Published: 6 January 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution 4.0 International License (CC BY)
Abstract
Context: Species classification disputes can be resolved using integrative taxonomy, which involves the use of both phenotypic and genetic information to determine species boundaries.
Aims: Our aim was to clarify species boundaries of two commercially important cryptic species of halfbeak (Hemiramphidae), whose distributions overlap in south-eastern Australia, and assist fisheries management.
Methods: We applied an integrative taxonomic approach to clarify species boundaries and assist fisheries management.
Key results: Mitochondrial DNA and morphological data exhibited significant differences between the two species. The low level of mitochondrial DNA divergence, coupled with the lack of difference in the nuclear DNA, suggests that these species diverged relatively recently (c. 500 000 years ago) when compared with other species within the Hyporhamphus genus (>2.4 million years ago). Genetic differences between the species were accompanied by differences in modal gill raker counts, mean upper-jaw and preorbital length, and otolith shape.
Conclusions: On the basis of these genetic and morphological differences, as well as the lack of morphological intergradation between species along the overlapping boundaries of their geographical distributions, we propose that Hyporhamphus australis and Hyporhamphus melanochir remain valid species.
Implications: This study has illustrated the need for an integrative taxonomic approach when assessing species boundaries and has provided a methodological framework for studying other cryptic fish species in a management context.
Keywords: biogeography, diversity, fish, fisheries, garfish, genetics, otoliths, speciation.
Introduction
Systematics forms the backbone of many ecological fields, which allows policymakers to build conservation practices around species boundaries and identify endangered populations (Haig et al. 2006). Despite the importance of species classification, the existence of competing species concepts complicates the separation of similar species. Many widely accepted species concepts use competing diagnostic criteria that may establish different definitions of species boundaries and can result in contradictory species diagnoses (De Queiroz 2007). These conflicting opinions can result in taxonomic inflation, and surveys have shown that studies using the phylogenetic species concept hold a 48% higher species count than those using non-phylogenetic species concepts (Agapow et al. 2004). Problems with determining species boundaries are especially important for cryptic species, whose morphological forms are almost indistinguishable. Delimiting cryptic species boundaries is important because similar-looking species with different life-history traits may require different conservation or management tactics (Bickford et al. 2007). Cryptic fish species are common in marine ecosystems where their distributions can overlap (Knowlton 2000; Takahashi et al. 2020), and the management implications of cryptic species discrimination are crucial for commercially exploited marine fishes, as stock management practices may affect species in different ways.
The unified species concept defines a species simply as a ‘separately evolving metapopulation lineage’ and uses multiple diagnostic criteria to delimit species boundaries (De Queiroz 2007). By accepting the many different characteristics a lineage can acquire along the course of divergence as different lines of evidence to assess a species status, the unified species concept provides a solution to the discrepancies in cryptic species delimitation through integrative taxonomy (De Queiroz 2007, 2011). Integrative taxonomy aims to determine units of life from multiple complementary perspectives rather than relying on traditional morphological or more recent phylogenetic methods of species discrimination alone (Dayrat 2005). Integrative methods, combining both morphological and molecular traits, are increasingly being used to resolve the taxonomy of cryptic fish species (Delrieu-Trottin et al. 2022; Liggins et al. 2022). Examining the differences between cryptic fish species using multiple lines of evidence not only aids in resolving species concept disputes but can also confirm stock subdivision and connectivity from multiple standpoints (Izzo et al. 2017), thus ensuring that management practices are tailored to species-specific needs. For example, the cryptic species diagnosis of two commercially targeted wobbegong species in eastern Australia, namely Orectolobus ornatus and Orectolobus halei (Huveneers 2006), has allowed species-specific information regarding differences in distribution, reproduction, age, growth, and genetic structure to be incorporated into fisheries management (Corrigan et al. 2008, 2016; Huveneers et al. 2013).
Recent advances in genetic and morphological species-discrimination techniques, such as DNA barcoding and otolith shape analysis, can aid in resolving cryptic fish-species boundaries. DNA barcoding usually involves a comparison of the mitochondrial cytochrome c oxidase subunit I (COI) gene between different species, with the number of base-pair differences between COI sequences acting as an indication of the level of genetic separation between different species (Hebert et al. 2003). Indeed, this is possible because of the utility of the ‘barcoding gap’, where interspecific sequence variability for congeneric COI sequences is almost always greater than is intraspecific sequence variability (Meyer and Paulay 2005). Although COI barcoding can provide a clear means of cryptic-species discrimination when morphological evidence is sparse, the use of genetic evidence alone to confirm cryptic-species status without thorough taxonomic assessment has been criticised (Moritz and Cicero 2004) and barcoding is more widely accepted when used in conjunction with other lines of genetic, morphological, behavioural, or ecological data (Padial et al. 2010; DeSalle and Goldstein 2019). Otolith shape analysis, involving the comparison of inner-ear bone morphology, is well established as a stock discrimination technique to compare different populations within a single species group (Stransky 2014), and has become another tool employed alongside genetic and morphological data to better distinguish cryptic fish species (Nielsen et al. 2010; Zhuang et al. 2015).
Cryptic species are common within the globally distributed garfish (or ‘halfbeak’) genus Hyporhamphus. Of the 40 species within the Hyporhamphus genus, many can be distinguished only on the basis of slight differences in body shape or number of gill rakers (Supplementary Table S1). The small size of gill raker structures in Hyporhamphus species makes them difficult to count in the field, which poses problems for the discrimination of commercially exploited species and could hinder the implementation of species-specific management practices. One pair of commercially important cryptic Hyporhamphus species that face this diagnostic dilemma includes the Australian-endemic garfish Hyporhamphus australis (Steindachner) and Hyporhamphus melanochir (Valenciennes). At present, gill raker count is the only diagnostic criterion for delimiting them as unique species (Collette 1974). This makes it difficult for recreational and commercial fisheries to distinguish between H. australis and H. melanochir in southern New South Wales (NSW) waters where their distribution is thought to overlap (Collette 1974). Species-level classification is important for stock management, given documented life-history differences between the two species (Supplementary Table S2). Hyporhamphus australis is a smaller but faster-growing species that reaches maturity at a smaller size and younger age than H. melanochir (Jones et al. 2002; Stewart et al. 2005). Current minimum mesh-size restrictions for ocean garfish lampara nets in NSW protect H. australis juveniles below 20.1-cm fork length (FL) (Stewart et al. 2004); however, these management practices are not designed to protect immature H. melanochir stocks that may also be found in NSW waters.
Whereas the level of genetic differentiation between H. australis and H. melanochir has not been thoroughly explored (but see Noell et al. 2001 for brief notes on mitochondrial DNA or mtDNA comparison), newer methods of discrimination such as DNA barcoding and otolith shape analysis could clarify their species boundaries. Using a nuclear DNA marker alongside COI barcoding can further confirm the extent of their genetic separation and, if diagnostic base pair differences are present, identify hybrid individuals (Pavan-Kumar et al. 2016). Hybridisation near the town of Eden (37°S, southern NSW), where the distributions of H. australis and H. melanochir are likely to overlap, has been hypothesised from the collection of a single specimen with intermediate morphological traits (Collette 1974). However, further analysis of the levels of gene exchange between the species is necessary to support this assumption.
This study aims to explore genetic and morphological differences between H. melanochir and H. australis to determine the best method of discrimination between the two species and confirm their overlapping distributions in southern NSW. Using whole fish and tissue samples collected throughout the mainland Australian distributions of both species, we assessed differences in the body morphology and otolith shape between the two species alongside genetic differences in the mtDNA COI and nuclear TMO-4C4 regions. We hypothesise that this integrated taxonomic study will identify new genetic and morphological methods to better discriminate between H. australis and H. melanochir, and that an examination of the COI and TMO-4C4 regions between the species may indicate the presence of hybrid individuals.
Materials and methods
Sample collection and preparation
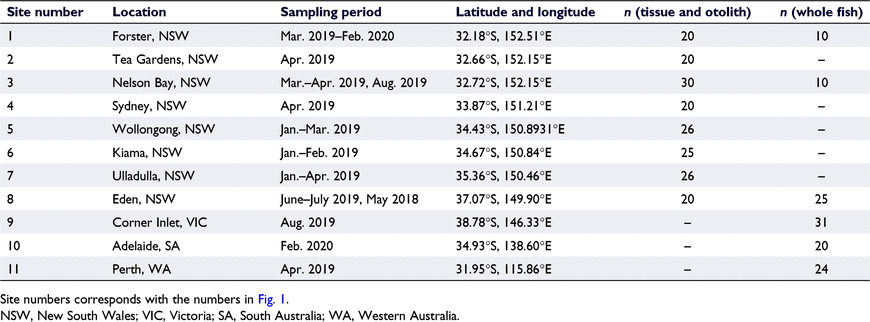
In total, 120 whole fish specimens were obtained from recreational and commercial catches across the mainland Australian distribution of H. australis and H. melanochir (Fig. 1, Table 1, Supplementary Table S3). All specimens were stored frozen at −20°C prior to analysis. An additional set of tissue and otolith samples was acquired from 187 fish (Table 1, Supplementary Table S4) collected by NSW Department of Primary Industry (NSW DPI) as part of their port-monitoring program between January and July of 2019. These samples were chosen to ensure a balanced mix of individuals from different age, sex, and size classes from each catch. All whole fish were processed at the Australian Museum (detailed methods and voucher numbers are outlined in the ‘Supplemental detailed sample preparation’ section in the Supplementary material).
|
|
|
All fish samples were obtained from the NSW DPI commercial-fishery port-monitoring program. The program is exempt from requiring Animal Care and Ethics Approval as all fish sampled have been captured and retained for sale by licenced commercial fishers working under approved fishing practices.
Genetic analysis – mitochondrial COI
DNA was extracted from a total of 307 tissue samples by using the ISOLATE II Genomic DNA Kit (Meridian Bioscience, Cincinnati, OH, USA) following the manufacturer’s protocol.
A 609-base pair (bp) segment of the mtDNA COI gene was amplified with a combination of Fish F1/F2 and Fish R2 primers (Supplementary Table S5) following polymerase chain reaction (PCR) conditions from Ward et al. (2005). All mtDNA amplification by PCR, gel visualisation, and Sanger sequencing were performed by the Ramaciotti Centre for Genomics at the University of New South Wales in Sydney, Australia. The DNA sequences were aligned, edited, and trimmed to the same length by using Geneious Prime (ver. 2020.2.4, see https://www.geneious.com) and deposited in the National Centre for Biotechnology Information (NCBI) GenBank nucleotide database (GenBank accession numbers MZ575768–MZ576070).
To determine a species ID for each fish and divide samples into species groups for downstream analysis, the trimmed and edited COI sequences were compared with reference sequences from GenBank for H. australis (GenBank accession number KX781932.1) and H. melanochir (GenBank accession number HQ956051.1) by using the Basic Local Alignment Search Tool (BLAST; Altschul et al. 1990). Initial comparison of these two sequences showed four consistent diagnostic base differences between the species. Only sequences with a 100% match to either of these reference sequences at these four diagnostic bases were assigned to a species group. Twelve sequences appeared to have mixed identity on the basis of these four diagnostic bases, including three sequences with double peaks on their chromatogram traces only at these base pair positions, and thus could not be assigned definitively to either species.
All sequences were exported in FASTA format and collapsed into haplotypes before conversion into ARLEQUIN and NEXUS file formats by using the online tool FaBox (see http://www.birc.au.dk/software/fabox; Villesen 2007). Unique haplotypes were then imported into jModelTest (ver. 2.1.10, see http://evomics.org/learning/phylogenetics/jmodeltest/; Guindon and Gascuel 2003; Darriba et al. 2012), which was used with an Akaike information criteria (AIC; Bozdogan 1987) to determine the best nucleotide substitution model for the sequence data. For the mtDNA haplotypes, the Tamura–Nei (TrN; Tamura and Nei 1993) model was selected for use in subsequent genetic analysis as required.
To test for differences in genetic diversity between H. australis and H. melanochir, ARLEQUIN (ver. 3.5.2.2, see http://cmpg.unibe.ch/software/arlequin35; Excoffier and Lischer 2010) was used to calculate haplotype (h) and nucleotide (π) diversity. These diversity indices were then used in an analysis of molecular variance (AMOVA) to determine global ΦST values with non-parametric permutations (n = 99 999). Pairwise ΦST statistics were generated to test the significance of differences between locations and then adjusted to correct for false discovery rate (as per Narum 2006). Fu’s Fs values (Fu 1997) were also calculated to test for deviations from neutral sequence evolution, with significance being tested using 99 999 permutations.
To visualise the relationship between haplotypes, the genetics software PopART (ver. 1.7, see https://popart.maths.otago.ac.nz/; Leigh and Bryant 2015) was used to construct a median-joining network, which was produced using algorithms and settings according to Bandelt et al. (1999).
The corrected average Kimura two-parameter (K2P) pairwise difference between the COI haplotypes of both species was calculated in ARLEQUIN and divided by the number of base pairs in the sequence to determine the mean K2P percentage difference between the COI sequences of both species. This difference was then used to estimate the time of divergence on the basis of a molecular-clock calibration rate for fish of 1.2% COI divergence per million years. This calibration rate is rooted in differences between the COI regions of sister taxa separated by the rise of the Isthmus of Panama c. 3.8 million years ago (Bermingham et al. 1997) and has been used to estimate the time of divergence for a variety of marine fish species (Lessios 2008; Tea et al. 2019; Delrieu-Trottin et al. 2022). To confirm that the K2P mean difference and time of divergence were supported by the model of nucleotide substitution that was chosen in jModelTest to best fit our species (i.e. Tamura–Nei), this approach was repeated using the corrected average TrN pairwise difference calculated in ARLEQUIN, which produced identical results (Supplementary Table S6).
To contextualise genetic differences within the genus, the mean K2P percentage difference and approximate time of divergence was then calculated for all other species within the Hyporhamphus genus with available reference sequences on GenBank and compared to our two study species. All COI sequences over 600 bp long for 12 Hyporhamphus species were downloaded from GenBank, then aligned and trimmed in Geneious Prime (Supplementary Table S7). Aligned sequences were then exported, collapsed into haplotypes, and converted to ARLEQUIN file format by using FaBox. This sequence-editing workflow was repeated for each species to construct a pairwise table of mean K2P percentage difference and approximate time since divergence between each species. These methods were then repeated using the corrected average TrN pairwise difference as above (Table S6).
Genetic analysis – nuclear TMO-4C4
A 450-bp segment of the nuclear DNA TMO-4C4 gene was amplified using primers TMO_f1_5 and TMO_r1_3 and PCR conditions outlined in Streelman and Karl (1997) prior to sequencing at the Ramaciotti Centre for Genomics (see the ‘Supplemental detailed genetic methods’ section in the Supplementary material and Table S5). All TMO-4C4 sequences were aligned, trimmed and visually inspected for base ambiguities in Geneious Prime and deposited in the NCBI GenBank nucleotide database (GenBank accession numbers MZ580145–MZ580399). Any alignments with ambiguous base calls were edited against their reverse complement for base confirmation, and all alignments with <70% high-quality bases were excluded from further analysis (n = 16). After exporting sequences in FASTA format, the allelic states of these nuclear sequences were inferred using the Bayesian algorithm PHASE (ver. 2.1, see https://stephenslab.uchicago.edu/phase/download.html; Stephens et al. 2001; Stephens and Donnelly 2003) implemented within the software DnaSP (ver. 6.12.03, see http://www.ub.edu/dnasp/; Rozas et al. 2017). Three runs in PHASE (100 000 iterations) with a burn-in of 10 000 all returned consistent allelic identities, and PHASE was able to resolve most alleles with 100% certainty. Individuals with <80% certainty at single nucleotide positions were removed from further analysis (n = 34 individuals, 13% of total samples). Allelic sequences were then exported in FASTA file format, collapsed into unique alleles, and converted into ARLEQUIN file format with FaBox prior to genetic analysis.
AIC analysis of unique TMO sequences in jModelTest also resulted in the selection of the TrN model, which was used in ARLEQUIN to calculate diversity indices (h and π), Fu’s Fs-values and AMOVA statistics to complement those performed for the mitochondrial COI gene.
Morphological analysis – body morphometrics and meristics
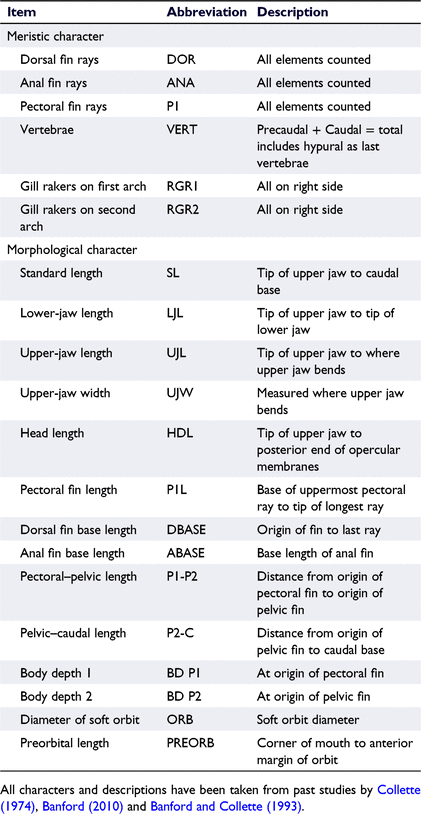
Following formalin preservation, 14 morphometric measurements and 6 meristic counts were recorded for each whole fish. All characters measured were taken from past comprehensive studies of other Hyporhamphus species (Collette 1974; Banford 2010; Table 2). Measurements were recorded to the nearest 0.1 mm with digital callipers, and vertebrae were counted from the X-rays taken of each specimen at the Australian Museum, visualised using Adobe Illustrator. X-Rays were also used to corroborate the dorsal and anal fin ray counts taken directly from specimens.
|
All data were partitioned into species groups for analysis in R (ver. 4.0.4, R Foundation for Statistical Computing, Vienna, Austria, see https://www.R-project.org/), on the basis of the species diagnosis obtained from the COI analysis. Any samples that could not be diagnosed as either species were excluded from further morphological analysis.
Multivariate principal component analysis (PCA) was performed on morphometric and meristic data separately, to visually assess the degree of morphological difference between each species and identify which morphological characters had the greatest variation. To reduce the effect of allometric differences among individuals and species, all morphometric measurements were log-transformed and regressed against the corresponding log-transformed standard length to obtain residual values for use in PCA. Logarithmic transformations were performed to reduce the scaling effects between standard length and residual variance. Scores for the first two principal components were visualised as bivariate plots, and factor loadings of each variable were used to assess their contribution to PC variation (see Tea et al. (2019) for a similar transformation and analysis).
To assess significant differences in the meristic counts of each species, histograms and summary tables of the most important variables contributing to factor loadings along the first axis were constructed. The size-adjusted mean morphometric variables for each species were compared by performing an analysis of covariance (ANCOVA) on each variable. All measurements were log-transformed to meet assumptions of normality and reduce scaling effects, and any observations whose standardised residuals were >3 were considered outliers and excluded from that analysis. Bonferroni adjustments were applied to the resulting P-values to adjust for multiple testing. Morphological variables identified as significantly (P ≤ 0.003) different were then visualised as boxplots to further explore differences among species and locations.
Morphological analysis – otolith shape
All otoliths were photographed using a Leica IC80 HD camera mounted on a Leica M125 dissecting microscope. Left-side sagittal otoliths were positioned distal side up with their rostrum horizontally aligned, on a black background (Fig. S1). Any otoliths that had broken in storage were pieced together under the microscope by using tweezers. Adobe Photoshop (ver. 22.2.0, Adobe Inc., San Jose, CA, USA) was used to align and stitch together these fragments to reconstruct the shape outline. In cases where the left-side otolith was missing or broken and the right-side one was intact, the right-side otolith was photographed, and Adobe Photoshop was used to flip the image to align horizontally with the other samples (see Steer and Fowler 2015 for similar methods on H. melanochir otoliths). Data were partitioned into species groups prior to analysis on the basis of the results from COI barcoding.
To extract otolith contour outlines and transform them into independent coefficients by using discrete wavelet analysis, all photographs were processed using the R package ShapeR (ver. 0.1.5, see https://cran.r-project.org/package=shapeR; Libungan and Pálsson 2015). The mean otolith shape for each species was plotted, and coefficients that showed an interaction with fish length (n = 6) were removed to adjust each otolith shape with respect to allometric relationships with fish length. To analyse the variation in otolith shape between the two species, a permutational multivariate analysis of variance (PERMANOVA) was applied to the length standardised wavelet coefficients by using the R package vegan (ver. 2.5.6, J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, and H. Wagner, see https://cran.r-project.org/web/packages/vegan/). Assumptions of equal variance among groups were checked with a dispersion test by using the function ‘betadisper()’. No differences were detected between the outlines of the photoshopped and non-photoshopped files (PERMANOVA, P > 0.1 for all locations, Supplementary Table S8). Random forest classification analysis was then performed on these coefficients by using the R package randomForest (ver. 4.6.14, see https://cran.r-project.org/package=randomForest; Liaw and Wiener 2002) to determine the error rate of classification of individuals to their species group.
Species discrimination
To assess the potential use of morphological variables as diagnostic markers, classification trees were constructed using the R packages rpart (ver. 4.1.15, T. Therneau and B. Atkinson, see https://CRAN.R-project.org/package=rpart) and rpart.plot (ver. 3.0.9, S. Milborrow, see https://CRAN.R-project.org/package=rpart.plot). Ratios were constructed among key variables identified from PCA and ANCOVA to account for differences in standard length among samples and to attempt to increase the strength of differences in variables among the species. Trees with diagnostic error rates were then constructed separately for meristic counts and ratios of key measurements to compare the probability of correct diagnosis between both methods. To reduce the impact of differences in sample size between species on probability estimates and account for potential geographic variation in H. melanochir morphology, only data from NSW and Victoria were used to construct classification trees (see Table 1, Sites 1–9).
Results
Genetic analysis – mitochondrial COI
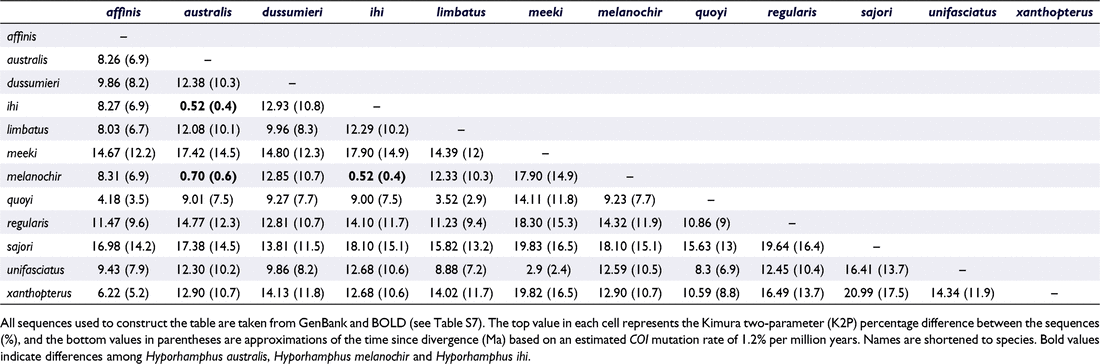
The mitochondrial COI fragment for H. australis amplified in this study differed from that of H. melanochir by 0.66%. On the basis of a 1.2% per million year rate of divergence calibrated with the COI gene and the rise of the Isthmus of Panama (Bermingham et al. 1997), our study suggests that the two species may have separated c. 500 000 years ago. Comparison of this pairwise genetic difference with other species in the genus indicates that, of the 12 Hyporhamphus species found on GenBank, H. australis, H. melanochir and the New Zealand endemic H. ihi are the most closely related on the basis of COI (Table 3). Because there was only a single haplotype present on GenBank for H. australis and H. melanochir, the difference between these samples was slightly higher than the results from our overall sampling efforts (0.7% difference, indicating separation c. 600 000 years ago, Table 3). Average conspecific (within-species) differentiation at COI was only 0.03% for H. australis and 0.04% for H. melanochir.
|
The low genetic divergence observed among and within species groups is supported by the median-joining haplotype network (Fig. 2a). Within each species group, one major haplotype was dominant (Fig. 2a). Haplotypes among species consistently differed at the four diagnostic nucleotides, whereas haplotypes of conspecifics differed by no more than two nucleotide substitutions. No haplotypes attributed to H. australis were found outside of NSW; however, the dominant H. melanochir haplotype (Fig. 2a) was found in NSW regions outside of their overlapping distribution at Eden (n = 4 samples, one each from Tea Gardens, Nelson Bay, Kiama, and Ulladulla). Aside from these four samples, Eden was the only location with a complete overlap in haplotypes from both species (Fig. 2a). The COI sequence data showed 13 haplotypes for H. australis and six for H. melanochir, with a small number of intermediate haplotypes (n = 3) that could not be attributed to either species group. One haplotype was found across multiple locations (Tea Gardens, Eden, Victoria, and Adelaide), and two were found only in Eden. Of the two intermediate haplotypes unique to Eden, one haplotype shared by three individuals had double peaks on their chromatogram traces at all diagnostic bases between the two species.
|
|
AMOVA confirmed that there were significant genetic differences between the two species that explained 95% of the variation in haplotype diversity (overall ΦST = 0.95, P < 0.001, Supplementary Table S9). Population pairwise tests among all samples grouped by location showed that Eden was the only population that differed significantly from populations at all other geographic sites (Fig. 3, Supplementary Table S10). Victorian, South Australian, and Western Australian populations were significantly different from all NSW populations, but not from each other. All NSW populations (except for Eden) were also not significantly different from one another. Indeed, within each species group, there were no significant differences among locations (H. australis: overall ΦST = 0.005, P = 0.153; H. melanochir: overall ΦST = 0.04, P = 0.057; Table S9).
|
|
When comparing across all locations, the haplotype diversity (h) of H. melanochir was approximately 50% higher than that of H. australis, whereas nucleotide diversity (π) was the same for both species (Supplementary Table S11). Tests of neutrality showed negative and significant Fu’s Fs values for both species; within each species group, values of h, π and Fs varied across locations (Table S11). Within the H. australis group, the populations sampled at Forster, Nelson Bay, Wollongong, and Eden all had higher haplotype diversity and significant Fs values (Table S11). Within the H. melanochir group, Eden had the highest haplotype and nucleotide diversity, but significant Fs values were seen only in the Victoria and South Australia populations (Table S11).
Genetic analysis – nuclear TMO-4C4
The nuclear TMO-4C4 sequences of 221 garfish individuals showed 15 alleles, of which 8 were found in both species, with approximately equal allelic and nucleotide diversities (Table S11). Hyporhamphus melanochir from Eden and Western Australia were the only groups that contained unique alleles, and had significant and negative Fs values (Fs = −3.24 and −3.28 respectively; Table S11).
AMOVA analysis and visualisation of the median-joining network based on the TMO-4C4 alleles indicated that there was no significant genetic difference between species (P = 0.09) or locations (P = 0.47; Supplementary Table S12, Fig. 2b). Two major alleles dominated at all locations across both species, which differed by only one nucleotide substitution (Fig. 2b).
Morphological analysis – body morphometrics and meristics
PCA showed clear distinctions between the meristic characters of the two species (Fig. 4a). Individuals were separated into species groups along the PC 1 axis, which accounted for 41.4% of the total variance (Fig. 4a). Gill raker counts on the first and second arch contributed the most to the factor loadings of PC 1 (−0.8 and −0.87 respectively; Supplementary Table S13).
|
|
The PCA of morphometric characters indicated separation between the species but with a considerable amount of overlap along the PC 2 axis, which accounted for 16.4% of the total variance (Fig. 4b). Preorbital length and upper-jaw length both contributed the most to factor loadings of PC 2 (−0.74 and −0.65 respectively; Supplementary Tables S14, S15).
Mean gill raker counts differed between the two species, with H. australis presenting higher mean gill raker counts on both the first and second gill arches (Fig. 5, Table 4). Mean vertebral counts also differed between the species; however, H. melanochir showed a significant amount of within-species variation among locations (Fig. 5). Vertebral counts were lower across the entire range of H. australis v. those for H. melanochir individuals from the geographically closest locations (Eden and Victoria), but there was no clear difference between vertebral counts of H. australis and H. melanochir from more distant locations (South Australia and Western Australia; Fig. 5, Table 4). Although dorsal fin rays were identified in PCA as contributing to factor loadings along the PC 1 axis, there was a high degree of overlap between the two species (Fig. 5).
|
|

|
ANCOVA showed that, of the 13 morphometric measurements examined, there were significant differences between the group means of only three measurements, namely, head length, upper-jaw length, and preorbital length (P < 0.003; Supplementary Tables S16, S17). Hyporhamphus australis was found to have a larger head size than was H. melanochir, which was driven by its longer upper-jaw and preorbital length (Fig. 6). There was no significant difference in the orbital diameter or distance between the orbit and the posterior end of the opercular membrane between the species (P = 0.5 and 0.08 respectively; Tables S16, S17). ANCOVA also showed a significant difference in the mean pectoral fin length between the species (P < 0.003; Tables S16, S17), but owing to the small effect size and high degree of overlap in the range of lengths for each group, this trait was deemed to be an unsuitable diagnostic feature (ges = 0.08, Tables S16, S17, Fig. S2).
|
|
In general, both species maintained these differences in characteristics even within areas where their distributions overlapped (Eden, Fig. 6). Difference in upper-jaw length was more pronounced between species in Eden than between species from more geographically distant populations, but there was more overlap in preorbital length between species in Eden than between species from other locations (Fig. 6). Although the mean upper-jaw and preorbital lengths were significantly different between the two species, there was still a considerable amount of overlap in these characters (Fig. 6).
Morphological analysis – otolith shape
PERMANOVA showed significant variation in otolith shape between the two species (P = 0.001; Supplementary Table S18). Wavelet reconstruction of mean otolith shape showed that the otoliths of H. australis tend to have a slightly longer, more pointed rostrum than do those of H. melanochir (Fig. 7).
Random forest classification based on wavelet coefficients resulted in assignment to species with a high degree of success (∼88%; Supplementary Table S19). The within-class error was substantially lower for H. australis than H. melanochir (5.49 and 28.99% respectively; Table S19), suggesting a higher variability in H. melanochir otolith shape.
Species discrimination
Classification trees using data from NSW and VIC populations showed that, although it is possible to distinguish between the species on the basis of ratios of head characteristics, gill raker counts remain the most accurate method of diagnosis between the two species (Fig. 8). The number of gill rakers on the second arch alone were enough to distinguish between the species with ∼92% accuracy, whereas ratios of four separate measurements had a lower accuracy rate of ∼89% (Fig. 8). These classification trees provide a more accurate species diagnosis than does otolith shape (Table S19); however, owing to the higher degree of overlap in morphological measurements between the species (see Fig. 6), gill rakers should be used to distinguish between species wherever possible.
|
|
Discussion
This study has demonstrated the use of an integrative taxonomic approach to separate two cryptic halfbeak species with overlapping distributions. Hyporhamphus australis and H. melanochir are a cryptic halfbeak species pair with consistent differences in their mtDNA COI gene regions that align with differences in the number of gill rakers, head length, and otolith shape. Although these differences are modest, they reinforce previous findings (Collette 1974) and confirm the taxonomic status of both species. Our study has provided new methods to discriminate between the species and has confirmed that their distributions overlap in southern NSW. The identification of both H. melanochir and H. australis in landings from southern NSW has important implications for fisheries management, both providing new morphological methods to determine the species caught in landings around Eden and highlighting the potential need for changes into catch regulation to accommodate the growth rates and life-history strategies of H. melanochir stocks in NSW.
Genetic differences
The shallow divergence in the mtDNA COI gene region and the lack of difference in the nuclear TMO-4C4 gene suggests that these two species have only recently diverged from each other relative to other species in the Hyporhamphus genus. A relatively short period of evolutionary isolation may not have allowed enough time for differences to develop in the nuclear genome since mtDNA has an effective population size approximately one-quarter that of nuclear markers (Hurst and Jiggins 2005) and, therefore, mutates at a faster rate. The low levels of genetic divergence identified among H. australis, H. melanochir and the New Zealand endemic species H. ihi suggest that these three species have only recently diverged from one another on an evolutionary time scale.
The estimated time point of divergence c. 500 000 years ago is consistent with the assumed allopatric speciation of H. australis and H. melanochir across the Bass Strait (Collette 1974). During periods of low sea level associated with Plio-Pleistocene glacial maxima, the Bassian Isthmus is thought to have acted as a land bridge connecting Tasmania to south-eastern Victoria (Dartnall 1974; Lambeck and Chappell 2001). The hypothesis that the Bassian Isthmus created a biogeographical barrier fragmenting marine populations (Hedley 1904) is supported by genetic signatures of historical isolation in other marine species (DiBattista et al. 2014; Wilson et al. 2017). Since complete submergence of the Isthmus c. 14 000 years ago (Lambeck and Chappell 2001), a combination of demersal spawning and contemporary oceanographic features may have sustained these patterns of historical vicariance. The complex convergence of the East Australian Current (EAC) and South Australian Boundary Current has been shown to limit colonisation and dispersal in other marine species separated historically by the Bassian Isthmus (Waters 2008; Colton and Swearer 2012). Both H. melanochir and H. australis are demersal spawners, and their substrate-adhesive eggs and well developed larvae may increase the likelihood of early life-stage retention within seagrass habitats (Jones et al. 2002; Stewart et al. 2005), thus likely limiting passive dispersal by coastal currents.
Both H. australis and H. melanochir are found concurrently in southern NSW (Eden), making this an area of secondary contact between these cryptic species. The formation of hybrid zones resulting from secondary contact is not as rare as previously considered, especially in closely related marine species at the borders of biogeographic provinces (Hobbs et al. 2009, 2022; Montanari et al. 2014). Eden is located on the extremities of two such biogeographic provinces, namely, the eastern Peronian and southern Maugean, in an area of transition between warm and cool temperate waters (Poore and O’Hara 2007). Although little research has been conducted into the hybridisation of other marine fish around Eden, genetic signatures of introgression resulting from secondary contact after isolation have been discovered within the cuttlefish species Sepia apama in southern NSW (Kassahn et al. 2003). Eden is also an area of overlap of two distinct stocks of the commercially important snapper Chrysophrys auratus, with the suggestion of admixture and interbreeding (Morgan et al. 2018) around this area. Hybridisation at Eden was first suggested by Collette (1974) and is supported by the presence of individuals in this study with intermediate mtDNA haplotypes in Eden and adjacent areas of NSW. Four samples identified as H. melanochir from NSW may also represent backcrossed individuals, but this cannot be confirmed because of the lack of diagnostic nuclear DNA differences between the species. Future studies should focus on the development of a panel of single nucleotide polymorphism (SNPs) markers, because their high abundance and genomic coverage often enable the detection of fine-scale genetic differences, hybridisation, and introgression between overlapping species (Gaither et al. 2015).
Species discrimination
The largely allopatric distributions of H. australis and H. melanochir means that species identity can almost always be determined from an individual’s location of origin. However, the confirmation of a spatial and temporal species distribution overlap from the 56 km of coastline surrounding Eden, as well as the presence of a H. melanochir specimen from ∼500 km north of their previous known range, makes diagnostic criteria essential for species identification.
Counting gill rakers remains the most reliable morphological method to distinguish these two species, and gill raker numbers should be retained as their standard diagnostic feature. These small structures are difficult to count without training and the aid of a microscope, but provide higher accuracy than morphological ratios. We recommend that gill raker counts should be integrated into the current landings monitoring conducted by NSW DPI, to better understand the proportions of both species from catches in southern NSW. Morphological ratios can also be used to discriminate between specimens in circumstances (e.g. in the field) where gill rakers may be difficult to accurately count. However, these ratios should be used with caution because of the unknown level of phenotypic plasticity in the species’ morphological traits and their larger degree of overlap. Owing to the low number of individuals in this study from Eden (n = 25), the decision trees contain data from samples across NSW and Victoria; therefore, future studies should aim to use a much larger sample size taken across all seasons in the area of overlapping distributions.
Morphological difference
The slight morphological differences found between the species and the lack of integration of characters from the closest geographic populations support a hypothesis of recent divergence and reinforce previously published findings (Collette 1974). Most meristic and morphological traits in fish are heritable, but they are also influenced by environmental factors affecting diet, ontogeny, and growth (Swain et al. 2005). Differences in gill raker number, jaw length, and skull structure in fish species are often attributed to adaptive divergence and radiation, because these functional morphological traits are directly related to feeding behaviour and trophic niche use (Harrod et al. 2010; Parsons et al. 2011). Both H. australis and H. melanochir are omnivorous, feeding on Zostera seagrass and hyperbenthic crustaceans (Table S2), and differences in prey size and availability across their distributions may be a factor driving the differences in their gill raker counts, and upper-jaw, and preorbital lengths.
Vertebrae numbers are highly heritable in fish but are also influenced by environmental conditions during ontogeny (Swain et al. 2005). The high variability in vertebral counts within H. melanochir may be a plastic response to abiotic differences across the species latitudinal distribution, because differences found in vertebral counts are consistent with an increase in the number of fish vertebrae with an increasing latitude (i.e. Jordan 1891). The mechanism behind this pattern is unknown, but it is likely to be due to a combination of a longer ontogenetic development and larger body sizes in cooler waters (reviewed by McDowall 2008).
Our study also found significant differences in the mean otolith shapes between the two species. As is the case for other morphological features, both environmental and genetic influences have been shown to induce changes in otolith shape (Vignon and Morat 2010). This may be due to the direct and indirect effects of these factors on growth rates, which have been shown to significantly affect otolith shape in other fish species (Stransky 2014; Rodgveller et al. 2017). Differences in growth rates between the species may therefore contribute to their different otolith shapes, with H. australis having a faster growth rate than has H. melanochir (230 mm FL and 160–180 mm FL at 1 year of age respectively; Jones et al. 2002; Stewart and Hughes 2007; Table S2).
Taxonomic considerations
The corroborative genetic and morphological differences between H. australis and H. melanochir are reinforced by differences in their growth and life history. Hyporhamphus australis achieves smaller maximum sizes overall but grows faster and reaches maturity at smaller sizes and younger ages than does H. melanochir (Jones et al. 2002; Stewart et al. 2004; Table S2). Both species also have seasonal differences in their reproductive peaks, although there is a small degree of overlap owing to their protracted spawning seasons (Jones et al. 2002; Stewart and Hughes 2007; Table S2). Together, these differences clearly show that H. australis and H. melanochir are two distinct groups of fish for management purposes; however, the question remains as to whether these fish should be considered different species.
There are three reasons that could potentially be used to justify the revised classification of these populations as subspecies or stocks rather than species. First, the degree of genetic differentiation between this species pair (average K2P percent difference = 0.66%) is much lower than that of other species in the Hyporhamphus genus (Table 2) and among other congeneric species found in Australia (average K2P percentage difference = 9.93%; Ward et al. 2005). Second, the presence of intermediate COI haplotypes suggests hybridisation and potential introgression between the species, which goes against the criteria of reproductive isolation, which is needed to delimit species boundaries under the biological species concept (Mayr 1942). Finally, the morphological features that do distinguish the species have very small degrees of difference and exhibit some overlap.
However, a subspecies classification of H. australis and H. melanochir does not consider the multiple lines of evidence found in our study that separate them, and this reclassification would go against standards employed across fish taxonomy and within the Hyporhamphus genus. Although the average K2P percentage difference between H. australis and H. melanochir is relatively low, their between-species difference (0.66%) is ∼18× higher than their average intraspecific differences (0.03 and 0.04%, respectively). Comparing these interspecific and intraspecific differences can provide a threshold to delimit potential species boundaries. Hebert et al. (2004) proposed a standard threshold of interspecific difference at 10× that of intraspecific difference to flag provisional species, which would suggest H. australis and H. melanochir are different species, but a threshold of intraspecific to interspecific difference alone is not enough evidence to justify their species status (Moritz and Cicero 2004). The taxonomic status of this cryptic species pair is instead confirmed by the corroboration between their genetic separation, differences in morphology and life history, as well as the maintenance of distinct traits even in geographically proximate locations. Subspecies classification would be an unsuitable assignment in this case because it is rarely used in fish taxonomy and is usually based only on discrete (i.e. allopatric) distributions (Collette 1974; Haig et al. 2006).
Implications
The distributional overlap of H. australis and H. melanochir in Eden has significant implications for local fisheries management. Although the increase in lampara net mesh size from 25 to 28 mm and a reduction in fishing effort in NSW between 2004 and 2015 has successfully reduced the fishing mortality of H. australis (Broadhurst et al. 2018), these practices are not designed to accommodate the different age and size at maturity of H. melanochir (Table S2). The study that determined the optimal mesh size for H. australis also found that a larger 32-mm mesh size selected 50% of fish at 24.5 cm FL, which would effectively prevent the retention of juveniles from both species; however, this mesh type also resulted in lowered catch rates and damage to export quality fish (Stewart et al. 2004). Going forward, fisheries management in Eden will need to weigh the potential benefits of further increasing the minimum mesh-size limits to protect immature H. melanochir against the negative effect this could have on fishery catch rates.
Our study was able to show the presence of H. melanochir in Eden only in winter, and further research is needed to establish whether this presence is maintained year-round and explore the potential for spawning overlap. Future studies should also aim to examine traits from freshly caught fish, because the lower jaws of most of our study specimens snapped while frozen and these measurements were unable to be included in analysis. Collette (1974) noted that H. australis has a relatively longer lower jaw than does H. melanochir at larger sizes, but did not find a significant difference in their mean lengths.
The impact of these management implications may seem localised, but they could become much broader in the face of potential climate change-driven shifts in species distributions. The warming rates of western boundary currents such as the EAC are two to three times faster than that of the global sea-surface mean, and current flows are also intensifying (Wu et al. 2012; Malan et al. 2021). Changing EAC flows could shift the range of H. australis, which already moves up and down the NSW coast following seasonal fluctuations in the strength and temperature of the EAC (Stewart and Hughes 2007), which pushes the species further south across management jurisdictions. Similar range shifts have already been observed for many other species in this region (reviewed by Gervais et al. 2021). Although the smaller size and age at maturity of H. australis would likely mean that Tasmania’s current management strategies for H. melanochir are conservative enough to protect the juveniles of both species, their introduction may have unforeseen impacts on the genetic or population structuring of local stocks. Even if separate management strategies are difficult or ineffective to implement within a mixed fishery, exploring the potential presence of both species in both Eden and Tasmania can give a better assessment of the true sustainability of the fishery to all stocks present, as is currently recommended for other fisheries that contain cryptic species complexes (Huveneers 2006; Saha et al. 2017).
Our study demonstrated the need for discrimination of cryptic species at multiple levels to validate species-level taxonomic assignment. When comparing these two species at a morphological and functional level, there are consistent differences between them that validate a distinct taxonomic classification. As De Queiroz (2007) showed in advocating for a unified species concept, these differences are properties that divergent species acquire in the process of lineage separation, which together provide evidence of speciation regardless of traditional species concepts. Studies into cryptic species should adopt similar integrative methods to those employed here, to reinforce species boundaries and aid in conservation and management.
Supplementary material
Supplementary material is available online.
Data availability
The genetic sequence data underlying this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/genbank/ (accession numbers MZ575768–MZ576070 and MZ580145–MZ580399). All other data are available from GitHub at https://github.com/Indy-Riley/Hyporhamphus_Data_Repository.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This project was supported by the New South Wales Department of Primary Industry.
Acknowledgements
We thank Jackie Chan at the Ramaciotti Centre for Genomics (UNSW) for all Sanger Sequencing conducted in this study. We also extend our gratitude to Amanda Hay, Kerryn Parkinson and Sally Reader, from the Australian Museum Ichthyology Section, for guidance in the morphological examination of these specimens and assistance with fish X-rays and preservation, and to Anthony Gould and Nick Meadows from NSW DPI for logistical assistance. This is Sydney Institute of Marine Science (SIMS) publication #301.
References
Agapow, P-M, Bininda-Emonds, ORP, Crandall, KA, Gittleman, JL, Mace, GM, Marshall, JC, and Purvis, A (2004). The impact of species concept on biodiversity studies. The Quarterly Review of Biology 79, 161–179.| The impact of species concept on biodiversity studies.Crossref | GoogleScholarGoogle Scholar |
Altschul, SF, Gish, W, Miller, W, Myers, EW, and Lipman, DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215, 403–410.
| Basic local alignment search tool.Crossref | GoogleScholarGoogle Scholar |
Bandelt, HJ, Forster, P, and Röhl, A (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48.
| Median-joining networks for inferring intraspecific phylogenies.Crossref | GoogleScholarGoogle Scholar |
Banford, HM (2010). Hyporhamphus collettei, a new species of inshore halfbeak (Hemiramphidae) endemic to Bermuda, with comments on the biogeography of the Hyporhamphus unifasciatus species group. Proceedings of the Biological Society of Washington 123, 345–358.
| Hyporhamphus collettei, a new species of inshore halfbeak (Hemiramphidae) endemic to Bermuda, with comments on the biogeography of the Hyporhamphus unifasciatus species group.Crossref | GoogleScholarGoogle Scholar |
Banford, HM, and Collette, BB (1993). Hyporhampus meeki, a new species of halfbeak (Teleostei: Hemiramphidae) from the Atlantic and Gulf coasts of the United States. Proceedings of the Biological Society of Washington 106, 369–384.
Bermingham E, McCafferty SS, Martin AP (1997) Fish biogeography and molecular clocks: perspectives from the Panamanian Isthmus. In ‘Molecular systematics of fishes’. (Eds TD Kocher, CA Stepien) pp. 113–128. (Academic Press: San Diego, CA, USA)
Bickford, D, Lohman, DJ, Sodhi, NS, Ng, PKL, Meier, R, Winker, K, Ingram, KK, and Das, I (2007). Cryptic species as a window on diversity and conservation. Trends in Ecology & Evolution 22, 148–155.
| Cryptic species as a window on diversity and conservation.Crossref | GoogleScholarGoogle Scholar |
Bozdogan, H (1987). Model selection and Akaike’s Information Criterion (AIC): the general theory and its analytical extensions. Psychometrika 52, 345–370.
| Model selection and Akaike’s Information Criterion (AIC): the general theory and its analytical extensions.Crossref | GoogleScholarGoogle Scholar |
Broadhurst, MK, Kienzle, M, and Stewart, J (2018). Natural and fishing mortalities affecting eastern sea garfish, Hyporhamphus australis inferred from age-frequency data using hazard functions. Fisheries Research 198, 43–49.
| Natural and fishing mortalities affecting eastern sea garfish, Hyporhamphus australis inferred from age-frequency data using hazard functions.Crossref | GoogleScholarGoogle Scholar |
Collette, BB (1974). The garfishes (Hemiramphidae) of Australia and New Zealand. Records of the Australian Museum 29, 11–105.
| The garfishes (Hemiramphidae) of Australia and New Zealand.Crossref | GoogleScholarGoogle Scholar |
Colton, MA, and Swearer, SE (2012). Locating faunal breaks in the nearshore fish assemblage of Victoria, Australia. Marine and Freshwater Research 63, 218–231.
| Locating faunal breaks in the nearshore fish assemblage of Victoria, Australia.Crossref | GoogleScholarGoogle Scholar |
Corrigan, S, Huveneers, C, Schwartz, TS, Harcourt, RG, and Beheregaray, LB (2008). Genetic and reproductive evidence for two species of ornate wobbegong shark Orectolobus spp. on the Australian east coast. Journal of Fish Biology 73, 1662–1675.
| Genetic and reproductive evidence for two species of ornate wobbegong shark Orectolobus spp. on the Australian east coast.Crossref | GoogleScholarGoogle Scholar |
Corrigan, S, Huveneers, C, Stow, A, and Beheregaray, LB (2016). A multilocus comparative study of dispersal in three codistributed demersal sharks from eastern Australia. Canadian Journal of Fisheries and Aquatic Sciences 73, 406–415.
| A multilocus comparative study of dispersal in three codistributed demersal sharks from eastern Australia.Crossref | GoogleScholarGoogle Scholar |
CSIRO (2009a) Australian National Fish Expert Distributions – Hyporhamphus australis. Available at https://www.marine.csiro.au/data/caab/taxon_report.cfm?caab_code=37234014 [Verified 12 February 2022]
CSIRO (2009b) Australian National Fish Expert Distributions – Hyporhamphus melanochir. Available at https://www.marine.csiro.au/data/caab/taxon_report.cfm?caab_code=37234001 [Verified 12 February 2022]
Darriba, D, Taboada, GL, Doallo, R, and Posada, D (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772.
| jModelTest 2: more models, new heuristics and parallel computing.Crossref | GoogleScholarGoogle Scholar |
Dartnall AJ (1974) Littoral biogeography. In ‘Biogeography and ecology in Tasmania. Monographiae Biologicae’. (Ed. WD Williams) Vol. 25, pp. 171–194. (Springer: Dordrecht, Netherlands) https://doi.org/10.1007/978-94-010-2337-5_8
Dayrat, B (2005). Towards integrative taxonomy. Biological Journal of the Linnean Society 85, 407–415.
| Towards integrative taxonomy.Crossref | GoogleScholarGoogle Scholar |
De Queiroz, K (2007). Species concepts and species delimitation. Systematic Biology 56, 879–886.
| Species concepts and species delimitation.Crossref | GoogleScholarGoogle Scholar |
De Queiroz, K (2011). Branches in the lines of descent: Charles Darwin and the evolution of the species concept. Biological Journal of the Linnean Society 103, 19–35.
| Branches in the lines of descent: Charles Darwin and the evolution of the species concept.Crossref | GoogleScholarGoogle Scholar |
Delrieu-Trottin, E, Hartmann-Salvo, H, Saenz-Agudelo, P, Landaeta, MF, and Pérez-Matus, A (2022). DNA reconciles morphology and colouration in the drunk blenny genus Scartichthys (Teleostei: Blenniidae) and provides insights into their evolutionary history. Journal of Fish Biology 100, 507–518.
| DNA reconciles morphology and colouration in the drunk blenny genus Scartichthys (Teleostei: Blenniidae) and provides insights into their evolutionary history.Crossref | GoogleScholarGoogle Scholar |
DeSalle, R, and Goldstein, P (2019). Review and interpretation of trends in DNA barcoding. Frontiers in Ecology and Evolution 7, 302.
| Review and interpretation of trends in DNA barcoding.Crossref | GoogleScholarGoogle Scholar |
DiBattista, JD, Randall, JE, Newman, SJ, and Bowen, BW (2014). Round herring (genus Etrumeus) contain distinct evolutionary lineages coincident with a biogeographic barrier along Australia’s southern temperate coastline. Marine Biology 161, 2465–2477.
| Round herring (genus Etrumeus) contain distinct evolutionary lineages coincident with a biogeographic barrier along Australia’s southern temperate coastline.Crossref | GoogleScholarGoogle Scholar |
Excoffier, L, and Lischer, HEL (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567.
| Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows.Crossref | GoogleScholarGoogle Scholar |
Fu, Y-X (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
| Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection.Crossref | GoogleScholarGoogle Scholar |
Gaither, MR, Bernal, MA, Coleman, RR, Bowen, BW, Jones, SA, Simison, WB, and Rocha, LA (2015). Genomic signatures of geographic isolation and natural selection in coral reef fishes. Molecular Ecology 24, 1543–1557.
| Genomic signatures of geographic isolation and natural selection in coral reef fishes.Crossref | GoogleScholarGoogle Scholar |
Gervais, CR, Champion, C, and Pecl, GT (2021). Species on the move around the Australian coastline: a continental-scale review of climate-driven species redistribution in marine systems. Global Change Biology 27, 3200–3217.
| Species on the move around the Australian coastline: a continental-scale review of climate-driven species redistribution in marine systems.Crossref | GoogleScholarGoogle Scholar |
Guindon, S, and Gascuel, O (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704.
| A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Crossref | GoogleScholarGoogle Scholar |
Haig, SM, Beever, EA, Chambers, SM, Draheim, HM, Dugger, BD, Dunham, S, Elliott-Smith, E, Fontaine, JB, Kesler, DC, Knaus, BJ, Lopes, IF, Loschl, P, Mullins, TD, and Sheffield, LM (2006). Taxonomic considerations in listing subspecies under the US Endangered Species Act. Conservation Biology 20, 1584–1594.
| Taxonomic considerations in listing subspecies under the US Endangered Species Act.Crossref | GoogleScholarGoogle Scholar |
Harrod, C, Mallela, J, and Kahilainen, KK (2010). Phenotype–environment correlations in a putative whitefish adaptive radiation. Journal of Animal Ecology 79, 1057–1068.
| Phenotype–environment correlations in a putative whitefish adaptive radiation.Crossref | GoogleScholarGoogle Scholar |
Hebert, PDN, Cywinska, A, Ball, SL, and deWaard, JR (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London – B. Biological Sciences 270, 313–321.
| Biological identifications through DNA barcodes.Crossref | GoogleScholarGoogle Scholar |
Hebert, PDN, Stoeckle, MY, Zemlak, TS, and Francis, CM (2004). Identification of birds through DNA barcodes. PLoS Biology 2, e312.
| Identification of birds through DNA barcodes.Crossref | GoogleScholarGoogle Scholar |
Hedley, C (1904). The effect of the Bassian Isthmus upon the existing marine fauna: a study in ancient geography. Proceedings of the Linnean Society of New South Wales 28, 876–883.
| The effect of the Bassian Isthmus upon the existing marine fauna: a study in ancient geography.Crossref | GoogleScholarGoogle Scholar |
Hobbs, J-PA, Frisch, AJ, Allen, GR, and Van Herwerden, L (2009). Marine hybrid hotspot at Indo-Pacific biogeographic border. Biology Letters 5, 258–261.
| Marine hybrid hotspot at Indo-Pacific biogeographic border.Crossref | GoogleScholarGoogle Scholar |
Hobbs, J-PA, Richards, ZT, Popovic, I, Lei, C, Staeudle, TM, Montanari, SR, and DiBattista, JD (2022). Hybridisation and the evolution of coral reef biodiversity. Coral Reefs 41, 535–549.
| Hybridisation and the evolution of coral reef biodiversity.Crossref | GoogleScholarGoogle Scholar |
Hurst, GDD, and Jiggins, FM (2005). Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proceedings of the Royal Society B: Biological Sciences 272, 1525–1534.
| Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts.Crossref | GoogleScholarGoogle Scholar |
Huveneers, C (2006). Redescription of two species of wobbegongs (Chondrichthyes: Orectolobidae) with elevation of Orectolobus halei Whitley 1940 to species level. Zootaxa 1284, 29–51.
| Redescription of two species of wobbegongs (Chondrichthyes: Orectolobidae) with elevation of Orectolobus halei Whitley 1940 to species level.Crossref | GoogleScholarGoogle Scholar |
Huveneers, C, Stead, J, Bennett, MB, Lee, KA, and Harcourt, RG (2013). Age and growth determination of three sympatric wobbegong sharks: how reliable is growth band periodicity in Orectolobidae? Fisheries Research 147, 413–425.
| Age and growth determination of three sympatric wobbegong sharks: how reliable is growth band periodicity in Orectolobidae?Crossref | GoogleScholarGoogle Scholar |
Izzo, C, Ward, TM, Ivey, AR, Suthers, IM, Stewart, J, Sexton, SC, and Gillanders, BM (2017). Integrated approach to determining stock structure: implications for fisheries management of sardine, Sardinops sagax, in Australian waters. Reviews in Fish Biology and Fisheries 27, 267–284.
| Integrated approach to determining stock structure: implications for fisheries management of sardine, Sardinops sagax, in Australian waters.Crossref | GoogleScholarGoogle Scholar |
Jones GK, Ye Q, Ayvazian S, Coutin P (2002) Fisheries biology and habitat ecology of southern sea garfish (Hyporhamphus melanochir) in southern Australian waters. FRDC Project 97/133. (Fisheries Research and Development Corporation and South Australian Research and Development Institute, SARDI) Available at http://www.frdc.com.au/Archived-Reports/FRDCProjects/1997-133-DLD.pdf
Jordan, DS (1891). Relations of temperature to vertebrae among fishes. Proceedings of the United States National Museum 14, 107–120.
| Relations of temperature to vertebrae among fishes.Crossref | GoogleScholarGoogle Scholar |
Kassahn, KS, Donnellan, SC, Fowler, AJ, Hall, KC, Adams, M, and Shaw, PW (2003). Molecular and morphological analyses of the cuttlefish Sepia apama indicate a complex population structure. Marine Biology 143, 947–962.
| Molecular and morphological analyses of the cuttlefish Sepia apama indicate a complex population structure.Crossref | GoogleScholarGoogle Scholar |
Knowlton, N (2000). Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420, 73–90.
| Molecular genetic analyses of species boundaries in the sea.Crossref | GoogleScholarGoogle Scholar |
Lambeck, K, and Chappell, J (2001). Sea level change through the last glacial cycle. Science 292, 679–686.
| Sea level change through the last glacial cycle.Crossref | GoogleScholarGoogle Scholar |
Leigh, JW, and Bryant, D (2015). PopART: full-feature software for haplotype network construction. Methods in Ecology and Evolution 6, 1110–1116.
| PopART: full-feature software for haplotype network construction.Crossref | GoogleScholarGoogle Scholar |
Lessios, HA (2008). The Great American Schism: divergence of marine organisms after the rise of the Central American Isthmus. Annual Review of Ecology, Evolution, and Systematics 39, 63–91.
| The Great American Schism: divergence of marine organisms after the rise of the Central American Isthmus.Crossref | GoogleScholarGoogle Scholar |
Liaw, A, and Wiener, M (2002). Classification and regression by randomForest. R News 2, 18–22.
Libungan, LA, and Pálsson, S (2015). ShapeR: an R package to study otolith shape variation among fish populations. PLoS ONE 10, e0121102.
| ShapeR: an R package to study otolith shape variation among fish populations.Crossref | GoogleScholarGoogle Scholar |
Liggins, L, Kilduff, L, Trnski, T, Delrieu-Trottin, E, Carvajal, JI, Arranz, V, Planes, S, Saenz-Agudelo, P, and Aguirre, JD (2022). Morphological and genetic divergence supports peripheral endemism and a recent evolutionary history of Chrysiptera demoiselles in the subtropical South Pacific. Coral Reefs 41, 797–812.
| Morphological and genetic divergence supports peripheral endemism and a recent evolutionary history of Chrysiptera demoiselles in the subtropical South Pacific.Crossref | GoogleScholarGoogle Scholar |
McDowall, RM (2008). Jordan’s and other ecogeographical rules, and the vertebral number in fishes. Journal of Biogeography 35, 501–508.
| Jordan’s and other ecogeographical rules, and the vertebral number in fishes.Crossref | GoogleScholarGoogle Scholar |
Malan, N, Roughan, M, and Kerrry, C (2021). The rate of coastal temperature rise adjacent to a warming western boundary current is nonuniform with latitude. Geophysical Research Letters 48, e2020GL090751.
| The rate of coastal temperature rise adjacent to a warming western boundary current is nonuniform with latitude.Crossref | GoogleScholarGoogle Scholar |
Mayr E (1942) ‘Systematics and the origin of species.’ (Columbia University Press: New York, NY, USA)
Meyer, CP, and Paulay, G (2005). DNA barcoding: error rates based on comprehensive sampling. PLoS Biolgy 3, e422.
| DNA barcoding: error rates based on comprehensive sampling.Crossref | GoogleScholarGoogle Scholar |
Montanari, SR, Hobbs, J-PA, Pratchett, MS, Bay, LK, and Van Herwerden, L (2014). Does genetic distance between parental species influence outcomes of hybridization among coral reef butterflyfishes? Molecular Ecology 23, 2757–2770.
| Does genetic distance between parental species influence outcomes of hybridization among coral reef butterflyfishes?Crossref | GoogleScholarGoogle Scholar |
Morgan, JA, Sumpton, WD, Jones, AT, Campbell, AB, Stewart, J, Hamer, P,, and Ovenden, JR (2018)). Assessment of genetic structure among Australian east coast populations of snapper Chrysophrys auratus (Sparidae). Marine and Freshwater Research 70, 964–976.
| Assessment of genetic structure among Australian east coast populations of snapper Chrysophrys auratus (Sparidae).Crossref | GoogleScholarGoogle Scholar |
Moritz, C, and Cicero, C (2004). DNA barcoding: promise and pitfalls. PLoS Biology 2, e354.
| DNA barcoding: promise and pitfalls.Crossref | GoogleScholarGoogle Scholar |
Narum, SR (2006). Beyond Bonferroni: less conservative analyses for conservation genetics. Conservation Genetics 7, 783–787.
| Beyond Bonferroni: less conservative analyses for conservation genetics.Crossref | GoogleScholarGoogle Scholar |
Nielsen, JR, Methven, DA, and Kristensen, K (2010). A statistical discrimination method using sagittal otolith dimensions between sibling species of juvenile cod Gadus morhua and Gadus ogac from the northwest Atlantic. Journal of Northwest Atlantic Fishery Science 43, 27–45.
| A statistical discrimination method using sagittal otolith dimensions between sibling species of juvenile cod Gadus morhua and Gadus ogac from the northwest Atlantic.Crossref | GoogleScholarGoogle Scholar |
Noell, CJ, Donnellan, S, Foster, R, and Haigh, L (2001). Molecular discrimination of garfish Hyporhamphus (Beloniformes) larvae in southern Australian waters. Marine Biotechnology 3, 509–514.
| Molecular discrimination of garfish Hyporhamphus (Beloniformes) larvae in southern Australian waters.Crossref | GoogleScholarGoogle Scholar |
Padial, JM, Miralles, A, De la Riva, I, and Vences, M (2010). The integrative future of taxonomy. Frontiers in Zoology 7, 16.
| The integrative future of taxonomy.Crossref | GoogleScholarGoogle Scholar |
Parsons, KJ, Cooper, WJ, and Albertson, RC (2011). Modularity of the oral jaws is linked to repeated changes in the craniofacial shape of African cichlids. International Journal of Evolutionary Biology 2011, 1–10.
| Modularity of the oral jaws is linked to repeated changes in the craniofacial shape of African cichlids.Crossref | GoogleScholarGoogle Scholar |
Pavan-Kumar A, Gireesh-Babu P, Jaiswar AK, Chaudhari A, Krishna G, Lakra WS (2016) DNA barcoding of marine fishes: prospects and challenges. In ‘DNA barcoding in marine perspectives: assessment and conservation of biodiversity’. (Eds S Trivedi, AA Ansari, SK Ghosh, H Rehman) (Springer International Publishing) https://doi.org/10.1007/978-3-319-41840-7
Poore GCB, O’Hara TD (2007) Marine biogeography and biodiversity of Australia. In ‘Marine ecology’. (Eds SD Connell, BM Gillanders) pp. 177–198. (Oxford University Press: Melbourne, Vic., Australia)
Rodgveller, CJ, Hutchinson, CE, Harris, JP, Vulstek, SC, and Guthrie, CM (2017). Otolith shape variability and associated body growth differences in giant grenadier, Albatrossia pectoralis. PLoS ONE 12, e0180020.
| Otolith shape variability and associated body growth differences in giant grenadier, Albatrossia pectoralis.Crossref | GoogleScholarGoogle Scholar |
Rozas, J, Ferrer-Mata, A, Sánchez-DelBarrio, JC, Guirao-Rico, S, Librado, P, Ramos-Onsins, SE, and Sánchez-Gracia, A (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34, 3299–3302.
| DnaSP 6: DNA sequence polymorphism analysis of large data sets.Crossref | GoogleScholarGoogle Scholar |
Saha, A, Hauser, L, Hedeholm, R, Planque, B, Fevolden, S-E, Boje, J, and Johansen, T (2017). Cryptic Sebastes norvegicus species in Greenland waters revealed by microsatellites. ICES Journal of Marine Science 74, 2148–2158.
| Cryptic Sebastes norvegicus species in Greenland waters revealed by microsatellites.Crossref | GoogleScholarGoogle Scholar |
Steer, MA, and Fowler, AJ (2015). Spatial variation in shape of otoliths for southern garfish Hyporhamphus melanochir – contribution to stock structure. Marine Biology Research 11, 504–515.
| Spatial variation in shape of otoliths for southern garfish Hyporhamphus melanochir – contribution to stock structure.Crossref | GoogleScholarGoogle Scholar |
Stephens, M, and Donnelly, P (2003). A comparison of bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics 73, 1162–1169.
| A comparison of bayesian methods for haplotype reconstruction from population genotype data.Crossref | GoogleScholarGoogle Scholar |
Stephens, M, Smith, NJ, and Donnelly, P (2001). A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 68, 978–989.
Stewart, J, and Hughes, JM (2007). Age validation and growth of three commercially important hemiramphids in south-eastern Australia. Journal of Fish Biology 70, 65–82.
| Age validation and growth of three commercially important hemiramphids in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Stewart, J, Walsh, C, Reynolds, D, Kendall, B, and Gray, C (2004). Determining an optimal mesh size for use in the lampara net fishery for eastern sea garfish, Hyporhamphus australis. Fisheries Management and Ecology 11, 403–410.
| Determining an optimal mesh size for use in the lampara net fishery for eastern sea garfish, Hyporhamphus australis.Crossref | GoogleScholarGoogle Scholar |
Stewart J, Hughes JM, Gray CA, Walsh C (2005) Life history, reproductive biology, habitat use and fishery status of eastern sea garfish (Hyporhamphus australis) and river garfish (H. regularis ardelio) in NSW waters. FRDC Project number 2001/027, final report series number 73. (NSW Department of Primary Industries Fisheries) https://doi.org/10.13140/RG.2.1.2688.6566
Stransky C (2014) Morphometric outlines. In ‘Stock identification methods: applications in fishery science’, 2nd edn. (Eds SX Cadrin, LA Kerr, S Mariani) pp. 129–140. (Elsevier) https://doi.org/10.1016/B978-0-12-397003-9.00007-2
Streelman, JT, and Karl, SA (1997). Reconstructing labroid evolution with single-copy nuclear DNA. Proceedings of the Royal Society of London – B. Biological Sciences 264, 1011–1020.
| Reconstructing labroid evolution with single-copy nuclear DNA.Crossref | GoogleScholarGoogle Scholar |
Swain DP, Hutchings JA, Foote CJ (2005) Environmental and genetic influences on stock identification characters. In ‘Stock identification methods: applications in fishery science’, 2nd edn. (Eds SX Cadrin, LA Kerr, S Mariani) pp. 45–85. (Elsevier) https://doi.org/10.1016/B978-012154351-8/50005-8
Takahashi, M, DiBattista, JD, Jarman, S, Newman, SJ, Wakefield, CB, Harvey, ES, and Bunce, M (2020). Partitioning of diet between species and life history stages of sympatric and cryptic snappers (Lutjanidae) based on DNA metabarcoding. Scientific Reports 10, 4319.
| Partitioning of diet between species and life history stages of sympatric and cryptic snappers (Lutjanidae) based on DNA metabarcoding.Crossref | GoogleScholarGoogle Scholar |
Tamura, K, and Nei, M (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10, 512–526.
| Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees.Crossref | GoogleScholarGoogle Scholar |
Tea, Y-K, Van Der Wal, C, Ludt, WB, Gill, AC, Lo, N, and Ho, SYW (2019). Boomeranging around Australia: historical biogeography and population genomics of the anti-equatorial fish Microcanthus strigatus (Teleostei: Microcanthidae). Molecular Ecology 28, 3771–3785.
| Boomeranging around Australia: historical biogeography and population genomics of the anti-equatorial fish Microcanthus strigatus (Teleostei: Microcanthidae).Crossref | GoogleScholarGoogle Scholar |
Vignon, M, and Morat, F (2010). Environmental and genetic determinant of otolith shape revealed by a non-indigenous tropical fish. Marine Ecology Progress Series 411, 231–241.
| Environmental and genetic determinant of otolith shape revealed by a non-indigenous tropical fish.Crossref | GoogleScholarGoogle Scholar |
Villesen, P (2007). FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes 7, 965–968.
| FaBox: an online toolbox for fasta sequences.Crossref | GoogleScholarGoogle Scholar |
Ward, RD, Zemlak, TS, Innes, BH, Last, PR, Paul, D, and Hebert, N (2005). DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society of London – B. Biological Sciences 360, 1847–1857.
| DNA barcoding Australia’s fish species.Crossref | GoogleScholarGoogle Scholar |
Waters, JM (2008). Marine biogeographical disjunction in temperate Australia: historical landbridge, contemporary currents, or both? Diversity and Distributions 14, 692–700.
| Marine biogeographical disjunction in temperate Australia: historical landbridge, contemporary currents, or both?Crossref | GoogleScholarGoogle Scholar |
Wilson, NG, Stiller, J, and Rouse, GW (2017). Barriers to gene flow in common seadragons (Syngnathidae: Phyllopteryx taeniolatus). Conservation Genetics 18, 53–66.
| Barriers to gene flow in common seadragons (Syngnathidae: Phyllopteryx taeniolatus).Crossref | GoogleScholarGoogle Scholar |
Wu, L, Cai, W, Zhang, L, Nakamura, H, Timmermann, A, Joyce, T, McPhaden, MJ, Alexander, M, Qiu, B, Visbeck, M, Chang, P, and Giese, B (2012). Enhanced warming over the global subtropical western boundary currents. Nature Climate Change 2, 161–166.
| Enhanced warming over the global subtropical western boundary currents.Crossref | GoogleScholarGoogle Scholar |
Zhuang, L, Ye, Z, and Zhang, C (2015). Application of otolith shape analysis to species separation in Sebastes spp. from the Bohai Sea and the Yellow Sea, Northwest Pacific. Environmental Biology of Fishes 98, 547–558.
| Application of otolith shape analysis to species separation in Sebastes spp. from the Bohai Sea and the Yellow Sea, Northwest Pacific.Crossref | GoogleScholarGoogle Scholar |


