Species delineation, phylogeography and conservation of temperate perches (Actinopterygii: Percichthyidae) from an endemism and climate change hotspot
Michael P. Hammer A , David L. Morgan B , Stephen J. Beatty B , Mark G. Allen B , Peter J. Unmack C D , Sean J. Buckley E F G , Luciano B. Beheregaray
C D , Sean J. Buckley E F G , Luciano B. Beheregaray  E , Jon Murphy H and Mark Adams
E , Jon Murphy H and Mark Adams  I J *
I J *
A
B
C
D
E
F
G
H
I
J
Abstract
The south-western corner of Australia is a biodiversity hotspot that includes a freshwater fauna with a high proportion of endemic species. The temperate perches comprise nearly half of the obligate freshwater fishes of the region, representing important components of local ecosystems and are of significant conservation concern.
Provide a spatially comprehensive molecular genetic assessment of species boundaries and major substructure for all local members of the family to better understand the interplay of ecology and environment across a common landscape.
Nuclear markers (allozymes) and matrilineal (cytb) datasets were generated to infer genetic groupings and any instances of hybridisation or introgression in relation to the current taxonomy, regional geography and ecological understanding.
There were contrasting patterns of diversification across genera, with Nannoperca housing four likely species-level splits, Nannatherina having three distinct geographically and ecologically separated subpopulations, and Bostockia comprising several refugial subpopulations that appear partially introgressed. Repeated genetic patterns were identified across particular biogeographic features, most notably the Margaret River and Shannon River.
This study highlighted the value of comparative range-wide molecular studies to inform taxonomy, ecology and conservation planning.
These analyses pave the way for taxonomic revision, management of key habitat refuges, and other conservation actions.
Keywords: allozymes, aquatic biodiversity, biogeography, conservation genetics, cryptic species, freshwater fishes, mtDNA, species delineation.
Introduction
Freshwater habitats support high biodiversity, but also face significant and increasing threatening processes, with, for example, one quarter of all freshwater fishes now being considered at risk of extinction (Arthington et al. 2016; Liu et al. 2017; Reid et al. 2019; Sayer et al. 2025). The temperate perches (Percichthyidae) are a group of obligate freshwater fishes of Australia and South America, being diverse in terms of distribution patterns, physical attributes and ecology (McDowall 1996; GR Allen et al. 2002; Arratia and Quezada-Romegialli 2019), and are of special conservation concern, with an alarming number of species (55%) being considered threatened with extinction (Lintermans et al. 2020; International Union for Conservation of Nature 2024). The family contains 22 valid species in 8 genera (Jerry et al. 2001; Near et al. 2012; Fricke et al. 2024), ranging from small (<10 cm) habitat specialists of streams and wetlands to large (>1 m) iconic top-order predators in riverine and floodplain habitat, and including species of significant cultural and recreational value (Lintermans 2007; Saddlier et al. 2013; Humphries 2023). Detailed molecular studies have found many taxa within this group to harbour cryptic species, some now having been formally described and others yet to receive taxonomic attention (Musyl and Keenan 1992; Ruzzante et al. 2006; Nock et al. 2010; Morgan et al. 2013; Unmack et al. 2013; Hammer et al. 2014; Buckley et al. 2018). Moreover, molecular data have also revived ongoing contentions around higher-level classification, such as the status of some genera, and resolution of two diadromous species now considered to belong to their own, yet to be defined, family (Jerry et al. 2001; Betancur-R et al. 2017; Arratia and Quezada-Romegialli 2019).
The bulk of the Percichthyidae in Australia includes 13 valid species in the temperate south-east of the continent, predominantly in close association with the Great Dividing Range, but with singular outliers in the Wet Tropics (small habitat pocket) and drier inland Lake Eyre Basin (Pusey and Kennard 2001; GR Allen et al. 2002; Beheregaray et al. 2017). A smaller contingent also occurs in the Mediterranean climatic region of south-western Australia, represented by three genera and four locally endemic species (Morgan et al. 2011). The monotypic nightfish (Bostockia porosa Castelnau, 1873), is the largest of these (up to 17 v. <10 cm for other perches in the region), being widespread across varied habitats and considered Near Threatened (Morgan 2020). The western pygmy perch (Nannoperca vittata Castelnau, 1873), is also a widespread generalist species that is considered stable but Vulnerable, having undergone major historical declines following land clearance and secondary salinisation (Morgan et al. 2003; Beatty et al. 2011; Morgan 2019). This contrasts markedly with two more restricted and narrow-range habitat specialists, the little pygmy perch (Nannoperca pygmaea Morgan, Beatty & Adams, 2013), and Balston’s pygmy perch (Nannatherina balstoni Regan, 1906), both considered Endangered because of either a range reduction or extent of occupancy as well as ongoing localised threats in stream and seasonal wetland habitats (Morgan et al. 1995, 2014a; Beatty et al. 2011; Beatty and Morgan 2019; Morgan and Beatty 2019; MG Allen et al. 2020).
The south-western corner of Australia is a unique, yet precarious, environment for freshwater biota. It has been historically well isolated from other aquatic systems as essentially a small fringe of stable coastal habitat in a higher-rainfall zone enveloped by hot arid conditions and the Southern and Indian Oceans. The region, consequently, boasts very high species diversity and endemism, being globally recognised as a biodiversity hotspot (Myers et al. 2000). This is largely based on terrestrial attributes, with the species richness of aquatic fauna low in comparison, but still displaying high endemism, especially for groups more tied to a reliance on aquatic habitat; namely obligate freshwater fishes (9 native species, 100% endemism), decapod crayfishes (n = 14, 100%), frogs (n = 27, 97%), turtles (n = 2, 100%), caddisflies, stoneflies and mayflies (n = 91, 75%), and isopods and amphipods (n = 28, 82%) (see full review in Davies and Stewart 2013). Although the regional context appears to have promoted endemism, the same geographic fixation and isolation leaves the biota susceptible to anthropogenic perturbations. In particular, the interplay of rapid climate change combined with the increasing industry of human land-use is seen as a significant threat to south-western freshwater fishes, with major declines in stream flow concomitant to reduced rainfall already observed (Morrongiello et al. 2011; Beatty et al. 2014; Ogston et al. 2016; MG Allen et al. 2020) and even more likely in the future as a region under elevated risk (Tims and Saupe 2023; Buckley et al. 2024).
Limited previous systematic treatment has been undertaken on the Percichthyidae within south-western Australia, with the alpha taxonomy being largely stable since the early 1900s (McCulloch 1929; Kuiter and Allen 1986; GR Allen et al. 2002). Broad work on a molecular phylogeny to understand continental east–west relationships affirmed the local genera and species endemism and flagged initial insights on cryptic diversity present as two major lineages within Nannoperca (Unmack et al. 2011). Nannoperca pygmaea was only recently discovered during environmental monitoring surveys as an obviously distinct phenotypic form hidden in a discrete area, and which was subsequently investigated and described (Morgan et al. 2013) using a combined-lines-of-evidence approach considering molecular, morphological and ecological data (Page et al. 2005; Hammer et al. 2013, 2018). This species was also included in subsequent phylogenomic assessments of pygmy perches (Buckley et al. 2018, 2024), as one of four proposed candidate species across the two major lineages previously observed (Unmack et al. 2011), with the selection of populations included by Buckley et al. (2018, 2024) being guided by the unpublished results of the current study. Herein, we present the first comprehensive spatial genetic assessment of all south-western percichthyids to map species boundaries and contemporary genetic connectivity, within a comparative focus to assess how levels and patterns of genetic divergence might be explained by ecological traits across genera and groups (Tibbets and Dowling 1996; Hughes et al. 2009; Hammer et al. 2019a). The baseline of ecological data to draw comparative observations from is significant owing to a sustained 30-year research program in the region (e.g. Morgan et al. 1998, 2004, 2011; Morgan and Gill 2000; Beatty et al. 2011, 2014; MG Allen et al. 2020).
The objectives of this study are to undertake a specific assessment of species boundaries across the south-western Australian percichthyids, and document patterns of divergence in relation to geography, including any signs of hybridisation or introgression (common in freshwater fishes and recorded in percichthyids: Unmack et al. 2011, 2013; Valenzuela-Aguayo et al. 2022). This study employs a combined molecular approach using multiple, moderately conservative nuclear genetic markers (allozyme loci) and a matrilineal gene (mitochondrial DNA, mtDNA; gene cytochrome b, cytb) to assess genetic and taxonomic diversity with a geographically comprehensive dataset. The generation of these data aims to pave the foundations for somewhat urgent conservation programs and follow-up studies in a biodiverse region experiencing increasing anthropogenic change.
Materials and methods
Sampling
The hydrological organisation for surface waters followed here is the Australian Hydrological Geospatial Fabric, which groups smaller single catchments or sub-basins into larger river basin reporting units (Atkinson et al. 2008). The temperate south-west of Australia is an expansive region occupying a coastal fringe of temperate river, stream and wetland habitats comprising three main sections relevant to obligate freshwater fish habitat. First, there is a series of ‘northern’ river basins running ~500 km, draining east–west to the coast, largely disembarking from the Darling Range, from the Arrowsmith River (Greenough River Basin) in the north, through the large Swan Coast–Avon River Basin covering the major city of Perth, and south to the Collie–Preston River Basin; there is a second series of ~200 km ‘southern’ flowing river basins between the Donnelly River Basin and the Angove River (Albany Coast River Basin); and wedged in between these regions is the small Busselton Coast River Basin (including the prominent Margaret River catchment), which occurs on a small peninsula (western flowing), plus the Blackwood River, which extends well inland and divides both northern and southern regions (Fig. 1). The region experiences a Mediterranean climate with strong seasonality in rainfall and stream flow (winter–spring dominated) that is regionally variable (mean annual rainfall ranges between 500 and 1200 mm) and highest along the southern coast (Morgan et al. 2002, 2014b; Buckley et al. 2024).
Map of sampling sites for south-western Australia’s percichthyids that were subjected to combined allozyme and mitochondrial DNA (mtDNA) genetic analysis (asterisks show that Sites a1–4 represent alcohol-preserved tissues only, i.e. mtDNA exclusive assessment). The hydrological framework of catchments grouped into river basins follows the Australian Hydrological Geospatial Fabric (Atkinson et al. 2008). Past sea levels are approximated from bathymetric data (General Bathymetric Chart of the Ocean 2024) and bathy rivers are reconstructed drainage patterns at low sea level after Unmack (2001).
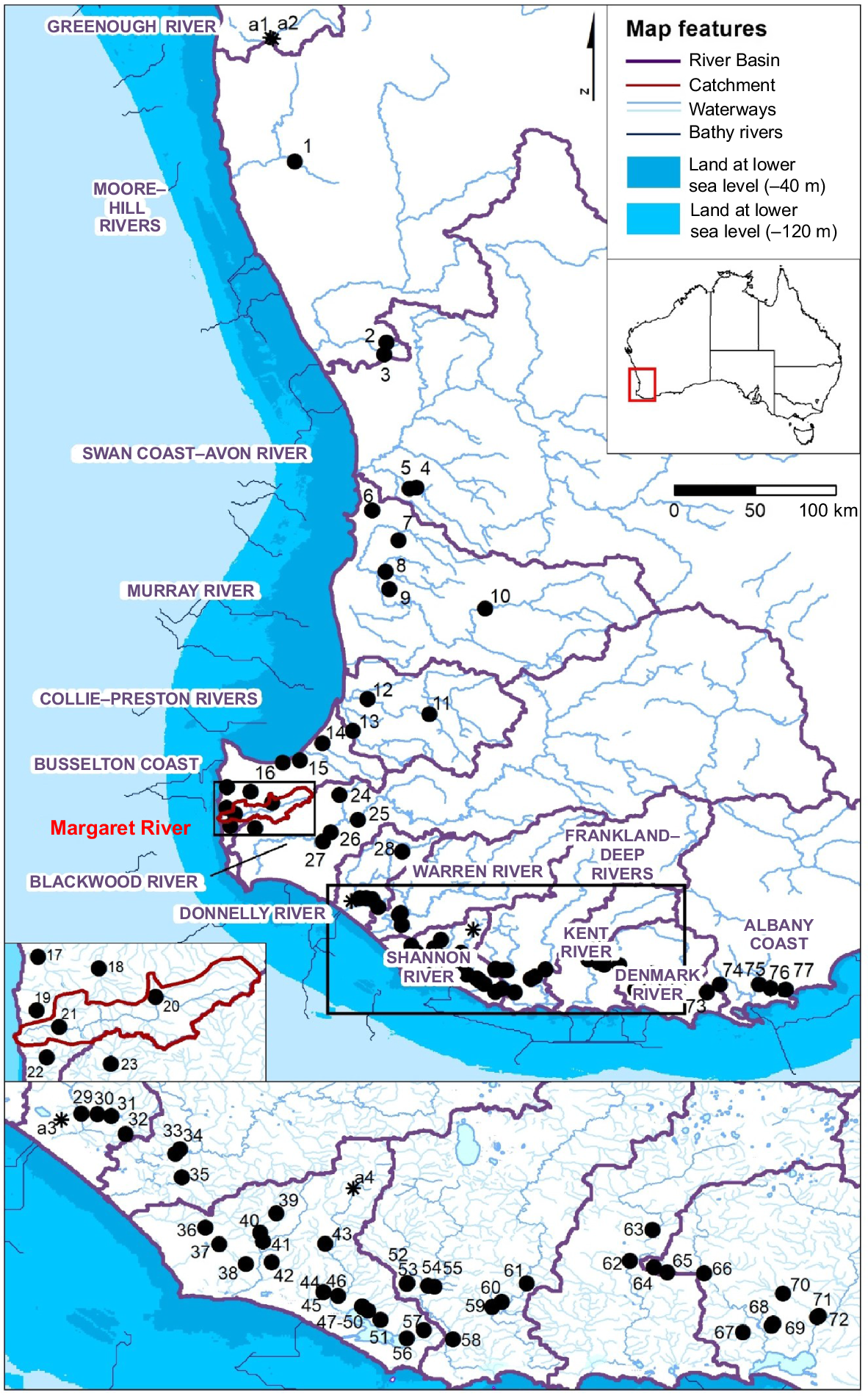
Percichthyids were sampled on multiple trips over 15 years by using a combination of methods from passive (traps and fyke nets) and active (dip net, seine net, backpack electrofishing) gear types. Sampling covered the extant range of all south-western percichthyids on the basis of review of historic records, baseline sampling and detailed regional investigations (e.g. Morgan et al. 2010, 2013, 2014b; Murphy 2010). Sampling effort thus represented a combination of dedicated surveys and value-adding to other projects to build up a bank of tissues for genetic assessment. Sampling was spatially comprehensive across the region, with material collected from some 81 sites between 1999 and 2014, representing different combinations of the three genera at each (Fig. 1, Table 1).
| Site | ABTC site code | Year | River Basin | Locality | Latitude | Longitude | NP | VWI | VMA | VSH | NTHB | BP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DM10-HI | 2010 | Moore–Hill | Hill River | −30.3050 | 115.4110 | 5/2 | ||||||
| 2 | PU09-30 | 2009 | Moore–Hill | Gingin Brook | −31.3120 | 115.9230 | 5/5 | 5/1 | |||||
| 3 | DM09-m04 | 2009 | Moore–Hill | Lennard Brook | −31.3800 | 115.9100 | 3/3 | ||||||
| 4 | MA08-64 | 2008 | Swan Coast–Avon | Canning River | −32.1229 | 116.0895 | 5/5 | ||||||
| 5 | DM02-m01 | 2002 | Swan Coast–Avon | Canning River #2 | −32.1300 | 116.0500 | 3/1 | 3/2 | |||||
| 6 | MA08-69 | 2008 | Murray | Brunswick River | −32.2504 | 115.8414 | 5/5 | ||||||
| 7 | MA08-68 | 2008 | Murray | Dirk Brook | −32.4173 | 115.9882 | 3/3 | 1/1 | |||||
| 8 | MA08-67 | 2008 | Murray | South Dandalup River | −32.5923 | 115.9166 | 5/5 | ||||||
| 9 | MA08-66 | 2008 | Murray | Murray River tributary | −32.6900 | 115.9375 | 3/3 | ||||||
| 10 | PU09-36 | 2009 | Murray | Hotham River | −32.7990 | 116.4720 | 1/1 | ||||||
| 11 | PU09-60 | 2009 | Collie–Preston | South Collie River | −33.3870 | 116.1630 | 5/5 | ||||||
| 12 | MA08-70 | 2008 | Collie–Preston | Collie River | −33.3027 | 115.8176 | 5/5 | ||||||
| 13 | MA08-72 | 2008 | Collie–Preston | Preston River | −33.4802 | 115.7335 | 5/5 | 3/1 | |||||
| 14 | MA08-73 | 2008 | Busselton Coast | Capel River | −33.5505 | 115.5643 | 1/1 | ||||||
| 15 | MA08-75 | 2008 | Busselton Coast | Abba River | −33.6462 | 115.4382 | 5/5 | 2/1 | |||||
| 16 | MA08-78 | 2008 | Busselton Coast | Vasse River | −33.6571 | 115.3422 | 5/4 | 4/1 | |||||
| 17 | MA08-80 | 2008 | Busselton Coast | Wilyabrup Brook | −33.7941 | 115.0313 | 5/4 | ||||||
| 18 | JM12-CACA | 2012 | Busselton Coast | Carbunup Brook | −33.8191 | 115.1623 | 4 | ||||||
| 19 | JM12-ELCA | 2012 | Busselton Coast | Ellen Brook | −33.9093 | 115.0286 | 1 | 3 | |||||
| 20(A) | PU09-58 | 2009 | Busselton Coast | Canebrake Pool 2009 | −33.8810 | 115.2830 | 4 A/4 | 1/1 | 3/1 | ||||
| 20(B) | JM12-CBMA | 2012 | Busselton Coast | Canebrake Pool 2012 | −33.8810 | 115.2830 | 8 | 3/1 | 5 | ||||
| 21 | MA08-79 | 2008 | Busselton Coast | Margaret River | −33.9446 | 115.0768 | 7/7 | 1/1 | |||||
| 22 | JM12-BOST | 2012 | Busselton Coast | Boojidup Brook | −34.0106 | 115.0500 | 3 | ||||||
| 23 | JM12-UCBDA | 2012 | Blackwood | Upper Chapman Brook | −34.0241 | 115.1879 | 4 | 3 | |||||
| 24 | JM12-MILBAK | 2012 | Blackwood | Mill Brook | −33.8376 | 115.6597 | 4 | 3 | |||||
| 25 | MA08-81 | 2008 | Blackwood | Blackwood River | −33.9760 | 115.7620 | 4/2 | ||||||
| 26 | JM12-MIBL | 2012 | Blackwood | Milyeannup Brk | −34.0454 | 115.6104 | 4 | 6 | 3 | ||||
| 27 | PU09-53 | 2009 | Blackwood | Milyeannup Brook | −34.0980 | 115.5680 | 4/4 | 1/1 | 4/1 | ||||
| 28 | MA08-82 | 2008 | Donnelly | Donnelly River | −34.1546 | 116.0088 | 3/3 | ||||||
| 29 | PU09-52 | 2009 | Donnelly | Donnelly River #2 | −34.4150 | 115.7720 | 4/4 | ||||||
| 30 | JM12-CARDO | 2012 | Donnelly | Carey Brook | −34.4158 | 115.8100 | 1 | 1 | |||||
| 31 | JM12-BEEDO | 2012 | Donnelly | Beedilup Brook | −34.4199 | 115.8422 | 1 | ||||||
| 32 | JM12-FLYD | 2012 | Donnelly | Fly Brook | −34.4623 | 115.8751 | 5 | 1 | |||||
| 33 | PU09-51 | 2009 | Warren | Treen Brook | −34.4970 | 116.0020 | 5/5 | ||||||
| 34 | JM12-WARV | 2012 | Warren | Warren River | −34.5078 | 115.9912 | 1 | 3 | |||||
| 35 | JM12-DOWA | 2012 | Warren | Dombakup Brook | −34.5635 | 116.0064 | 4 | 2 | |||||
| 36 | PU09-50 | 2009 | Shannon | Meerup River | −34.6800 | 116.0610 | 10/5 | 1 | |||||
| 37 | PU09-49 | 2009 | Shannon | Doggerup Creek | −34.7190 | 116.0930 | 5/5 | 3/1 | |||||
| 38 | MH00-m14 | 2000 | Shannon | near Chesapeake Brook | −34.7650 | 116.1560 | 7/2 | 2/2 | 3/2 | ||||
| 39 | PU09-48 | 2009 | Shannon | Boorara Brook | −34.6470 | 116.2270 | 4/3 | 3/1 | |||||
| 40 | JM12-BOOGA | 2012 | Shannon | Boorara Brook #2 | −34.6922 | 116.1887 | 5 | 3 | |||||
| 41 | JM12-CAGAR | 2012 | Shannon | Canterbury River | −34.7120 | 116.1950 | 5 | 1 | |||||
| 42 | JM12-BUGA | 2012 | Shannon | Buldania Creek | −34.7604 | 116.2162 | 5 | 3 | |||||
| 43 | JM12-SHNE | 2012 | Shannon | Shannon River #2 | −34.7180 | 116.3401 | 1 | ||||||
| 44 | JM12-CHCH | 2012 | Shannon | Chesapeake Brook #1 | −34.8305 | 116.3359 | 6 | 7 | 2 | 2 | |||
| 45 | JM13-CHBI | 2013 | Shannon | Chesapeake Brook #2 | −34.8305 | 116.3362 | 4 | 7 | 10 | ||||
| 46 | JM13-SHBI | 2013 | Shannon | Shannon River #3 | −34.8388 | 116.3706 | 5 | 2 | 4 | ||||
| 47 | JM13-4THBI | 2013 | Shannon | Fourth River | −34.8636 | 116.4259 | 18 | 4 | 4 | ||||
| 48 | JM12-4THR | 2012 | Shannon | 4th River | −34.8636 | 116.4259 | 10 | 4 | 1 | 1 | |||
| 49 | JM13-KIBI | 2013 | Shannon | Kingsman River | −34.8656 | 116.4288 | 7 | ||||||
| 50 | MH00-m15 | 2000 | Shannon | Broke Inlet | −34.8730 | 116.4400 | 2 | ||||||
| 51 | JM13-BIBI | 2013 | Shannon | Big Creek | −34.8946 | 116.4691 | 2 | 1 | 3 | ||||
| 52 | MH00-m16 | 2000 | Shannon | Deep River | −34.8110 | 116.5300 | 3/3 | 4/1 | |||||
| 53 | JM13-BPDE | 2013 | Shannon | Beardmore Pool | −34.8085 | 116.5319 | 2 | 12 B | 10 | ||||
| 54 | JM12-WEBE | 2012 | Shannon | Weld River | −34.8150 | 116.5793 | 5 | 5 | 1 | ||||
| 55 | JM12-DEFE | 2012 | Shannon | Fernhook Falls | −34.8170 | 116.5951 | 1 | 1 | |||||
| 56 | JM13-INBI | 2013 | Shannon | Inlet River | −34.9380 | 116.5307 | 1 | 2 | |||||
| 57 | JM12-IRSWH | 2012 | Shannon | Inlet River #2 | −34.9184 | 116.5697 | 3 | 6 | 3 | ||||
| 58 | JM12-DERXRA | 2012 | Shannon | Deep River #2 | −34.9396 | 116.6380 | 5 | ||||||
| 59 | JM13-ELFA | 2013 | Frankland–Deep | Elsie Brook | −34.8643 | 116.7292 | 13 | 5 | |||||
| 60 | JM13-WEFA | 2013 | Frankland–Deep | Wedding Brook | −34.8523 | 116.7519 | 1 | ||||||
| 61 | JM12-FACA | 2012 | Frankland–Deep | Frankland River | −34.8102 | 116.8097 | 7 | ||||||
| 62 | JM12-KEMI | 2012 | Kent | Kent River | −34.7572 | 117.0499 | 5 | ||||||
| 63 | DM14-m03 | 2014 | Kent | Kent River #2 | −34.6857 | 117.1027 | 7 | ||||||
| 64 | DM14-m01 | 2014 | Denmark | Dennmark River | −34.7718 | 117.1053 | 1 | ||||||
| 65 | DM14-m02 | 2014 | Denmark | Dennmark River #2 | −34.7841 | 117.1365 | 6 | ||||||
| 66 | JM13-DENM | 2013 | Denmark | Denmark River #3 | −34.7866 | 117.2231 | 6 | ||||||
| 67 | JM12-SCLI | 2012 | Denmark | Scotsdale Brook | −34.9232 | 117.3124 | 2 | ||||||
| 68 | DM09-m03 | 2009 | Denmark | Quickup River | −34.9090 | 117.3790 | 3/3 | ||||||
| 69 | DM09-m02 | 2009 | Denmark | Quickup Dam | −34.9030 | 117.3840 | 3/3 | ||||||
| 70 | PU09-37 | 2009 | Denmark | Mitchell River | −34.8340 | 117.4060 | 3/3 | ||||||
| 71 | DM09-m01 | 2009 | Denmark | Mitchell River #2 | −34.8880 | 117.4860 | 8 C/7 | 5/5 | 4/2 | 5/3 | |||
| 72 | JM13-MTHR | 2013 | Denmark | Hay River | −34.8847 | 117.4894 | 6 | ||||||
| 73 | PU09-38 | 2009 | Denmark | Marbelup Brook | −34.9400 | 117.7120 | 5/5 | 2 | |||||
| 74 | PU09-41 | 2009 | Albany Coast | King River | −34.8950 | 117.7830 | 4/4 | ||||||
| 75 | MA08-85 | 2008 | Albany Coast | Kalgan River | −34.8958 | 118.0023 | 4/4 | ||||||
| 76 | MH04-m02 | 2004 | Albany Coast | Goodga River | −34.9170 | 118.0670 | 3/3 | ||||||
| 77 | MH04-m01 | 2004 | Albany Coast | Angove River | −34.9230 | 118.1520 | 3/3 | 1 | |||||
| a1 | DM10-m02 | 2010 | Greenough | Arrowsmith #1 | −29.6184 | 115.2904 | 0/2 | ||||||
| a2 | DM10-m03 | 2010 | Greenough | Arrowsmith #2 | −29.5170 | 115.4410 | 0/1 | ||||||
| a3 | DM14-SMI | 2014 | Donnelly | Lake Smith | −34.4291 | 115.7260 | 0/5 | ||||||
| a4 | PU99-SHA | 1999 | Shannon | Shannon River | −34.5887 | 116.4063 | 0/1 | ||||||
| 22/12 | 255/119 | 19/11 | 62/10 | 89/9 D | 97/23 | ||||||||
Taxon abbreviations as follows: NP, Nannoperca pygmaea; VWI, VMA and VSH, codes designated in the text for the three candidate species of Nannoperca; NTHB, Nannatherina balstoni; BP, Bostockia porosa. Sample sizes per site are shown as allozymes/cytb (a single number means that no cytb sequences were generated).
All sampling was approved by the participants’ Animal Ethics committees under approval codes W2237/09, RW2521/12 and RW2793/15 (Murdoch University) plus IACUC 07-0403 (Brigham Young University), and conducted under wildlife collecting permits SF006431 and SF007216. Ethical approval was not required by the SA Museum because all samples were sourced from existing tissue collections and involved fish that were ethically collected under valid collecting permits. Most tissues (either whole fish or a portion of muscle from the anterior flank on one side) were snap frozen in liquid nitrogen for subsequent ultra-cold storage as part of the South Australian Museum’s Australian Biological Tissues Collection (ABTC). A few tissues were available only by preservation in 100% ethanol (and therefore not useable for allozymes). In most cases, formalin-fixed fish, either the same individual sampled for muscle or representatives from the same site, were retained as matching voucher specimens and lodged with the Western Australian and South Australian Museums or held in the Murdoch University reference collection. All vouchers can be identified on the basis of their field code (Table 1).
Choice of nuclear markers
Seeing as this study was conceived in the early 2000s and commenced in 2011 after suitable tissues became available (with an early pilot study: Hammer 2001), we chose allozymes as our primary nuclear genetic markers. Although largely supplanted by genomic datasets since then, allozymes have a long and successful history as reliable and proven genetic markers for delineating species, detecting between-species admixture and identifying major phylogeographic breaks within species (Richardson et al. 1986; Horner and Adams 2007; Adams et al. 2014). Attesting to this, allozyme datasets have provided comparable insights to a companion genomic dataset for these three above-mentioned systematic endeavours in every study we have undertaken or tracked thus far (Blom et al. 2016; Unmack et al. 2017, 2019, 2022, 2023; Thacker et al. 2022; Mossop et al. 2023; M. Adams and P. J. Unmack, unpubl. data), and can occasionally even match genomic data for showing fine-scale population structure (e.g. Jusaitis and Adams 2005; Brooks et al. 2022). Most importantly, allozymes have been shown to be pivotal in the identification of more than 50 candidate species thus far among the Australian freshwater fish fauna alone (Adams et al. 2023) and the systematic and phylogeographic framework they provide allows any follow-up genomic studies to optimally target key individuals and populations during sampling design.
Allozyme analyses
Three separate allozyme studies were undertaken, one for each percichthyid genus. Because it can be problematic to detect their hybrids a priori and distinguish juvenile Nannoperca from Nannatherina, each of these two studies also ‘accidentally’ included several examples of the other genus or a single F1 hybrid. Excluding these individuals, the allozyme sample sizes for each study were as follows: Nannoperca, 359 individuals from 67 sites; Nannatherina, 89 individuals from 21 sites; and Bostockia, 97 individuals from 38 sites (Fig. 1, Table 1).
Allozyme electrophoresis of muscle homogenates was conducted on cellulose acetate gels (Cellogel), according to standard principles and procedures (Richardson et al. 1986). All individuals were screened for the full suite of enzymes or non-enzymatic proteins employed in our previous studies on percichthyids (Hammer et al. 2010; Unmack et al. 2011, 2013; Morgan et al. 2013). Details of enzyme and locus abbreviations, enzyme commission numbers, electrophoretic conditions, stain recipes and allozyme nomenclature are presented in these papers or in Richardson et al. (1986).
For each generic dataset, the initial analysis involved the use of stepwise principal co-ordinates analysis (PCA) on individuals to identify discrete genetic lineages and any instances of likely admixture, independent of other taxonomic or geographic expectations. This procedure is always coupled with a comparison of the raw allozyme profiles of each major PCA cluster to assess cluster diagnosability (multiple fixed differences in sympatry being an unequivocal indication of the presence of two species at that site), detect obvious genetic heterogeneity at individual sites, and identify likely hybrids and individuals with significant admixture. The methods and protocols for conducting these stepwise PCAs are detailed elsewhere (Horner and Adams 2007).
Informed by our PCA characterisations of individuals, unique taxon–site combinations and primary genetic lineages, subsequent analyses for each genus involved calculating pairwise matrices of genetic distance and divergence among the chosen operational taxonomic units (whether individuals, sites, lineages or taxa). For assessments of species boundaries and lineage distinctiveness, we used the number of fixed differences (FDs) as the appropriate measure of genetic divergence and diagnosability. Consistent with the standard practice in morphological taxonomy of employing diagnostic characters that share character states at low frequency (Hammer et al. 2007; Horner and Adams 2007), our FD calculations allowed up to a 10% tolerance for the combined frequency of any rare shared alleles (as advocated by Adams et al. 2014).
We used two approaches to explore within-species genetic substructure for our ecological comparisons. Following standard methods (see Hammer et al. 2007; Horner and Adams 2007; Unmack et al. 2011), we constructed unrooted neighbour joining (NJ) networks by using Nei’s unbiased distance among sites (for species with adequate sample sizes at most sites, herein Nannoperca taxon ‘VWI’) or Rogers’ genetic distance among individuals (for Nannatherina and Bostockia). We also used STRUCTURE (ver. 2.3.4, see https://web.stanford.edu/group/pritchardlab/structure.html; Pritchard et al. 2000) to provide an independent assessment of population structure in all widespread candidate species plus one pair of parapatric taxa, with each input dataset limited to only those allozyme loci that were polymorphic in that species–taxon pair, after excluding loci with only rare alleles (i.e. sum of all rare alleles <0.05 in the metapopulation and <0.10 for each PCA and NJ cluster). Each STRUCTURE run employed the default settings for an admixture model with correlated allele frequencies and consisted of five replicates each for K = 1–8, with burn-in of 200,000 and 1,000,000 iterations. The results were then uploaded to the CLUMPAK website (see http://clumpak.tau.ac.il/; Kopelman et al. 2015) to provide estimates of the optimum K by using the ΔK method (Evanno et al. 2005) plus the raw input files required by CLUMPP (ver. 1.1.2, see https://rosenberglab.stanford.edu/clumpp.html; Jakobsson and Rosenberg 2007) to produce an optimum bar plot for each value of K. It should be noted here that, compared with genomic datasets, the numbers of informative allozyme loci are too low to provide any rigorous or detailed assessment of population structure in any taxon. Nevertheless, these initial explorations will help guide any future genomic studies aiming to provide such insights.
Mitochondrial DNA
For our matrilineal marker, we chose the cytb gene to build on an existing and informative dataset already available for pygmy perches (Unmack et al. 2011, 2013). In addition to sampling the geographic range of each genus, the choice of individuals to sequence was initially informed by the lineages identified in the allozyme datasets plus some preliminary mtDNA sequences (control region; Morgan et al. 2010; Murphy 2010), and, ultimately, by the phylogenetic and phylogeographic insights evident after the initial round of sequencing was completed for each group. Thus, there were considerably more cytb sequences generated for Nannoperca (four candidate species, structure evident within two of these) compared with the other two genera (little matrilineal structure evident and minimal or no support for the allozyme subpopulations). The final sample sizes for each genus were as follows (including sequences already on GenBank): Nannoperca, 152 individuals from 38 sites; Nannatherina, 9 individuals from 6 sites; and Bostockia, 23 individuals from 18 sites. All sequences were checked by amino acid coding to test for unexpected frame shift errors or stop codons.
All details regarding DNA isolation, amplification, sequencing and sequence editing are presented in Unmack et al. (2013). Phylogenetic analyses of the final sequence dataset employed maximum likelihood (ML), as implemented using IQ-TREE (ver. 1.6.12, see http://www.iqtree.org/; Nguyen et al. 2015) run on the W-IQ-TREE server (see http://iqtree.cibiv.univie.ac.at/; Trifinopoulos et al. 2016). These analyses employed the model selection procedure of IQ-TREE (-m TEST; Kalyaanamoorthy et al. 2017), resulting in the selection of the TIM2 + F + I + G4 model, followed by 10,000 replicates of ultrafast bootstrapping (Hoang et al. 2018).
Results
Identification of primary taxa and lineages in Nannoperca
The final nuclear dataset for Nannoperca comprised the allozyme genotypes of 359 fish at 57 putative loci. For simplicity, we have employed the following informal nomenclature throughout for the three diagnosable taxa currently assigned to N. vittata sensu lato: VWI, vittata ‘widespread’; VMA, vittata ‘Margaret River catchment’; and VSH, vittata ‘Shannon River Basin’. The allozyme profiles and heterozygosity measures for all pure taxa and two ‘hybrid’ individuals, along with a reference profile for N. balstoni, are presented in Supplementary Table S1.
An initial PCA on all individuals (Fig. 2a) displayed a primary split in the first dimension between taxa VWI + VMA (Lineage I) v. taxon VSH + N. pygmaea (Lineage II). Also evident was a single outlier individual whose allozyme profile and observed heterozygosity (HO) count were consistent with being a (VSH × VWI) × VSH backcross (Table S1) and was collected from a site harbouring both parental taxa (Site 53; Table 1). All four primary taxa were readily distinguishable by PCA once additional dimensions were considered (dimensions 2 and beyond for VWI v. VMA, inset follow-up PCA for VSH v. N. pygmaea; Fig. 2a) and all were unequivocally diagnosable from one another by multiple fixed allozyme differences (minimum five FDs, range 5–20; Fig. 2a, Table S1). These levels of diagnosability become even more substantial given there are two instances of sympatry among the four taxa (VWI and VSH at multiple sites; VWI and N. pygmaea; Table 1) plus taxon VMA is fully enveloped by VWI sites (Table 1; Fig. 1), although technically these two taxa are parapatric. For reference purposes, N. balstoni was diagnosable from the four Nannoperca taxa by 25–31 FDs (Table S1).
Scatterplots of principal co-ordinates analysis (PCA) scores in the first two dimensions for the three allozyme datasets. The relative contribution of each dimension is shown in parentheses (axes not scaled accordingly). Primary genetic groups are identified according to the colour key provided and likely admixed individuals are indicated by using a colour gradient representing their putative parental groups. Also shown are the number of fixed differences (FDs, as defined in the text) that diagnose each ‘pure’ primary group. (a) Initial PCA of all 359 Nannoperca; inset shows a second PCA focusing only N. pygmaea plus taxon VSH (n = 83). (b) Initial PCA for Nannatherina (n = 89). (c) Initial PCA for Bostockia (n = 97) with hypothesised ‘pure’ groups (PNC, PBL and PMA) labelled. Fish images © R. Swainston (see animafish.com), reproduced with permission.
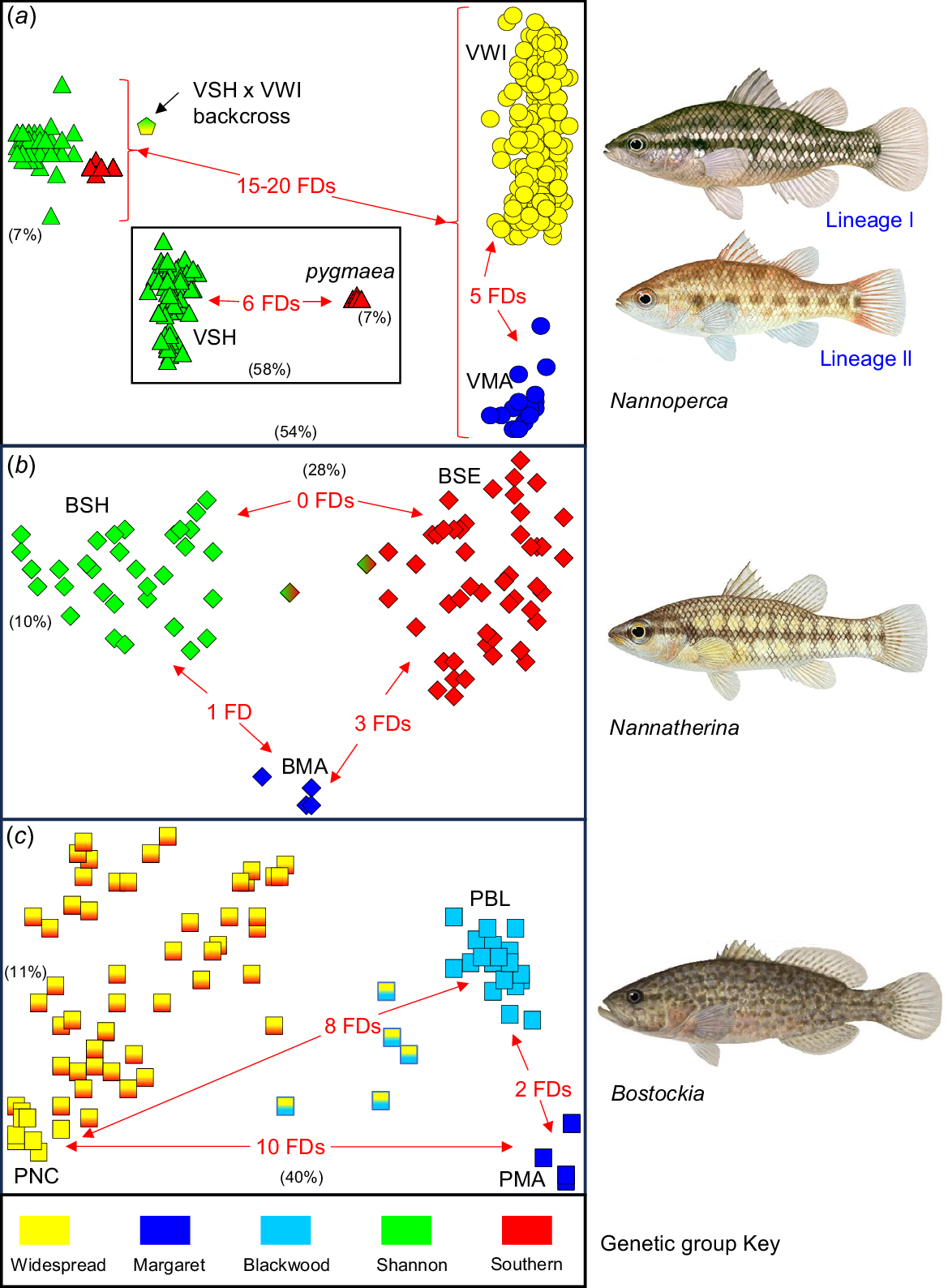
In addition to detecting the backcrossed individual at Site 53, allozyme profiling also showed the presence of a single F1 hybrid between N. pygmaea and N. balstoni at Site 71 (Table S1). This individual was highly heterozygous (HO > 0.5, compared with HO < 0.01 for both parental species), notably at all 27 diagnostic loci.
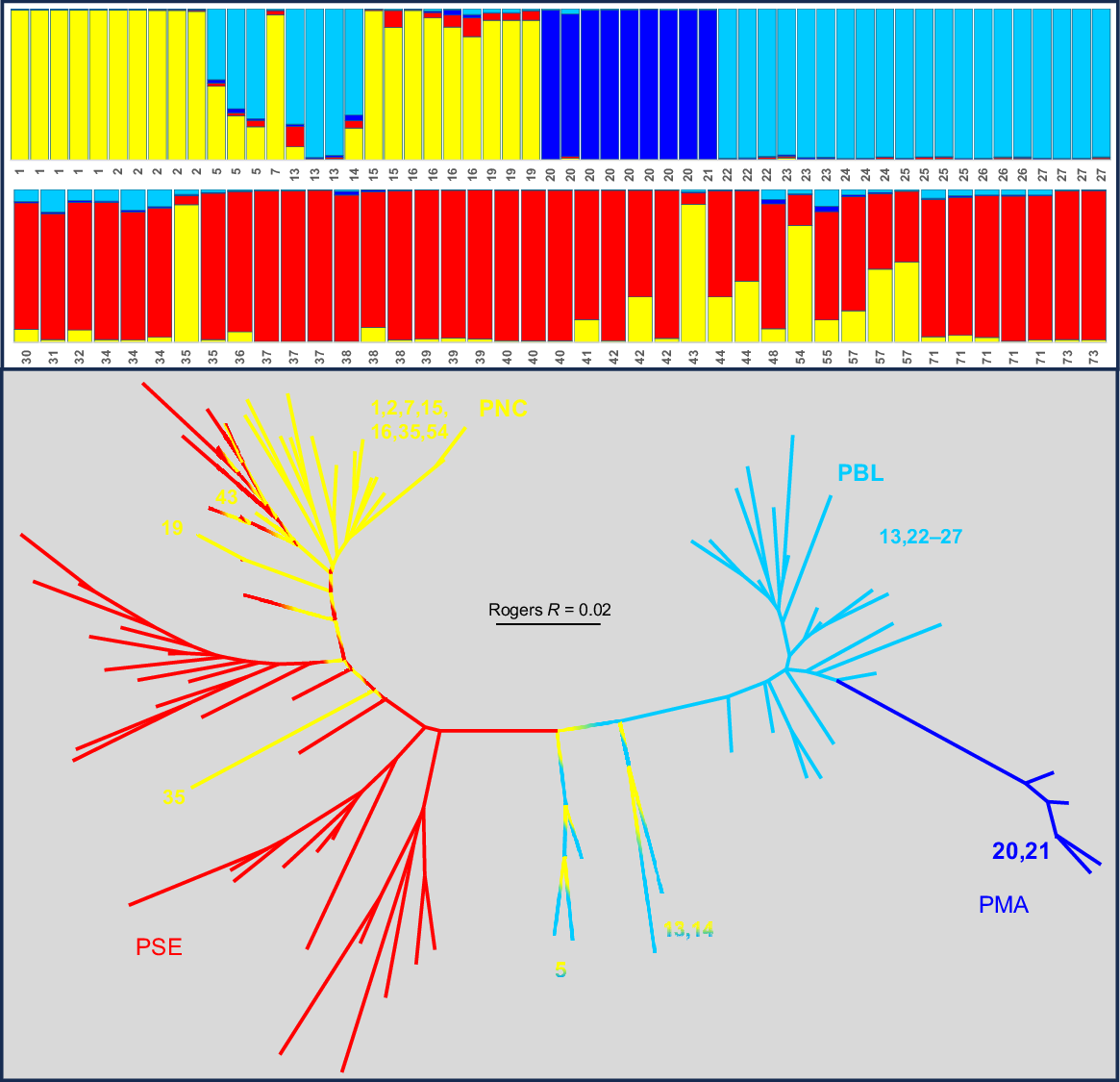
Population structure in taxon VWI
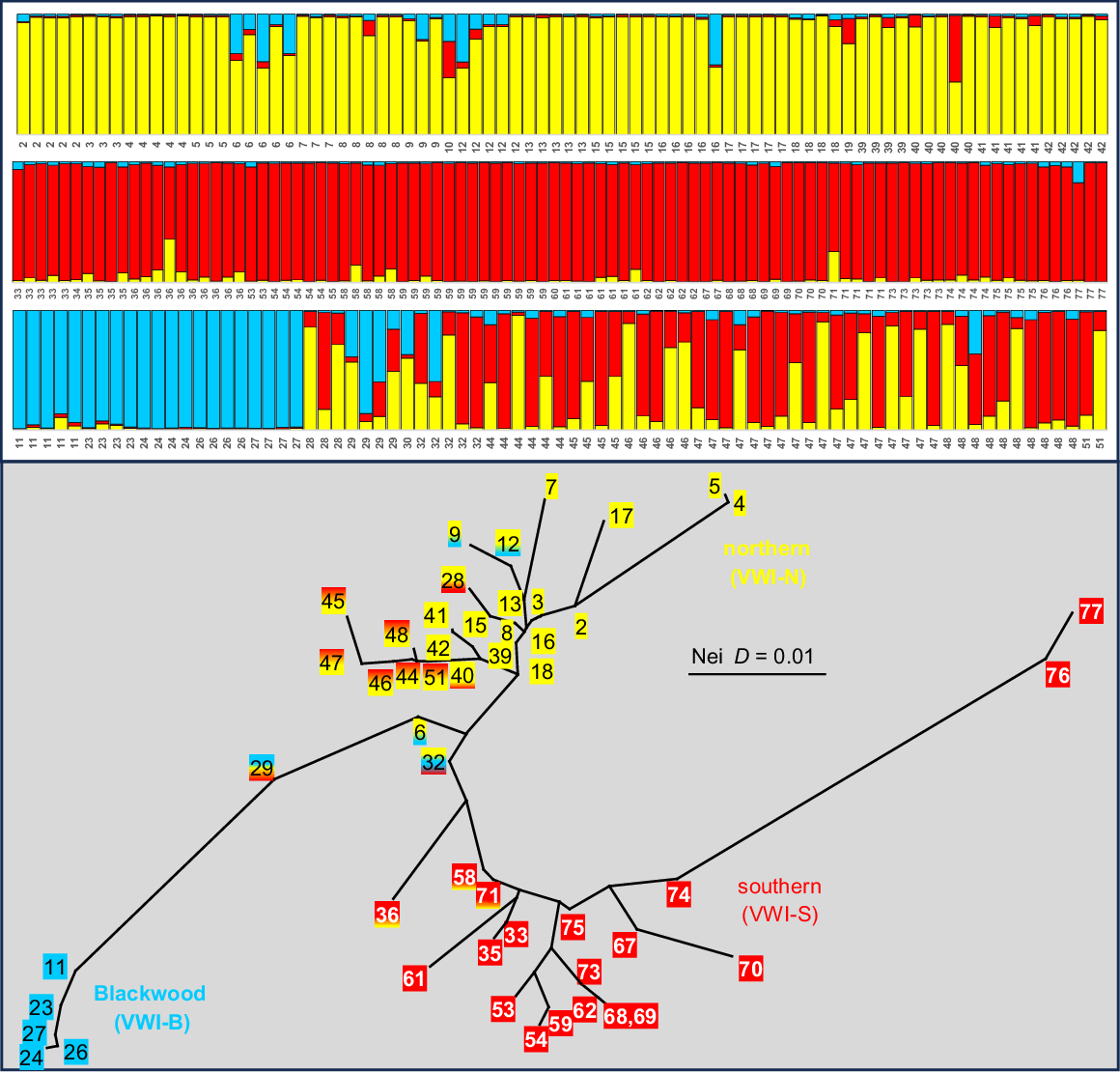
Of the four taxa evident in Nannoperca, only the broadly distributed VWI displayed sufficient within-taxon genetic heterogeneity to warrant an exploration of population structure. Fig. 3 presents the STRUCTURE plot for K = 3 (the optimum ΔK value), matched with a NJ network among those sites with n > 1. Both analyses are broadly concordant in identifying three subpopulations corresponding to ‘northern’, ‘Blackwood’ (all Blackwood sites plus Site 11 in the upper Collie), and ‘southern’, albeit with considerable admixture among all three subpopulations at a number of sites that mostly occupy geographically intermediate positions.
Companion STRUCTURE K = 3 plot (all individuals) and NJ network among sites (excluding those where n = 1) for Nannoperca taxon VWI. Individuals in the STRUCTURE plot and termini in the NJ network are labelled by site number (following Table 1 and Fig. 1) and display matching colours that broadly reflect the geographic template designated in Fig. 2.
Admixture between taxa VWI and VMA
Despite the presence of five FDs between the two parapatric taxa VWI and VMA, there was some indication of potential admixture in the initial PCA (Fig. 2a) and a follow-up PCA (not shown). A STRUCTURE plot (K = 4, the optimum ΔK value) involving all individuals of both taxa (Fig. 4) indicates that all but one VMA fish were pure exemplars of a distinctive VMA lineage. As expected, it is likely that the single anomalous individual had a hybrid ancestry involving pure VMA introgressed with VWI (presumably from a regional VWI population, with Site 19 being particularly close). Although this admixed individual (from Site 21 in the township of Margaret River itself) presented as the most prominent PCA outlier (Fig. 2a), its allozyme profile was not consistent with being an F1 hybrid, indicating that VWI × VMA F1 hybrids must be at least partially fertile.
STRUCTURE K = 4 plot for Nannoperca taxa VWI plus VMA (Sites 20 and 21). Individuals in the STRUCTURE plot are labelled by site number (following Table 1 and Fig. 1). Colours for VWI subpopulations match those used in Fig. 3. The single VMA individual displaying evidence of admixture is highlighted with a blue box.
Nannatherina allozyme analyses
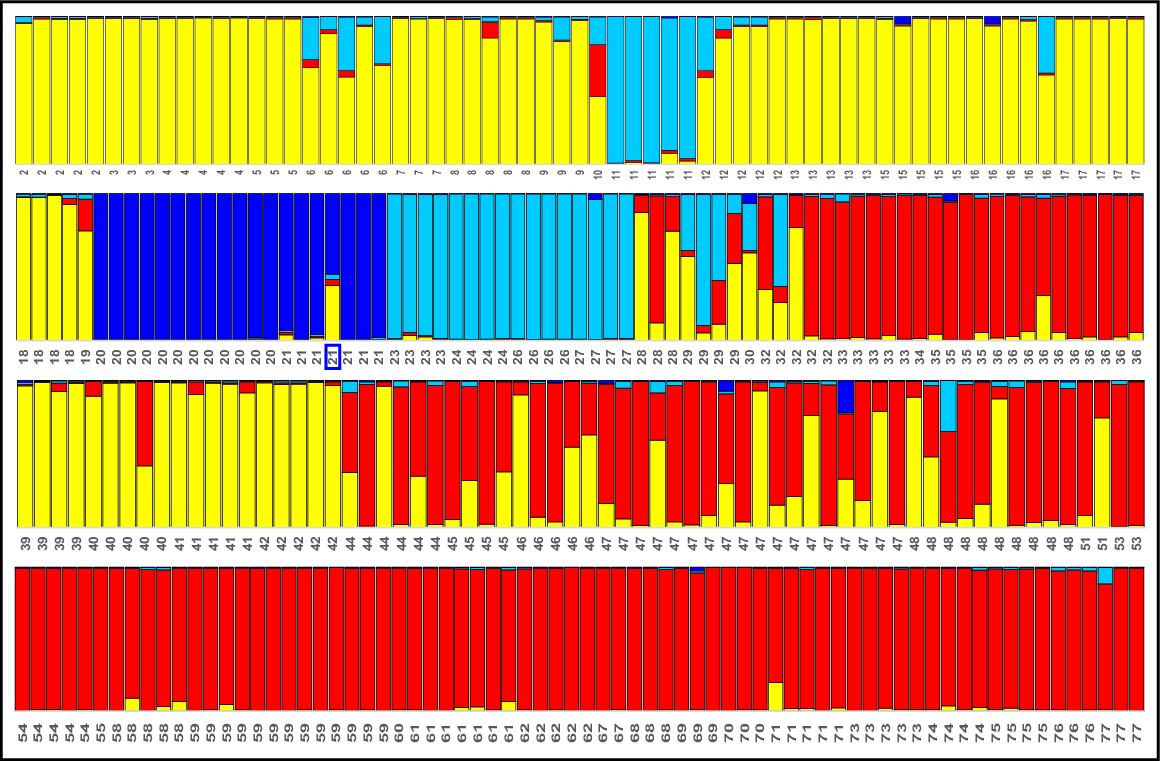
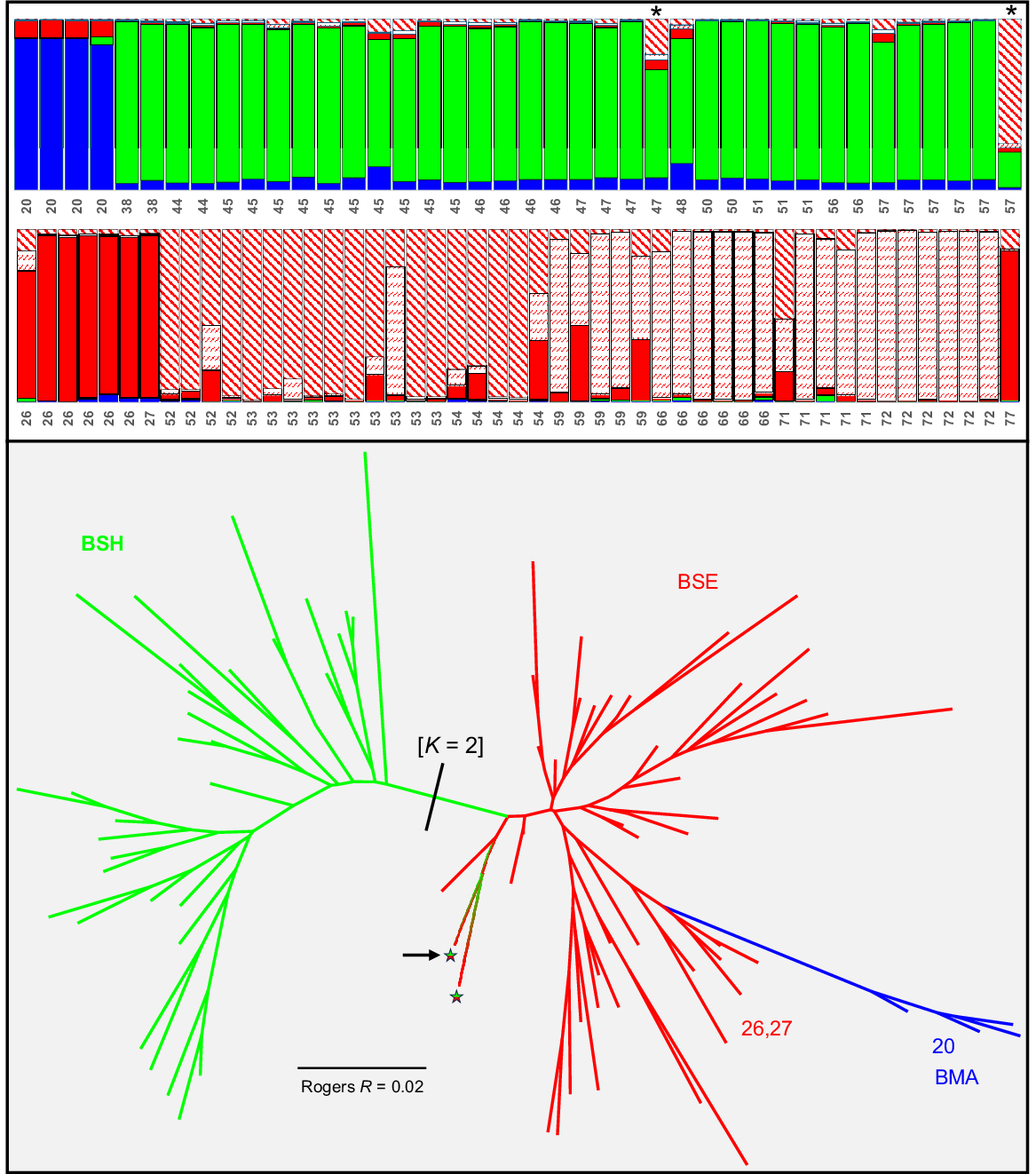
The final allozyme dataset for Nannatherina contained genotypes for 89 fish at 61 putative loci. An overview PCA of all individuals (Fig. 2b) inferred the existence of three primary genetic groups, namely (a) BMA (Site 20 in the Margaret River catchment, (b) BSH (Sites 38,44–48, 50, 51 and 57 in the Shannon River Basin), and (c) BSE (=‘southern’; Sites 26 and 27 in the Blackwood, all sites to the east of the Shannon, plus Sites 52–54 in the eastern portion of the Shannon). There was also some evidence of modest admixture at Sites 47 (1 of 4 fish) and 57 (1 of 6 fish) between BSH and BSE, an observation consistent with an absence of fixed differences. By contrast, BMA was diagnosable by the FD counts, most notably from the widespread BSE subpopulations (three FDs), despite occupying adjacent systems. However, such modest levels of diagnosability, coupled with small sample sizes and a lack of sympatry or shallow parapatry, are clearly insufficient to infer that these three groups warrant recognition as anything other than distinct subpopulations. The allozyme profiles and heterozygosity measures for the three genetic groups and the two outlier fish are presented in Table S2.
Further exploration of population structure in N. balstoni is summarised by the STRUCTURE analyses and NJ network (Fig. 5). The optimum ΔK estimate predicted K = 2 for STRUCTURE, corresponding to all BSH sites v. BMA plus BSE sites plus a single admixed at Site 57 (Fig. 5). Also presented is the detailed plot for K = 5, this being both the next most optimum value and the lowest value that consistently delineated the Margaret and Blackwood River sites from one another (as found by PCA and FD counts, Fig. 2b). Both this second analysis and the NJ network broadly supported the presence of the three primary PCA subpopulations plus two admixed individuals (Sites 47 and 57; individuals marked with an asterisk), although STRUCTURE further split the ‘southern’ BSE subpopulation into separate ‘Blackwood + Angove’, ‘Shannon’ and ‘east of Shannon’ subunits, with sporadic admixture between the latter two.
Companion STRUCTURE K = 5 plot and NJ network among individuals for Nannatherina. Individuals in the STRUCTURE plot are labelled by site number (following Table 1 and Fig. 1) and putative subpopulations are coloured to match those used in the NJ network (with each of the three ‘southern’ subpopulations bearing unique patterning). For the NJ network, the only individuals labelled are those for the Margaret (Site 20), Blackwood (Sites 26, 27), and the two individuals (marked with an asterisk) that were intermediate in the PCA of Fig. 2b; instead, branches are colour coded to reflect the geographic template used in Fig. 2. Also shown is the primary STRUCTURE split evident using the optimum value of K = 2 and the single Site 57 individual (arrowed) inferred to be admixed.
Bostockia allozyme analyses
For Bostockia, the final allozyme dataset involved 97 fish profiled at 56 putative loci. An initial PCA on all individuals (Fig. 2c) presented a complex geographic scenario. Reflecting the FD counts, we formulated a working hypothesis of three pure groups (diagnosable from one another by 2–10 FDs), namely (a) ‘upper northern coastal’ (PNC, Sites 1 and 2), (b) ‘Blackwood’ (PBL, Sites 22–27 plus two individuals from Site 13 in the Preston), and (c) ‘Margaret’ (PMA, Sites 20 and 21), plus two broad but still separable clusters of admixed individuals. One of these clusters was consistent (i.e. significantly more heterozygous and only 0–1 FDs from parental subpopulations; Table S3) with the five individuals concerned (Sites 5 and 14 plus one fish at Site 13) being contemporary ‘hybrids’ between PNC and PBL, whereas the other comprised a geographically diverse assemblage of individuals (all other sites) that displayed varying levels of presumably ongoing or ‘historic’ admixture between PBL and PNC. As for Nannatherina, the allozyme data are too limited in sampling and genomic intensity to conclude here that Bostockia comprises more than a single species. The allozyme profiles and heterozygosity measures for all hypothesised pure and admixed subpopulations are presented in Table S3.
Fig. 6 presents the STRUCTURE results and companion NJ network among individuals for Bostockia. Again, we chose K = 4 for the STRUCTURE plot rather than that predicted by ΔK (K = 2), this being the lowest value that consistently delineated the PMA and PBL subpopulations from one another (as shown by both PCA and FD counts, Fig. 2c). Broadly consistent with our PCA-based expectations, the STRUCTURE analysis delineated all three hypothesised ‘pure’ subpopulations and supported our prediction of five ‘hybrid’ fish involving PBL and PNC. However, it also provided an alternative viewpoint on population structure, predicting an expanded distribution for subpopulation PNC (allocating the more southerly Sites 7, 15, 16 and 19), and inferring that a fourth ‘southern’ subpopulation (PSE, pure Sites 30–34, 36–40, 71–73) is the parental subpopulation contributing to our hypothesised admixture with PNC (admixed Sites 35, 41–44, 48, 54, 55 and 57; Fig. 6) rather than the PBL subpopulation. The companion NJ network (Fig. 6), although less informative, is consistent with both alternative viewpoints.
Companion STRUCTURE K = 4 plot and NJ network among individuals for Bostockia. Individuals in the STRUCTURE plot are labelled by site number (following Table 1 and Fig. 1) and putative subpopulations are coloured to match those used in the NJ network. For the NJ network, only selected OTUs were labelled for cross-referencing with the PCA (Fig. 2c) and STRUCTURE plot; instead, branches are colour-coded to broadly reflect the geographic template used in Fig. 2.
Mitochondrial gene tree across all genera
The final cytb dataset consisted of 1141 bp of sequence data for 184 individuals (including 10 previously deposited in Genbank) across all three genera. In addition to positioning each of the three genera as very distinctive clades, the resultant ML tree (-ln score of −4642.492695, Fig. 7) strongly supports the existence of the four Nannoperca species identified by our allozyme analyses. As predicted herein and found previously (Buckley et al. 2018, 2024), Nannoperca comprises two primary lineages, one for sister species N. pygmaea and VSH and the other containing the closely related candidate species VWI and VMA. Within-species structure is also evident for species VWI (full haplotype tree in Supplementary Fig. S1), which presents as two moderately well-supported (bootstrap value 75%) clades with broadly ‘northern’ v. ‘southern’ distributions, but with some overlap in the Donnelly, Warren and western Shannon River Basins plus one geographic anomaly (Site 7; Fig. 7, S1). Of note was verification in the mtDNA dataset of an additional population of N. pygmaea from Lake Smith (Site a3, Donnelly River Basin), well separated from other sites to the east.
Maximum likelihood tree for the 184 cytb sequences available across all three genera. For species VWI, only the two primary clades are shown, alongside a map of their comparative geographic distributions. The full VWI haplotype tree is presented in Fig. S1. Individuals are labelled by species code + site code (as per Table 1) + individual code (.1, .2 etc. for fish from the same site). Genbank sequences are denoted by HQ, AY or MR numbers. Bootstrap proportions are indicated for the earlier-branching nodes by asterisks (***, 00%; **, 95–99%; *, 85–94%). Also shown are molecular clock estimates (millions of years ± s.d.) for key nodes (A–E) as calculated by Buckley et al. (2018)by using r8s, being in line with those of Unmack et al. (2011) using BEAST.
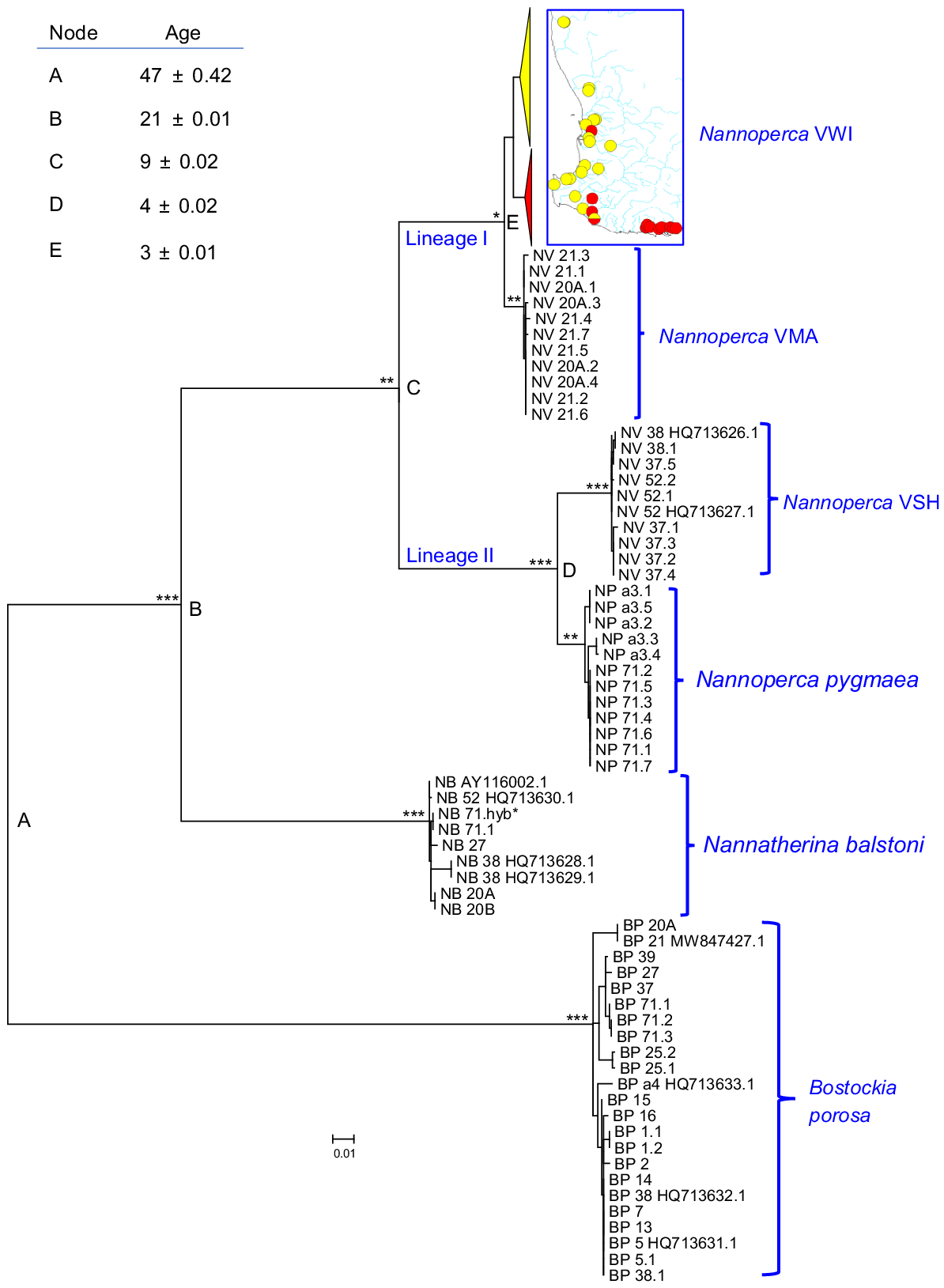
In stark contrast with Nannoperca, both Nannatherina and Bostockia displayed minimal cytb diversity across their entire geographic range that reflected all identified subpopulations (Fig. 7). This result further reinforces the hypothesis that neither harbours cryptic taxa. It also indicates that both sets of subpopulations are likely to experience genetic connectivity over time whenever environmental conditions permit.
Fig. 7 also includes the best available molecular clock estimates for the key nodes in the south-western percichthyid phylogeny, as calculated by Buckley et al. (2018) by using r8s (ver. 1.81, see https://sourceforge.net/projects/r8s/) for 13,991 concatenated single-nucleotide polymorphism (SNP) loci and calibrated using the aridification of the Nullarbor region to date the loss of connectivity between eastern and western Nannoperca species. These dates are also consistent with those obtained for a combined analysis of cytb plus S7 plus RAG sequences (Unmack et al. 2011) by using BEAST (ver. 1.5.3, see http://beast.community/beast). For Nannoperca, divergence dates for both pairs of sister species fall within the Pliocene, with the primary split between Lineages I and II referable to the Miocene (Unmack 2001).
Landscape patterns
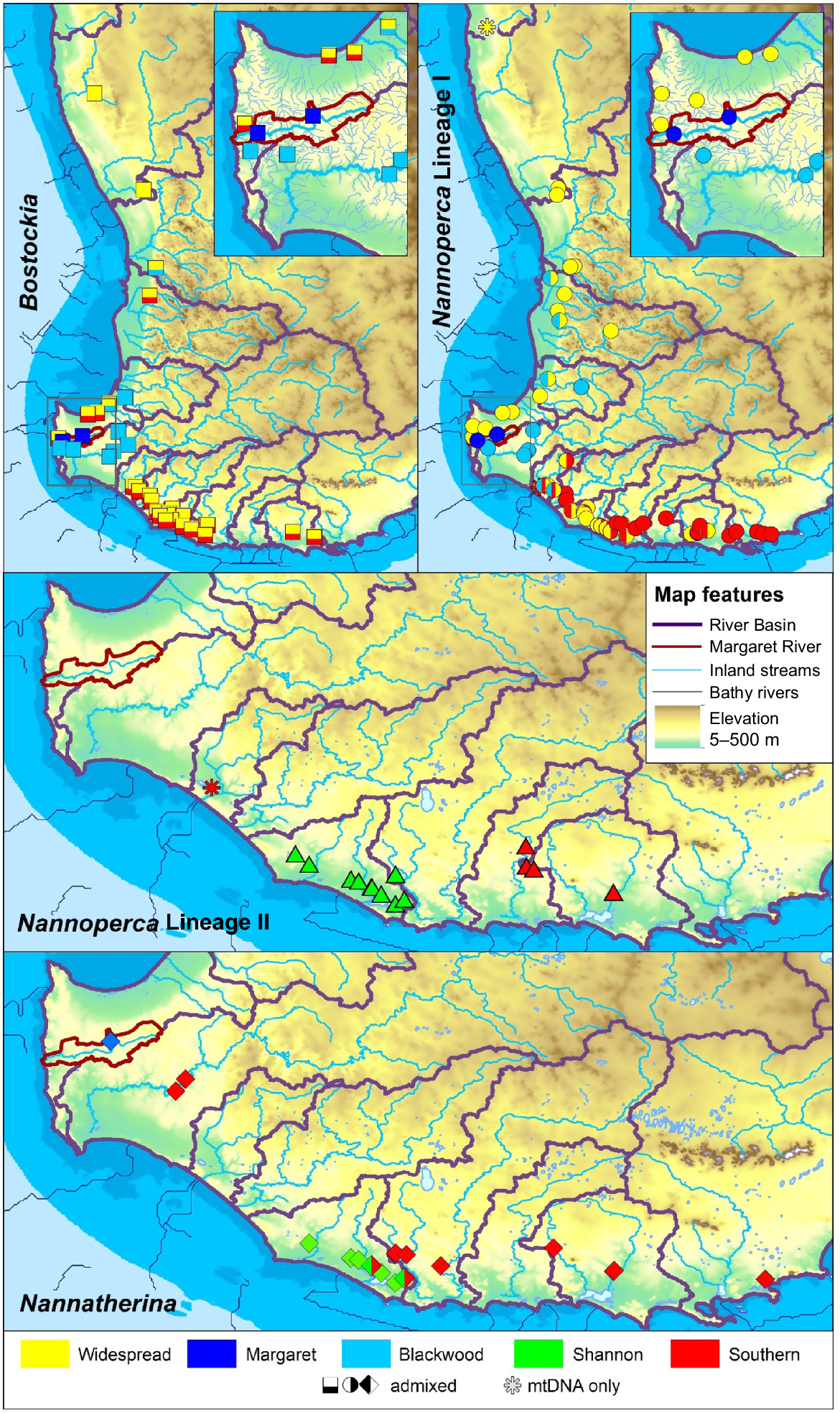
With comprehensive sampling and genetic screening across the extant distribution of the three target genera, it is possible to compare patterns observed against the broader common aquatic landscape. Table 2 summarises the comparative levels of geographically based genetic subdivision across the four south-western percichthyid lineages (i.e. including the two for Nannoperca), as either unique candidate species or subpopulations. This summary is mainly based on allozyme patterns, but is also largely congruent with the mtDNA data for Nannoperca. The spatial distribution of these groups is also mapped and compared in Fig. 8 against major hydrological units, elevation, and historic drainage patterns and low-sea levels (Unmack 2001; Atkinson et al. 2008; General Bathymetric Chart of the Ocean 2024).
| Region | Nannoperca I | Nannoperca II | Nannatherina | Bostockia | |
|---|---|---|---|---|---|
| Northern | Subpopulation (VWI-N) plus admixture | Absent | Absent | Subpopulation (PNC) plus admixture | |
| Margaret | Species level (VMA) | Absent | Subpopulation (BMA) | Subpopulation (PMA) | |
| Blackwood | Subpopulation plus upper Collie (VWI-B) | Absent | Subpopulation plus Angove (BSE-B) | Subpopulation (PBL) | |
| Shannon | Subpopulation with southern (VWI-S) plus admixture | Species level (VSH) | Subpopulation (BSH) | No specific substructure (admixed) | |
| Southern | Subpopulation (VWI-S) plus admixture | Species level (N. pygmaea) | Two possible subpopulations? (BSE-eastern) | No specific substructure (admixed) |
Taxon and subpopulation nomenclature and assignments broadly match those used in Fig. 2–6.
Composite map of geographic genetic groups across the three percichthyid data sets (see Fig. 1 for river basin and site details). Note that historically Nannatherina was more widespread occurring into the region of the Moore–Hill Rivers catchment, but assessment of the genetic affinity of this region was not possible because of extirpation.
Discussion
High endemism is often manifested through unique form and ecology, being both charismatic and irreplaceable (Allan and Flecker 1993; Ebner et al. 2016). The partitioned nature of freshwater environments can promote local isolation and adaptation as a forerunner to endemism, especially in dispersal-limited obligate freshwater fishes (Dudgeon et al. 2006; Reyjol et al. 2007). The south-western corner of Australia is a global biodiversity hotspot defined primarily for its terrestrial flora and fauna, but extending to a freshwater fauna that displays a very high proportion of endemic species (Morgan et al. 2011; Davies and Stewart 2013). The region is geographically isolated from other freshwater bioregions and features a heavily partitioned hydrological framework through a series of isolated river basins and subcatchments. Here, we explored the molecular systematics of temperate perches (family Percichthyidae) across this habitat templet, represented by the genera Bostockia, Nannoperca and Nannatherina, and discovered unique and contrasting patterns at and below the species level. These data have strong implications for management and conservation, thus providing a foundation for future taxonomic and biological studies.
Comparative ecology
Landscape-related genetic structure in freshwater fishes can be influenced by varying combinations of biological traits, such as dispersal ability, behaviour and habitat specialisation, all of which can interact with the physical aquatic environment to affect gene flow (Tibbets and Dowling 1996; Hughes et al. 2009; Hammer et al. 2019a). However, previous ecological assessments of south-western Australian percichthyids that were based on existing field data have been confounded by unrealised problems in their taxonomic foundation, i.e. the presence of cryptic species. The detection of a narrow-range candidate species nested within an otherwise widespread Nannoperca lineage (i.e. VMA + VWI), and a second narrow-range candidate species (VSH) forming a second lineage with N. pygmaea, implies that three different sets of biological attributes are likely at play within the current biological data available for N. vittata sensu lato.
Nevertheless, on the basis of a considered review of biological characteristics, we would expect that B. porosa, as a larger growing (i.e. potential better swimming ability) and more generalist species in terms of habitat occupancy (Pen and Potter 1990), might be less substructured. This was supported by mostly shallow substructure across the region, albeit with a strong hint of refuge contraction in dry evolutionary periods followed by subsequent wide dispersal (i.e. distinct subgroups, some of which showed signals of admixture). Clearly, a full elucidation of this complex biogeographic pattern is beyond the resolving power of allozymes and thus would require a comprehensive genomic dataset. Likewise, the widespread Nannoperca taxon VWI is likely to have fairly general habitat needs and relatively good dispersal ability (Pen and Potter 1991; data from a VWI population), as reflected in limited substructuring and signs of broad admixture across an expansive distribution from the Arrowsmith River through to the Angove River. By contrast, N. balstoni is a noted habitat specialist primarily in off-channel acidic pools with high structural habitat integrity (Morgan et al. 1995), and this is likely to restrict broad gene-flow as reflected by the presence of three distinct subpopulations within a relatively restricted distribution. Nannoperca pygmaea has also been shown to have highly specialised habitat requirements, including specific refuge needs, which may help explain its restricted distribution (MG Allen et al. 2020). However, our matrilineal data (mtDNA only for Smith Lake) indicated limited genetic substructure across two fragmented populations (Kent and Denmark v. Donnelly River Basins), which suggests a broader historic distribution and therefore dispersal opportunities prior to major human-mediated landscape change. Ongoing fine-scale field sampling and genomic data would help inform population genetics for this threatened species.
There was an overall pattern of strong genetic substructure within the targeted taxa, including the presence of cryptic species, a trend that is likely to apply to the remaining five endemic obligate freshwater fishes of south-western Australia, this being a general pattern for freshwater fishes in naturally occurring, heavily divided landscapes (Beheregaray and Caccone 2007; Hammer et al. 2013; Hending 2025). Comparative ecology studies in these habitats must be mindful to ensure that a sound systematic framework interrogated with modern taxonomic approaches is in place from the outset (Page et al. 2005; Feckler et al. 2014).
An additional ecological insight from this study was the detection of rare instances of hybridisation between taxa across both genera and lineages, which field ecologists should be mindful of when identifying samples. This issue is likely to become more problematic in the future, given that anthropogenic modifications of habitat often lead to an increase in hybridisation between species (Hasselman et al. 2014; McFarlane and Pemberton 2019).
Landscape patterns and conservation
The additional taxonomic complexity and spatial genetic substructure observed in south-western Australia’s percichthyids, a group with already high endemism and conservation concerns (Morgan et al. 2014a; Lintermans et al. 2020; International Union for Conservation of Nature 2024), clearly elevates the required focus on land and natural resource management in general, plus identifies several priority focal regions.
The Margaret River is of high evolutionary and conservation significance for aquatic biota (MG Allen et al. 2017). A major level of genetic distinctness was shown for each obligate freshwater fish examined from the system, ranging between a likely species-level split in N. vittata and distinctive subpopulations in Nannatherina and Bostockia. This distinctiveness implies that long-term refugial habitat has existed in this catchment, presumably reflecting longer-term biogeographic isolation from adjacent catchments (e.g. narrow continental shelf width limiting coastal connections at lower sea levels with distinctive eastern flow direction; also potentially aided by the presence of small waterfalls). The conservation implications are to protect the genetic integrity and population health of the Margaret River fish community, which has already been challenged by the introduction of eastern gambusia, Gambusia holbrooki, to key permanent water refuges in the upper catchment (a specific threatening process for the N. balstoni BMA subpopulation), and faces broad land management issues (MG Allen et al. 2017).
The significance and extinction risk of the Margaret River aquatic fauna is re-enforced by the example of the decapod crustacean group marron (genus Cherax). Genetic data, followed by combined morphological and ecological characterisation, documented two species in the lineage, the hairy marron (Cherax tenuimanus) endemic to the Margaret River, and a widespread species occurring across the south-west, the smooth marron, Cherax cainii (Austin and Ryan 2002). However, the narrow-range endemic declined rapidly through aggressive competition, predation and introgressive hybridisation following human-mediated dispersal of the widespread sister species into the Margaret River (Duffy et al. 2014; Guildea et al. 2015). In pygmy perches, the observed signal of potential introgression between the narrow-range Margaret endemic candidate species and the more widespread sister species in Nannoperca (VMA v. VWI) warrants urgent investigation and careful pro-active management to limit any human-mediated dispersal into and within the system. This should include both direct fish movement plus other indirect means such as transfer of aquatic vegetation, habitat modifications altering natural barriers, and even restoring passage to artificial barriers (i.e. consideration of altered ecological condition in goal setting). Additional unique aquatic species of conservation concern in the Margaret River include the narrow-range endemic Margaret River burrowing crayfish, Engaewa pseudoreducta (Morgan et al. 2011), and a potentially species-level lineage of the freshwater mussel genus, Westralunio (Klunzinger et al. 2021).
The Shannon River Basin was also noted as an important region of diversification in the south-western percichthyid fauna. An endemic candidate species of Nannoperca (VSH), sister to N. pygmaea, was entirely restricted to the Shannon River Basin. Likewise for a distinct subpopulation of Nannatherina, which maintains parapatry with a separate subpopulation in surrounding areas (BSH v. BSE), where a low level of gene flow appears to be potentially balanced by some form of strong selection (and therefore worthy of further spatial and temporal genomic investigation). The Shannon River Basin itself (Shannon and Gardner Rivers) has low drainage divides with adjoining systems along the coast and would have converged with other systems at low sea levels. Hence, given the patterns of gene flow observed, a high level of habitat specificity is likely to be involved, possibly associated with a distinctive, low-gradient habitat of coastal swamp or similar. Other southern coast river basins are also clearly important areas for percichthyids, such as the Donnelly, Kent and Denmark River Basins being habitat of the Endangered N. pygmaea and of general refuge value for a diverse fauna (Morgan and Beatty 2008; MG Allen et al. 2020).
It is important to note that it was not possible to fully assess the genetic affinity of fish from some regions, owing to large declines from the historic distribution. The most notable example was northern populations of Nannatherina that ranged northward to at least the Swan Coastal Plain, a region now heavily affected by agricultural development, urbanisation and introduced fishes (WA Museum record P.27025, Gingin Brook, 1981: Morgan et al. 1998, 2014a; Hourston et al. 2014). Other extirpated populations of Nannoperca and Bostockia well inland would also be of interest for assessing genetic patterns, such as from the now heavily salinised upper Blackwood River (e.g. WAM P.3109, 3111 and 3112, Dumbleyung Lake area, 1947: Morgan et al. 2003). These and other now extirpated populations with voucher material remain a potential target for historic DNA techniques focusing on museum specimens (Appleyard et al. 2021; Tims et al. 2024).
Taxonomic considerations
The suggestion of two additional candidate species within the genus Nannoperca necessitates an urgent taxonomic assessment by using combined lines of evidence for species description and morphological diagnosis (i.e. undertaking a full complementary data assessment of morphological traits across different taxa and populations) (Morgan et al. 2013; Hammer et al. 2019b). Thereafter, a chart of action to address differing taxonomic issues needs to start by matching available names (i.e. old-type material) to morphological traits or geographic boundaries of candidate species. Both aspects can be challenging owing to the generally poor physical condition and limited information accompanying early museum vouchers, and given that historic sampling data have shown that distributions were broader in the 1800s, perhaps reflecting other patterns unrealised through lack of data.
The available names for species in the Nannoperca complex are N. vittata, with a type locality listed simply as ‘fresh waters of the interior of Western Australia by Rev. Bostock’ (a large type specimen in reasonable condition which should facilitate comparison; Bostock was based in Perth and Fremantle, so collection from this region is likely) and Nannoperca viridis Castelnau, 1873, with type locality ‘interior of King George Sound by Mr Maxwell’ (two small <2 cm and fairly damaged types, which may make assignment difficult). Both were assessed in the same paper (Castelnau 1873), where N. vittata is the senior synonym following assignment in a review of the taxonomy of the pygmy perches by Kuiter and Allen (1986), should they prove to be conspecific. Kuiter and Allen (1986) conducted the first modern data assessment for N. vittata; however, these data unknowingly included both Nannoperca lineages (VWI and VSH candidate species), with their raw summary being repeated as comparative N. vittata data in the description of N. pygmaea (Morgan et al. 2013: the Lake Smith, Donnelly catchment population was not known at the time). Clearly, a complete revision will be required, including the need to re-diagnose both N. vittata and N. pygmaea.
The allozyme data are powerful in diagnosing four candidate species in Nannoperca. Nevertheless, additional site sampling and genomic data for the Margaret River would assist in the confidence of taxonomic assessment for the VMA taxon, especially considering the small geographic distances involved and the finding of a single admixed fish at one of the two sites surveyed (which could represent natural low-level interactions in parapatry or more disturbing signs of human-mediated gene flow). Preliminary genomic data (Buckley et al. 2024) comparing four populations covering the full geographic range of candidate species VWI plus one VMA site have already demonstrated a wealth of potential diagnostic genetic markers for delineating these two sister taxa (871–1223 SNPs; Fig. S2), plus shown great potential for exploring genetic substructure in VWI (over 300 SNPs differ between VWI-N and VWI-S subpopulations; Fig. S2).
Conclusions
The south-western corner of Australia is a unique region of high significance for biodiversity that faces considerable future challenges, not least of which is advancing climate change that will worsen key aquatic habitat availability and quality issues. The current study represents the first spatially comprehensive genetic assessment of a key group of aquatic fauna for the region, the obligate freshwater percichthyids. Although the data have been a while in preparation, their availability is very timely to tie into identification of natural assets and threat assessment within land and resource management, identifying urgent issues around narrow-range endemic candidate species and subpopulations in two key areas (Margaret and Shannon). Moreover, recent advances in genomic data now offer a means whereby many of the interesting questions unable to be fully addressed by allozyme datasets can be readily investigated and expanded using the broad biogeographic patterns presented here.
Data availability
The raw allozyme and aligned cytb data are available on Zenodo (Adams and Hammer 2024). All novel cytb sequences have also been deposited in GenBank (Accession numbers PQ472737–PQ472909).
Conflicts of interest
Peter Unmack is an Associate Editor of Marine and Freshwater Research. Despite this relationship, Peter took no part in the review and acceptance of this manuscript, in line with the publishing policy. The authors declare that they have no further conflicts of interest.
Declaration of funding
The South Australian Museum, Murdoch University and University of Canberra provided funds to support the project.
Acknowledgements
The authors acknowledge the Noongar people who are the Traditional Custodians of the land on which this research took place, and we recognise their ongoing connection to country, water and fishes. This work formed part of PhD research by the late Jon Murphy, and we pay tribute to his contributions to aquatic biology. Various people provided field or other assistance including Chris Burridge, David Galeotti and Alan Lymbery. Roger Swainston (see animafish.com) kindy provided permission to reproduce his percichthyid illustrations. We also thank Glenn Moore (Western Australian Museum) and Ralph Foster (South Australian Museum) for general support with museum data or specimen lodgement.
References
Adams M, Hammer M (2024) Species delineation, phylogeography, and conservation of WA perches. Zenodo 2024, ver. 1 [Dataset, published 24 October 2024].
| Crossref | Google Scholar |
Adams M, Raadik TA, Burridge CP, Georges A (2014) Global biodiversity assessment and hyper-cryptic species complexes: more than one species of elephant in the room? Systematic Biology 63(4), 518-533.
| Crossref | Google Scholar | PubMed |
Adams M, Hammer MP, Unmack PJ, Raadik TA, Jense C, Burridge CP (2023) Multi-gene insights into the taxonomy and conservation of Tasmania’s galaxiid fishes. Marine and Freshwater Research 74(13), 1113-1128.
| Crossref | Google Scholar |
Allan JD, Flecker AS (1993) Biodiversity conservation in running waters: identifying the major factors that threaten destruction of riverine species and ecosystems. BioScience 43(1), 32-43.
| Crossref | Google Scholar |
Allen MG, Beatty SJ, Morgan DL (2017) Aquatic fauna refuges in Margaret River and Cape to Cape region of Australia’s Mediterranean-climatic Southwestern Province. FiSHMED Fishes in Mediterranean Environments 2, 1-27.
| Google Scholar |
Allen MG, Morgan DL, Close PG, Beatty SJ (2020) Too little but not too late? Biology of a recently discovered and imperilled freshwater fish in a drying temperate region and comparison with sympatric fishes. Aquatic Conservation: Marine and Freshwater Ecosystems 30(7), 1412-1423.
| Crossref | Google Scholar |
Appleyard SA, Maher S, Pogonoski JJ, Bent SJ, Chua X-Y, McGrath A (2021) Assessing DNA for fish identifications from reference collections: the good, bad and ugly shed light on formalin fixation and sequencing approaches. Journal of Fish Biology 98(5), 1421-1432.
| Crossref | Google Scholar | PubMed |
Arratia G, Quezada-Romegialli C (2019) The South American and Australian percichthyids and perciliids. What is new about them? Neotropical Ichthyology 17(1), e180102.
| Crossref | Google Scholar |
Arthington AH, Dulvy NK, Gladstone W, Winfield IJ (2016) Fish conservation in freshwater and marine realms: status, threats and management. Aquatic Conservation: Marine and Freshwater Ecosystems 26(5), 838-857.
| Crossref | Google Scholar |
Austin CM, Ryan SG (2002) Allozyme evidence for a new species of freshwater crayfish of the genus Cherax Erichson (Decapoda: Parastacidae) from the south-west of Western Australia. Invertebrate Systematics 16(3), 357-367.
| Crossref | Google Scholar |
Beatty S, Morgan DL (2019) Little pygmy perch Nannoperca pygmaea. In ‘The IUCN Red List of Threatened Species 2019’. e.T122906197A123382291. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/122906197/123382291
Beatty SJ, Morgan DL, Rashnavadi M, Lymbery AJ (2011) Salinity tolerances of endemic freshwater fishes of south-western Australia: implications for conservation in a biodiversity hotspot. Marine and Freshwater Research 62(1), 91-100.
| Crossref | Google Scholar |
Beatty SJ, Morgan DL, Lymbery AJ (2014) Implications of climate change for potamodromous fishes. Global Change Biology 20(6), 1794-1807.
| Crossref | Google Scholar | PubMed |
Beheregaray LB, Caccone A (2007) Cryptic biodiversity in a changing world. Journal of Biology 6, 9.
| Crossref | Google Scholar | PubMed |
Beheregaray LB, Pfeiffer LV, Attard CRM, Sandoval-Castillo J, Domingos FMCB, Faulks LK, Gilligan DM, Unmack PJ (2017) Genome-wide data delimits multiple climate-determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua). Molecular Phylogenetics and Evolution 111, 65-75.
| Crossref | Google Scholar | PubMed |
Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Orti G (2017) Phylogenetic classification of bony fishes. BMC Evolutionary Biology 17, 162.
| Crossref | Google Scholar |
Blom MPK, Horner P, Moritz C (2016) Convergence across a continent: adaptive diversification in a recent radiation of Australian lizards. Proceedings of the Royal Society of London – B. Biological Sciences 283(1832), 20160181.
| Crossref | Google Scholar |
Brooks E, Slender AL, Cu S, Breed MF, Stangoulis JCR (2022) A range-wide analysis of population structure and genomic variation within the critically endangered spiny daisy (Acanthocladium dockeri). Conservation Genetics 23(6), 1027-1037.
| Crossref | Google Scholar |
Buckley SJ, Domingos FMCB, Attard CRM, Brauer CJ, Sandoval-Castillo J, Lodge R, Unmack PJ, Beheregaray LB (2018) Phylogenomic history of enigmatic pygmy perches: implications for biogeography, taxonomy and conservation. Royal Society Open Science 5(6), 172125.
| Crossref | Google Scholar | PubMed |
Buckley SJ, Brauer CJ, Unmack PJ, Hammer MP, Adams M, Beatty SJ, Morgan DL, Beheregaray LB (2024) Long-term climatic stability drives accumulation and maintenance of divergent freshwater fish lineages in a temperate biodiversity hotspot. Heredity 133, 149-159.
| Crossref | Google Scholar |
Castelnau FL (1873) Contribution to the ichthyology of Australia. Number III. Supplement to the fishes of Victoria. Proceedings of the Zoological and Acclimatisation Society of Victoria 2, 37-58.
| Google Scholar |
Davies PM, Stewart BA (2013) Aquatic biodiversity in the Mediterranean climate rivers of southwestern Australia. Hydrobiologia 719, 215-235.
| Crossref | Google Scholar |
Dudgeon D, Arthington AH, Gessner MO, Kawabata Z-I, Knowler DJ, Lévêque C, Naiman RJ, Prieur-Richard A-H, Soto D, Stiassny MLJ, Sullivan CA (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biological Reviews 81(2), 163-182.
| Crossref | Google Scholar | PubMed |
Duffy R, Ledger J, Dias J, Snow M (2014) The critically endangered hairy marron, Cherax tenuimanus Smith, 1912: a review of current knowledge and actions required to prevent extinction of a species. Journal of the Royal Society of Western Australia 97, 297-306.
| Google Scholar |
Ebner BC, Morgan DL, Kerezsy A, Hardie S, Beatty SJ, Seymour JE, Donaldson JA, Linke S, Peverell S, Roberts D, Espinoza T, Marshall N, Kroon FJ, Burrows DW, McAllister RRJ (2016) Enhancing conservation of Australian freshwater ecosystems: identification of freshwater flagship fishes and relevant target audiences. Fish and Fisheries 17(4), 1134-1151.
| Crossref | Google Scholar |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14(8), 2611-2620.
| Crossref | Google Scholar | PubMed |
Feckler A, Zubrod JP, Thielsch A, Schwenk K, Schulz R, Bundschuh M (2014) Cryptic species diversity: an overlooked factor in environmental management? Journal of Applied Ecology 51(4), 958-967.
| Crossref | Google Scholar |
Fricke R, Eschmeyer WN, Van der Laan R (2024) Escmeyer’s catalog of fishes. Electronic version. (California Academy of Sciences) Available at http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp [Verified 12 October 2024]
General Bathymetric Chart of the Ocean (2024) Bathymetric data services. (GBCO) Available at https://download.gebco.net/ [Verified 12 October 2024]
Guildea C, Hitchen Y, Duffy R, Dias PJ, Ledger JM, Snow M, Kennington WJ (2015) Introgression threatens the survival of the critically endangered freshwater crayfish Cherax tenuimanus (Decapoda: Parastacidae) in the wild. PLoS ONE 10(3), e0121075.
| Crossref | Google Scholar | PubMed |
Hammer MP, Adams M, Unmack PJ, Walker KF (2007) A rethink on Retropinna: conservation implications of new taxa and significant genetic sub-structure in Australian smelts (Pisces: Retropinnidae). Marine and Freshwater Research 58(4), 327-341.
| Crossref | Google Scholar |
Hammer MP, Unmack PJ, Adams M, Johnson JB, Walker KF (2010) Phylogeographic structure in the threatened Yarra pygmy perch Nannoperca obscura (Teleostei: Percichthyidae) has major implications for declining populations. Conservation Genetics 11, 213-223.
| Crossref | Google Scholar |
Hammer MP, Unmack PJ, Adams M, Raadik TA, Johnson JB (2014) A multigene molecular assessment of cryptic biodiversity in the iconic freshwater blackfishes (Teleostei: Percichthyidae: Gadopsis) of south-eastern Australia. Biological Journal of the Linnean Society 111(3), 521-540.
| Crossref | Google Scholar |
Hammer MP, Allen GR, Martin KC, Adams M, Ebner BC, Raadik TA, Unmack PJ (2018) Revision of the Australian Wet Tropics endemic rainbowfish genus Cairnsichthys (Atheriniformes: Melanotaeniidae), with description of a new species. Zootaxa 4413(2), 271-294.
| Crossref | Google Scholar | PubMed |
Hammer MP, Adams M, Thacker CE, Johnson JB, Unmack PJ (2019a) Comparison of genetic structure in co-occurring freshwater eleotrids (Actinopterygii: Philypnodon) reveals cryptic species, likely translocation and regional conservation hotspots. Molecular Phylogenetics and Evolution 139, 106556.
| Crossref | Google Scholar |
Hammer MP, Allen GR, Martin KC, Adams M, Unmack PJ (2019b) Two new species of dwarf rainbowfishes (Atheriniformes: Melanotaeniidae) from northern Australia and southern New Guinea. Zootaxa 4701(3), 201-234.
| Crossref | Google Scholar |
Hasselman DJ, Argo EE, McBride MC, Bentzen P, Schultz TF, Perez-Umphrey AA, Palkovacs EP (2014) Human disturbance causes the formation of a hybrid swarm between two naturally sympatric fish species. Molecular Ecology Resources 23(5), 1137-1152.
| Crossref | Google Scholar |
Hending D (2025) Cryptic species conservation: a review. Biological Reviews 100(1), 258-274.
| Crossref | Google Scholar | PubMed |
Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2), 518-522.
| Crossref | Google Scholar | PubMed |
Horner P, Adams M (2007) ‘A molecular-systematic assessment of species boundaries in Australian Cryptoblepharus (Reptilia: Squamata: Scincidae): a case study for the combined use of allozymes and morphology to explore cryptic biodiversity. Supplement 3. The Beagle.’ (Museums and Art Galleries of the Northern Territory)
Hourston M, Ledger J, Vercoe P, Lawrence C (2014) Native and non-native fishes in wetlands of the Swan Coastal Plain, Western Australia. Journal of the Royal Society of Western Australia 97(2), 331-341.
| Google Scholar |
Hughes JM, Schmidt DJ, Finn DS (2009) Genes in streams: a molecular approach to understanding movement of freshwater fauna and their riverine habitat. BioScience 59(7), 573-583.
| Crossref | Google Scholar |
International Union for Conservation of Nature (2024) The IUCN Red List of Threatened Species. Version 2024-1. (IUCN) Available at http://www.iucnredlist.org
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14), 1801-1806.
| Crossref | Google Scholar | PubMed |
Jerry DR, Eliphinstone MS, Baverstock PR (2001) Phylogenetic relationships of Australian members of the family Percichthyidae inferred from mitochondrial 12S rRNA sequence data. Molecular Phylogenetics and Evolution 18(3), 335-347.
| Crossref | Google Scholar | PubMed |
Jusaitis M, Adams M (2005) Conservation implications of clonality and limited sexual reproduction in the endangered shrub Acanthocladium dockeri (Asteraceae). Australian Journal of Botany 53(6), 535-544.
| Crossref | Google Scholar |
Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14(6), 587-589.
| Crossref | Google Scholar | PubMed |
Klunzinger MW, Lopes-Lima M, Gomes-dos-Santos A, Froufe E, Lymbery AJ, Kirkendale L (2021) Phylogeographic study of the West Australian freshwater mussel, Westralunio carteri, uncovers evolutionarily significant units that raise new conservation concerns. Hydrobiologia 848(12), 2951-2964.
| Crossref | Google Scholar |
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15(5), 1179-1191.
| Crossref | Google Scholar | PubMed |
Kuiter RH, Allen GR (1986) A synopsis of the Australian pygmy perches (Percichthyidae) with the description of a new species. Revue Francaise Aquariologie 12(4), 109-116.
| Google Scholar |
Lintermans M, Geyle HM, Beatty S, Brown C, Ebner BC, Freeman R, Hammer MP, Humphreys WF, Kennard MJ, Kern P, Martin K, Morgan DL, Raadik TA, Unmack PJ, Wager R, Woinarski JCZ, Garnett ST (2020) Big trouble for little fish: identifying Australian freshwater fishes in imminent risk of extinction. Pacific Conservation Biology 26(4), 365-377.
| Crossref | Google Scholar |
Liu C, Comte L, Olden JD (2017) Heads you win, tails you lose: life-history traits predict invasion and extinction risk of the world’s freshwater fishes. Aquatic Conservation: Marine and Freshwater Ecosystems 27(4), 773-779.
| Crossref | Google Scholar |
McCulloch AR (1929) A check-list of the fishes recorded from Australia. Australian Museum Memoirs 5(1–2), 1-329.
| Google Scholar |
McFarlane SE, Pemberton JM (2019) Detecting the true extent of introgression during anthropogenic hybridization. Trends in Ecology & Evolution 34(4), 315-326.
| Crossref | Google Scholar |
Morgan DL (2019) Western pygmy perch Nannoperca vittata. In ‘The IUCN Red List of Threatened Species 2019’. e.T123358592A123382821. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/123358592/123382821
Morgan DL (2020) Nightfish Bostockia porosa. In ‘The IUCN Red List of Threatened Species 2020’. e.T123358443A123382766. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/123358443/123382766
Morgan DL, Beatty SJ (2008) The Donnelly River catchment: an important refuge for all of south-western Australia’s endemic freshwater fishes and the pouched lamprey (Geotria australis). Western Australian Naturalist 26(2), 112-127.
| Google Scholar |
Morgan DL, Beatty S (2019) Balston’s pygmy perch Nannatherina balstoni. In ‘The IUCN Red List of Threatened Species 2019’. e.T14320A123378416. (International Union for Conservation of Nature and Natural Resources) Available at https://www.iucnredlist.org/species/14320/123378416
Morgan DL, Gill HS (2000) Fish associations within the different inland habitats of lower south-western Australia. Records of the Western Australian Museum 20(1), 31-37.
| Google Scholar |
Morgan DL, Gill HS, Potter IC (1995) Life cycle, growth and diet of Balston’s pygmy perch in its natural habitat of acidic pools in south-western Australia. Journal of Fish Biology 47(5), 808-825.
| Crossref | Google Scholar |
Morgan DL, Gill HS, Potter IC (1998) Distribution, identification and biology of freshwater fishes in south-western Australia. Records of the Western Australian Museum 56, 1-97.
| Google Scholar |
Morgan DL, Hambleton SJ, Gill HS, Beatty SJ (2002) Distribution, biology and likely impacts of the introduced redfin perch (Perca fluviatilis) (Percidae) in Western Australia. Marine and Freshwater Research 53(8), 1211-1221.
| Crossref | Google Scholar |
Morgan DL, Thorburn DC, Gill HS (2003) Salinization of southwestern Western Australian rivers and the implications for the inland fish fauna – the Blackwood River, a case study. Pacific Conservation Biology 9(3), 161-171.
| Crossref | Google Scholar |
Morgan DL, Gill HS, Maddern MG, Beatty SJ (2004) Distribution and impacts of introduced freshwater fishes in Western Australia. New Zealand Journal of Marine and Freshwater Research 38(3), 511-523.
| Crossref | Google Scholar |
Morgan DL, Beatty SJ, Adams M (2013) Nannoperca pygmaea, a new species of pygmy perch (Teleostei: Percichthyidae) from Western Australia. Zootaxa 3637(4), 401-411.
| Crossref | Google Scholar |
Morgan DL, Beatty SJ, Allen MG, Keleher JJ, Moore GI (2014a) Long live the King River perchlet (Nannatherina balstoni). Journal of the Royal Society of Western Australia 97, 307-312.
| Google Scholar |
Morgan DL, Unmack PJ, Beatty SJ, Ebner BC, Allen MG, Keleher JJ, Donaldson JA, Murphy J (2014b) An overview of the ‘freshwater fishes’ of Western Australia. Journal of the Royal Society of Western Australia 97(2), 263-278.
| Google Scholar |
Morrongiello JR, Beatty SJ, Bennett JC, Crook DA, Ikedife DNEN, Kennard MJ, Kerezsy A, Lintermans M, McNeil DG, Pusey BJ, Rayner T (2011) Climate change and its implications for Australia’s freshwater fish. Marine and Freshwater Research 62(9), 1082-1098.
| Crossref | Google Scholar |
Mossop KD, Lemmon AR, Lemmon EM, Eytan R, Adams M, Unmack PJ, Date KS, Morales HE, Hammer MP, Wong BBM, Chapple DG (2023) Phylogenomics and biogeography of arid-adapted Chlamydogobius goby fishes. Molecular Phylogenetics and Evolution 182, 107757.
| Crossref | Google Scholar | PubMed |
Musyl MK, Keenan CP (1992) Population genetics and zoogeography of Australian freshwater golden perch, Macquaria ambigua (Richardson 1845) (Teleostei: Percichthyidae), and electrophoretic identification of a new species from the Lake Eyre Basin. Australian Journal of Marine and Freshwater Research 43(6), 1585-1601.
| Crossref | Google Scholar |
Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403(6772), 853-858.
| Crossref | Google Scholar | PubMed |
Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proceedings of the National Academy of Sciences 109(34), 13698-13703.
| Crossref | Google Scholar |
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1), 268-274.
| Crossref | Google Scholar | PubMed |
Nock CJ, Elphinstone MS, Rowland SJ, Baverstock PR (2010) Phylogenetics and revised taxonomy of the Australian freshwater cod genus, Maccullochella (Percichthyidae). Marine and Freshwater Research 61(9), 980-991.
| Crossref | Google Scholar |
Ogston G, Beatty SJ, Morgan DL, Pusey BJ, Lymbery AJ (2016) Living on burrowed time: aestivating fishes in south-western Australia face extinction due to climate change. Biological Conservation 195, 235-244.
| Crossref | Google Scholar |
Page TJ, Choy SC, Hughes JM (2005) The taxonomic feedback loop: symbiosis of morphology and molecules. Biology Letters 1(2), 139-142.
| Crossref | Google Scholar | PubMed |
Pen LJ, Potter IC (1990) Biology of nightfish, Bostockia porosa Castelnau, in a south-western Australian River. Australian Journal of Marine and Freshwater Research 41(5), 627-645.
| Crossref | Google Scholar |
Pen LJ, Potter IC (1991) Biology of the western pygmy perch, Edelia vittata, and comparisons with two other teleost species endemic to south-western Australia. Environmental Biology of Fishes 31, 365-380.
| Crossref | Google Scholar |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2), 945-959.
| Crossref | Google Scholar | PubMed |
Pusey BJ, Kennard MJ (2001) Guyu wujalwujalensis, a new genus and species (Pisces: Percichthyidae) from north-eastern Queensland. Ichthyological Exploration of Freshwaters 12(1), 17-28.
| Google Scholar |
Reid AJ, Carlson AK, Creed IF, Eliason EJ, Gell PA, Johnson PTJ, Kidd KA, MacCormack TJ, Olden JD, Ormerod SJ, Smol JP, Taylor WW, Tockner K, Vermaire JC, Dudgeon D, Cooke SJ (2019) Emerging threats and persistent conservation challenges for freshwater biodiversity. Biological Reviews 94(3), 849-873.
| Crossref | Google Scholar | PubMed |
Reyjol Y, Hugueny B, Pont D, Bianco PG, Beier U, Caiola N, Casals F, Cowx I, Economou A, Ferreira T, Haidvogl G, Noble R, De Sostoa A, Vigneron T, Virbickas T (2007) Patterns in species richness and endemism of European freshwater fish. Global Ecology and Biogeography 16(1), 65-75.
| Crossref | Google Scholar |
Ruzzante DE, Walde SJ, Cussac VE, Dalebout ML, Seibert J, Ortubay S, Habit E (2006) Phylogeography of the Percichthyidae (Pisces) in Patagonia: roles of orogeny, glaciation, and volcanism. Molecular Ecology 15(10), 2949-2968.
| Crossref | Google Scholar | PubMed |
Saddlier S, Koehn JD, Hammer MP (2013) Let’s not forget the small fishes – conservation of two threatened species of pygmy perch in south-eastern Australia. Marine and Freshwater Research 64(9), 874-886.
| Crossref | Google Scholar |
Sayer CA, Fernando E, Jimenez RR, Macfarlane NBW, Rapacciuolo G, Böhm M, Brooks TM, Contreras-MacBeath T, Cox NA, Harrison I, Hoffmann M, Jenkins R, Smith KG, Vié J-C, Abbott JC, Allen DJ, Allen GR, Barrios V, Boudot J-P, Carrizo SF, Charvet P, Clausnitzer V, Congiu L, Crandall KA, Cumberlidge N, Cuttelod A, Dalton J, Daniels AG, De Grave S, De Knijf G, Dijkstra K-DB, Dow RA, Freyhof J, García N, Gessner J, Getahun A, Gibson C, Gollock MJ, Grant MI, Groom AER, Hammer MP, Hammerson GA, Hilton-Taylor C, Hodgkinson L, Holland RA, Jabado RW, Juffe Bignoli D, Kalkman VJ, Karimov BK, Kipping J, Kottelat M, Lalèyè PA, Larson HK, Lintermans M, Lozano F, Ludwig A, Lyons TJ, Máiz-Tomé L, Molur S, Ng HH, Numa C, Palmer-Newton AF, Pike C, Pippard HE, Polaz CNM, Pollock CM, Raghavan R, Rand PS, Ravelomanana T, Reis RE, Rigby CL, Scott JA, Skelton PH, Sloat MR, Snoeks J, Stiassny MLJ, Tan HH, Taniguchi Y, Thorstad EB, Tognelli MF, Torres AG, Torres Y, Tweddle D, Watanabe K, Westrip JRS, Wright EGE, Zhang E, Darwall WRT (2025) One-quarter of freshwater fauna threatened with extinction. Nature 638(8049), 138-145.
| Crossref | Google Scholar | PubMed |
Thacker CE, Geiger DL, Unmack PJ (2022) Species delineation and systematics of a hemiclonal hybrid complex in Australian freshwaters (Gobiiformes: Gobioidei: Eleotridae: Hypseleotris). Royal Society Open Science 9, e220201.
| Crossref | Google Scholar |
Tibbets CA, Dowling TE (1996) Effects of intrinsic and extrinsic factors on population fragmentation in three species of North American minnows (Teleostei: Cyprinidae). Evolution 50(3), 1280-1292.
| Crossref | Google Scholar | PubMed |
Tims AR, Saupe EE (2023) Forecasting climate-driven habitat changes for Australian freshwater fishes. Diversity and Distributions 29(5), 641-653.
| Crossref | Google Scholar |
Tims AR, Unmack PJ, Hammer MP, Brown C, Adams M, McGee MD (2024) Museum genomics reveals the hybrid origin of an extinct crater lake endemic. Systematic Biology 73(3), 506-520.
| Crossref | Google Scholar |
Trifinopoulos J, Nguyen L-T, Von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1), W232-W235.
| Crossref | Google Scholar | PubMed |
Unmack PJ (2001) Biogeography of Australian freshwater fishes. Journal of Biogeography 28(9), 1053-1089.
| Crossref | Google Scholar |
Unmack PJ, Hammer MP, Adams M, Dowling TE (2011) A phylogenetic analysis of pygmy perches (Teleostei: Percichthyidae) with an assessment of the major historical influences on aquatic biogeography in southern Australia. Systematic Biology 60(6), 797-812.
| Crossref | Google Scholar | PubMed |
Unmack PJ, Hammer MP, Adams M, Dowling TE, Johnson JB (2013) The role of continental shelf width in determining freshwater phylogeographic patterns in south-eastern Australian pygmy perches (Teleostei: Percichthyidae). Molecular Ecology 22(6), 1683-1699.
| Crossref | Google Scholar | PubMed |
Unmack PJ, Sandoval-Castillo J, Hammer MP, Adams M, Raadik TA, Beheregaray LB (2017) Genome-wide SNPs resolve a key conflict between sequence and allozyme data to confirm another threatened candidate species of river blackfishes (Teleostei: Percichthyidae: Gadopsis). Molecular Phylogenetics and Evolution 109, 415-420.
| Crossref | Google Scholar | PubMed |
Unmack PJ, Adams M, Bylemans J, Hardy CM, Hammer MP, Georges A (2019) Perspectives on the clonal persistence of presumed ‘ghost’ genomes in unisexual or allopolyploid taxa arising via hybridization. Scientific Reports 9(1), 4730.
| Crossref | Google Scholar | PubMed |
Unmack PJ, Adams M, Hammer MP, Johnson JB, Gruber B, Gilles A, Young M, Georges A (2022) Plotting for change: an analytical framework to aid decisions on which lineages are candidate species in phylogenomic species discovery. Biological Journal of the Linnean Society 135(1), 117-137.
| Crossref | Google Scholar |
Unmack PJ, Cook BD, Johnson JB, Hammer MP, Adams M (2023) Phylogeography of a widespread Australian freshwater fish, western carp gudgeon (Eleotridae: Hypseleotris klunzingeri): cryptic species, hybrid zones, and strong intra-specific divergences. Ecology and Evolution 13(11), e10682.
| Crossref | Google Scholar | PubMed |
Valenzuela-Aguayo F, McCracken GR, Diaz G, Manosalva A, Habit E, Ruzzante DE (2022) Connectivity, diversity, and hybridization between two endemic fish species (Percilia spp.) in a complex temperate landscape. Conservation Genetics 23(1), 23-33.
| Crossref | Google Scholar |


