Beef cattle breeding in Australia with genomics: opportunities and needs
D. J. Johnston A B , B. Tier A and H.-U. Graser AA Animal Genetics and Breeding Unit1, University of New England, Armidale, NSW 2351, Australia.
B Corresponding author. Email: djohnsto@une.edu.au
Animal Production Science 52(3) 100-106 https://doi.org/10.1071/AN11116
Submitted: 20 June 2011 Accepted: 9 December 2011 Published: 6 March 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
Opportunities exist in beef cattle breeding to significantly increase the rates of genetic gain by increasing the accuracy of selection at earlier ages. Currently, selection of young beef bulls incorporates several economically important traits but estimated breeding values for these traits have a large range in accuracies. While there is potential to increase accuracy through increased levels of performance recording, several traits cannot be recorded on the young bull. Increasing the accuracy of these traits is where genomic selection can offer substantial improvements in current rates of genetic gain for beef. The immediate challenge for beef is to increase the genetic variation explained by the genomic predictions for those traits of high economic value that have low accuracies at the time of selection. Currently, the accuracies of genomic predictions are low in beef, compared with those in dairy cattle. This is likely to be due to the relatively low number of animals with genotypes and phenotypes that have been used in developing genomic prediction equations. Improving the accuracy of genomic predictions will require the collection of genotypes and phenotypes on many more animals, with even greater numbers needed for lowly heritable traits, such as female reproduction and other fitness traits. Further challenges exist in beef to have genomic predictions for the large number of important breeds and also for multi-breed populations. Results suggest that single-nucleotide polymorphism (SNP) chips that are denser than 50 000 SNPs in the current use will be required to achieve this goal. For genomic selection to contribute to genetic progress, the information needs to be correctly combined with traditional pedigree and performance data. Several methods have emerged for combining the two sources of data into current genetic evaluation systems; however, challenges exist for the beef industry to implement these effectively. Changes will also be needed to the structure of the breeding sector to allow optimal use of genomic information for the benefit of the industry. Genomic information will need to be cost effective and a major driver of this will be increasing the accuracy of the predictions, which requires the collection of much more phenotypic data than are currently available.
Introduction
The revolution in genotyping provided by high-density SNP chips and the associated reduction in cost has resulted in large numbers of individuals with genome-wide genotypic data. This supports the development of genomic selection as outlined by Meuwissen et al. (2001) through the use of genomic predictions or genomic estimated breeding values (GEBVs) developed using results from genome-wide association studies (GWAS). The availability of such genomic information on large numbers of individuals is radically changing dairy cattle breeding in many countries (Hayes et al. 2009a) and also has the potential to change the way beef cattle are recorded and selected in Australia. GEBVs will increase the accuracy of EBVs for traits in beef cattle that currently have little recorded information, thus enabling significantly increased rates of genetic gain (Van Eenennaam et al. 2011). However, for the beef industry to capture the benefits of genomic selection there is a need to increase the accuracy of the GEBVs beyond current levels (Johnston et al. 2010). This is particularly the case for important traits in the breeding objective that have a low accuracy at the time of selection because they cannot be recorded on young bulls. In the longer term, there might be opportunities to change the way beef cattle are selected but this is likely to require the recording of a large number of animals with both genotypes and phenotypes, particularly on difficult- or costly-to-measure traits. The present paper outlines the current and future needs and opportunities that genomics will offer the Australian beef industry.
Current opportunities and needs
Lift rates of genetic progress
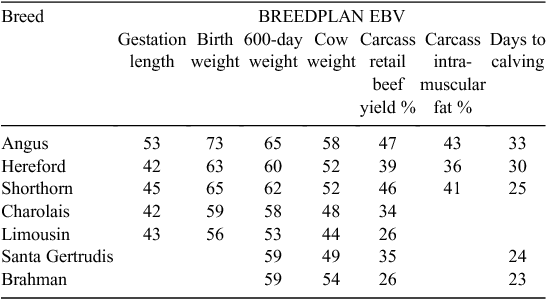
Opportunities exist to lift the rates of genetic progress in beef by using genomics, primarily through the increased accuracy of EBVs on young candidate bulls. In beef, many of the selection-criteria traits can be measured on the young by 18 months of age when selection is commonly practiced. These include calving ease, birthweight, gestation length, early weights, ultrasound carcass scans and scrotal size. However, there are some traits where only mid-parent EBVs or correlated trait information are available at the time of selection. These traits commonly have low accuracies because they require records on progeny (e.g. daughters or steers) and include abattoir carcass traits and maternal traits including maternal calving ease, maternal weaning weight, female reproduction and mature-cow size. There are additional traits in some beef-breeding objectives that have very low accuracies because no direct selection criteria exist. These include traits that are expensive to record such as feed efficiency or where no recording is practiced (e.g. cow survival). Table 1 lists the average BREEDPLAN EBV accuracy for the latest crop of young bulls (i.e. the current candidates for selection born in 2008 or 2009) from a range of breeds for selection criteria correlated with key breeding-objective traits in the main Australian beef breeds. Average accuracy reflects levels of recording, pedigree contributions and breed-specific variance components. For growth traits, the accuracies on these young bulls are in the 52–73% range; however, for abattoir carcass traits, the accuracies are all lower than 48%. For the female reproduction trait, days to calving, the accuracies are even lower, especially in the two tropically adapted breeds.
|
Current GEBV accuracies in beef
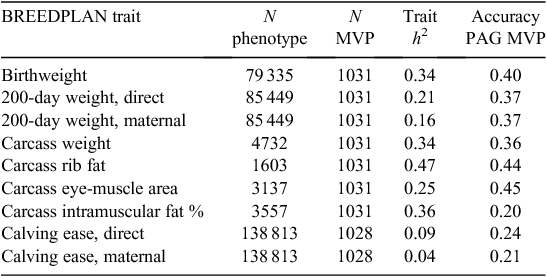
Increasing the accuracy of EBVs using genomic information requires genomic predictions that explain a significant proportion of the additive genetic variance of a trait. In the Australian beef industry, there has been a progression of commercial gene-marker products available over the past decade. The early GeneSTAR (Catapult Genetics Australia, Albion, Qld, Australia) gene markers were shown to generally have low accuracies to predict their target traits, with the exception of three tenderness markers (Johnston and Graser 2010). Progression to genomic predictions with 56 SNPs added relatively little to accuracy for those traits (Beef CRC 2009). Recently, predictions by Pfizer Animal Genetics (Albion, Qld, Australia) using the Illumina Bovine SNP50 BeadChip (50K) array (Illumina Inc., Hayward, CA, USA) have become available for Angus only. Australian results (Johnston et al. 2010) for traits included in BREEDPLAN showed accuracies of 0.20–0.45 (Table 2), while a study in US Angus (MacNeil et al. 2010) reported accuracies of 0.50–0.65 for a range of carcass traits from subsets of SNPs from the 50K panel. Recently, the American Angus Association (Northcutt 2011) reported accuracies for genomic predictions from Pfizer and Igenity (Merial, Duluth, GA, USA) ranging from 0.24 to 0.65 for early growth and carcass traits.
|
In dairy cattle, accuracies of GEBV of 0.7 averaged across 27 traits have been reported, compared with mid-parent accuracy of 0.5 (VanRaden et al. 2009). Hayes et al. (2009a) also reported significant improvements in accuracies from Australian dairy studies and predicted that the impact of genomic selection in the dairy industry will be a doubling of the rate of genetic gain. It is not clear whether there are similar opportunities in beef cattle to increase the accuracies of 50K predictions. Goddard (2009) and Goddard et al. (2010a) proposed that the theoretical accuracy of GEBVs was dependent on two main parameters, namely the proportion of genetic variation explained by the SNPs and the accuracy of estimating the SNP effects. The proportion of genetic variation explained by the SNPs is due to the SNPs being in linkage disequilibrium (LD) with the causal mutations and can be approximated by M/NeL, where M = density of SNP markers, L = length of the genome, and Ne = effective population size. The accuracy of estimating the SNP effects can be approximated by Th2/NeL, where T = number of animals with genotypes and phenotypes and h2 = trait heritability.
The most likely reasons for the difference in GEBV accuracies between dairy and beef cattle are differences in T, h2 and Ne between the two types of cattle. When traits of similar h2 are considered, the difference reduces to T and Ne. The beef industry can increase accuracy of GEBVs by increasing T, i.e. the number of animals in training populations. Dairy scientists typically perform genomic selection using data from thousands of highly accurate (>0.95) progeny-tested sires and are usually working with the dominant Holstein breed, which has a relatively small effective population size. In beef, far fewer sires have high-accuracy EBVs, there are many more breeds, and effective population sizes are expected to be larger for most of these breeds (The Bovine HapMap Consortium 2009). The accuracy of beef GEBVs are consequently lower. However, the accuracy can now be improved in beef by increasing M, thus enabling the detection of SNPs that are in higher LD with the causal mutations. This may allow data to be pooled across breeds, although there will be a trade-off from higher Ne.
The need for additional data
In beef cattle, there is a need to increase numbers of animals with high-density genotypes and key phenotypes. For current beef genetic evaluations, key phenotypes include traits that can be recorded only on daughters (e.g. maternal calving ease, days to calving, maternal weaning weight, and mature cow weight) or steer progeny (e.g. abattoir carcass and meat quality) or those traits costly to measure on the animal itself (e.g. feed intake). The numbers of animals in training populations will need to be greater for important traits such as female reproduction and calving ease, given their low heritabilities. The need to increase the number of animals recorded in an environment of limited testing resources presents a challenge as to which breeds should be included and which traits to measure, given the diverse range of breeds and production–marketing systems that exist in the Australian beef industry. The need for large datasets is partially being met by the Beef CRC phenotypic databases, but these include only eight breeds. Currently, this resource (N > 7000) has been genotyped with the 50K chip and the GWAS performed have focussed on female-reproduction and feed-intake traits across temperate and tropically adapted breeds. Most animals have been recorded for one or more trait complexes, including carcass and meat quality (n = 3670), feed intake and efficiency (n = 2520), female reproduction (n = 3950) and male reproduction (n = 1100). The majority of the animals also have comprehensive weight and live animal carcass ultrasound-scan records, along with a variety of other traits, including temperament.
Two further initiatives in Australia are addressing the need for animals with extensive phenotypes and DNA samples for genotyping. The first is the Beef Information Nucleus program (BIN) that has been implemented by five Australian beef breeds (Angus, Brahman, Charolais, Hereford and Limousin) through joint funding with Meat and Livestock Australia. These programs will jointly generate ~5700 progeny from 285 sires over three rounds of mating. Other breeds have also applied for funding to establish a BIN and, if implemented, these will almost double the total number of progeny generated. One key outcome is to create large amounts of phenotypic data to enable the accuracies of GEBVs to be determined from prediction equations developed by the Beef CRC or commercial genomic-research companies. These projects plan to collect the difficult-to-measure traits such as abattoir carcass traits, feed-intake, meat-quality and female-reproduction traits, depending on the breed. The BINs are based on a progeny test design and will produce ~20 progeny from high-index merit young sires, thus providing additional capacity to increase rates of genetic gain in the industry.
The second initiative to increase the number of genotyped and phenotyped animals is a project of the Beef CRC to genotype ~1450 industry sires from eight breeds with a range of BREEDPLAN trait EBVs with medium to high accuracies based on progeny data. Semen samples have been collated by the cooperating breed societies. The aim of this project is to provide a resource across the major breeds in Australia for the validation of genomic prediction equations developed by the Beef CRC for BREEDPLAN traits. The sires genotyped will represent a broad cross-section of each breed, and can be used in the future to construct genomic-relationship matrices for a one-step approach within and perhaps across breeds. All sires will be genotyped with the 50K chip, with a subset also genotyped with the Illumina BovineHD Beadchip containing more than 777 000 (800K) SNPs (Illumina Inc., Hayward, CA, USA). In the US, a similar project is underway (Garrick 2010) where a repository of DNA from more than 2000 influential sires or upcoming bulls across 16 breeds has been assembled and will be used to validate genomic prediction equations developed from their research populations. To increase the number of sires with high-density genotypes available in each country, exchange of genotypes between the Australian and USA resources is planned. In the future, these data as well as data from the BINs will be used to improve the existing prediction equations or in the construction of genomic relationship matrices (Misztal et al. 2009).
GEBVs and traditional EBVs
To make best use of genomic data, we require a commercially viable system to allow GEBVs to be combined with traditional sources of data in genetic evaluations such as BREEDPLAN (Graser et al. 2005), to generate genomically enhanced EBVs. Currently, three methods are being used or developed for incorporating genomic information into existing Australian and overseas evaluations. Swan et al. (2012) provided a more extensive review of the methods. Briefly, the first method uses the GEBVs as an additional trait in multi-trait BLUP evaluation (e.g. Kachman 2008; Johnston et al. 2009; Northcutt 2010). The second method uses a selection-index approach to combine GEBVs into dairy EBVs (Hayes et al. 2009a; VanRaden et al. 2009; Harris and Johnson 2010). This approach was recently used in beef to include nine Pfizer 50K MVPs genomic predictions into Angus BREEDPLAN EBVs (Johnston et al. 2010). The third approach is to use genomic data to build a genomic relationship matrix (GRM) to replace the existing pedigree-based relationship matrix (e.g. Hayes et al. 2009b; Legarra et al. 2009) for those animals genotyped, and augment it with the existing relationship matrix (Misztal et al. 2009). This approach has been trialled in an Australian sheep evaluation (Swan et al. 2011) and broiler chickens (Chen et al. 2011). Each of these methods are different in the type of genomic data, the need for additional information for incorporation (e.g. covariance matrices), number of processes involved (e.g. single versus multiple steps) and the computing time. Therefore, the immediate needs are to determine the most suitable method to include genomic information into beef evaluations, considering the type of genomic data available, the existing structure of the genetic evaluation and the commercial computing capacity. Computing EBVs for Australian beef cattle is conducted on a fully commercial basis, and the benefits of individual approaches have to be weighed against costs.
Future opportunities and needs
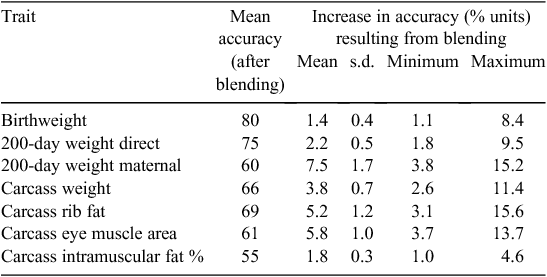
For genomic selection to have an impact in the beef industry, GEBVs that are more accurate at predicting breeding values across breeds or in multi-breed populations are required. This is critical, given the large number of key breeds in Australia that are unlikely, in the medium term, to have sufficiently large datasets for training genomic predictions. Table 3 presents the predicted increase in the accuracy of EBVs of a group of young Angus yearling bulls in a very well recorded herd if the currently available Pfizer Animal Genetic HD 50K GEBVs (their term is MVPs) had been available and blended with the existing EBVs. In this well recorded herd, the highest increase in accuracy would be for the 200-day weight maternal EBVs, with an average increase in accuracy of 7.5 percentage units. The largest increase in accuracy (15.6% units, i.e. from 0.35 to 0.51) for an individual would be for carcass rib fat.
|
GEBVs across breeds
Genomic estimated breeding values from prediction equations derived in one breed have considerably lower accuracy of prediction when applied to other breeds (de Roos et al. 2009) or in multi-breed beef populations (Weber et al. 2011). So, what is currently available for the Angus breed has little value in other Bos taurus breeds and is even less likely to be useful in Bos indicus breeds. This is well recognised by Pfizer Animal Genetics who are not using current Angus prediction equations to calculate MVPs for any other breed.
To overcome this problem, Goddard et al. (2010b) suggested an approach where breeds are pooled in training sets and animals are genotyped using higher-density SNP chips to increase the likelihood that markers have the same phase of LD between the SNPs and quantitative trait loci in divergent populations (de Roos et al. 2009). Studies have shown that 50K SNPs are not dense enough for divergent breeds such as Holstein, Jersey and Angus, and that accommodating prediction across such breeds will require greater than 300K SNP coverage (de Roos et al. 2008). This was also the conclusion of Kizilkaya et al. (2010) using simulation and of Weber et al. (2011). Therefore, research is now concentrating on increasing the levels of accuracy that GEBVs can achieve across breeds and crosses, using the highest-density chips available. In Australia, genotyping is underway by the Beef CRC by using the high-density 800K chip on a subset of animals (n = 1720) that have also been genotyped with the 50K chip. Given the cost of genotyping with the high-density chip, imputation will be used to increase the number of animals with high-density genotypes. In dairy, imputation of unobserved SNP genotypes has been shown to be very accurate from a smaller chip up to 50K (e.g. Weigel et al. 2010) and it is expected that 800K genotypes can be successfully imputed from all animals with 50K genotypes. This creates opportunities in Australian beef cattle to do GWAS with more than 7000 animals with imputed 800K genotypes. The expectation is that this will greatly increase the accuracy of the GEBVs, and research is underway to determine how successful this approach is to generate accurate GEBVs by pooling data from Bos taurus and Bos indicus breeds. However, according to estimates of Goddard and Hayes (2009), several million SNPs may be required to achieve the same LD phase across these subspecies of cattle.
New traits
There are opportunities in the future to have GEBVs for traits that are presently deemed too difficult or costly to measure in routine commercial operations. It may be possible to have genomic predictions for traits such as methane emissions, chemical attributes of meat, animal health, welfare and fitness. However, the development of predictions for these kinds of traits will require the collection of suitable phenotypes and will not come from the traditional seedstock recording sector. Therefore, considerable funding and cooperation will be required to collect these phenotypes. In Australia, it may be possible to use future BINs to fill this role.
Other applications of genomic information
There are also possibilities to use genomics beyond prediction of GEBVs. High-density chips are being used to determine the breed composition of individuals (Lewis et al. 2011) and this may have commercial application for sorting animals and may lead to methods for predicting expected levels of heterosis among individuals. It may also be possible to use SNP genotypes to manage inbreeding in breeding programs or to provide a method of predicting future recessive disorders (Goddard 2012). Whole genome sequences are becoming more readily available and less expensive as a result of recent developments in next-generation sequencing (Pérez-Enciso and Ferretti 2010). Not only will this allow genome sequence association studies, but the data can provide new information on copy number variants and RNA sequences. Gene expression arrays have been available to beef cattle but have had limited application. The availability of denser SNP chips and whole-genome sequences will lead to the discovery of genes and gene pathways and allow a far greater understanding of the genetic architecture of imprinting, dominance and epigenetic effects, although at this stage it is unclear how this will affect genetic evaluation or selection.
Improved genetic evaluation
Ongoing research will be required into the future to improve the methods for incorporating the ever-changing nature of genomic information into genetic evaluations systems. In the short term, there is a need to understand how the currently derived GEBVs will predict performance over subsequent generations. Goddard et al. (2010b) proposed that the rapid decline in genetic gain over generations, resulting from the use of GEBVs observed in simulation studies could be eliminated by identifying the quantitative trait locis. Also, the accuracy of genomic predictions has been shown to reduce as the additive genetic relationship to the initial training animals becomes more distant (Habier et al. 2010). If these effects cause significant reduction in the accuracy of prediction, then new methods will be required to account for this when including genomic information into EBVs. Alternatively, the statistical methods used to derive GEBVs may need to be further researched (Habier et al. 2010).
Opportunities also exist to improve genetic evaluation systems by using the high-density genotypes to validate pedigree information and correct the numerator relationship matrix used in standard BLUP evaluation. Even well recorded herds have a fraction of their calves (between 3% and 5%) identified with an incorrect pedigree. Over time, if genotyping becomes widespread, improved pedigrees could also contribute to higher heritability estimates. At a practical level, if the inclusion of genomic information becomes routine in beef evaluations, then it will benefit the accuracies of EBVs of animals that have not had their performance recorded or where fixed effects are unknown, especially of animals whose records are in single animal contemporary groups. The calculation of EBVs from genotypes does not require definition of management group, age adjustment or linkage across herds. This has possible applications beyond the seedstock sector (Kinghorn 2012), and genetic evaluation through genomic information could provide the commercial sector with new tools to manage and select cattle (e.g. replacement females).
An increase in the accuracy of EBVs of objective traits has the potential to increase rates of gains. But to make maximum genetic progress in profit also requires the correct weighting of traits in the breeding objective. Barwick et al. (2011) argued that current forms of genomic information should not require any fundamental changes to the development of breeding objectives, although as the technology develops, there may be the opportunity to more accurately define traits and to include genomic tests for genes of large effect (e.g. diseases, horns) directly into breeding objectives.
Database requirements
For genomic data to be included in genetic evaluation schemes will require databases for efficient storage and retrieval of extremely large volumes of genotypic data for the calculation of GEBVs or for the construction of GRMs. If regular re-estimation of prediction equations is necessary, then databases will need to store both genotypic and phenotypic records. Alternatively, if the GRM is the method for including genomic information, then only the genotypes would need to be stored. Storage of GEBV predictions from third parties (e.g. Pfizer Animal Genetics MVPs) will require correct unique animal identification, along with version details of the prediction equation used, because they are likely to change over time. A national genotype database has been developed in Australia as part of the Beef CRC, and it is being populated with genomic data, including 50K genotypes (N > 9000) and GEBVs. Significantly, over time, the capacities of this database will need to be expanded to allow storage of 800K genotypes and eventually whole-genome sequence data. The development of such genomic databases is not specific to the beef industry and collaboration with other livestock industries is critical to make the best use of research funds.
Cost-effective genotyping
The cost effectiveness of using genomic information in the beef industry relies on the accuracies of GEBVs and the price of genotyping. Results of Van Eenennaam et al. (2011) suggest that industry structure and strong price signals through the beef-production chain will be necessary to make genomic selection successful. From the breeding-industry perspective, there is a need to combine genotyping, single-gene tests and parentage tests onto a single chip. In the commercial sector, any use of genomics will be dictated by the price of genotyping because there is little opportunity to pass on the cost from an individual animal, unlike in the seedstock industry where a sire can have many hundreds of progeny that benefit. Some reduction in price may be achieved by using low-density chips, but the usefulness of these will still depend on the accuracy of prediction obtained for key commercial profit driver traits.
Breeding-industry changes
Genomic selection is rapidly changing breeding structures in the dairy industry and is also likely, over time, to have an impact on the beef industry bull-breeding sector. In dairy, genomic selection is having its greatest impact by reducing the number of young progeny-test sires selected and the proportion of cows mated to those young sires, thus reducing the cost and generation interval as originally suggested by Schaeffer (2006). Currently, the beef seedstock sector uses a combination of higher-accuracy artificial insemination sires and relatively low-accuracy young bulls. With the advent of higher-accuracy genomically enhanced EBVs, breeders will have the opportunity to increase rates of gain by selecting their own young bulls. However, genetic progress will still be dependent on generating increased selection differential in the sires or shortening the generation interval. Genomics offers increased accuracy of selection, but in beef, there is the issue of generating sufficient selection intensity, particularly in females. Accurate GEBVs on females would provide the opportunity to increase selection differentials and reduce generation intervals in dams by using elite females in multi-ovulation and embryo-transfer programs.
The greatest potential change, as GEBVs increase in accuracy, will be in recording practices. Breeders will be faced with issues of which animals should be phenotyped and/or genotyped. Availability of GEBVs could see performance recording contracted to relatively few breeders or cooperative groups of breeders, and their data may drive the development of genomic predictions for the breed. However, it is unclear how the costs of recording and genotyping would be shared across all beneficiaries.
In the commercial sector, natural service is likely to continue to dominate and thus the impact of genomic information will be through increased accuracy on young bulls, allowing more targeted matching of genetics with production–marketing systems. Genomic selection may have utility in the currently unrecorded bull-multiplier sector of pastoral companies in northern Australia, but again this will depend on the cost effectiveness of genotyping versus the accuracy of prediction.
Conclusions
For the beef industry to benefit from genomic selection it needs to invest heavily in the collection of many more phenotypes to enable a significant lift in accuracy of the current GEBVs, particularly female reproduction traits, abattoir carcass traits and feed efficiency. With more accurate GEBVs, breeds have the opportunity to make faster rates of genetic progress, but will require the information to be included in existing genetic evaluations. In the future, genomics offers even greater benefits, with higher-density chips and sequence data allowing further increases in accuracy and a far greater understanding of underlying genetic mechanisms. The beef industry will need to position itself by collecting large amounts of data, particularly on difficult-to-measure traits. Ultimately, the contribution of genomics to beef breeding will be driven by the cost-effectiveness of the technology and the willingness of the participants to embrace genetic improvement for the benefit of the industry. There is a danger that low and moderately accurate GEBVs will be seen as an ‘easy fix’ and lead to a reduction in performance recording in the more extensive sectors of the beef industry. This should be avoided.
Acknowledgements
The authors thank Meat and Livestock Australia for supporting this work (Project B.BFG.0050). We acknowledge each breed society for providing access to their data.
References
Barwick SA, Graser H-U, Swan AA, Hermesch S (2011) Experience in breeding objectives for beef cattle, sheep and pigs, new developments and future needs. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 19, 23–30.Beef CRC (2009) Australian marker panel evaluation. Available at http://www.beefcrc.com.au/Assets/572/1/DJ_Pfizer_MVP_Report-3toCRC.pdf [Verified 1 May 2011]
Chen CY, Misztal I, Aguilar I, Tsuruta S, Meuwissen THE, Aggrey SE, Wing T, Muir WM (2011) Genome-wide marker-assisted selection combining all pedigree phenotypic information with genotypic data in one step: an example using broiler chickens. Journal of Animal Science 89, 23–28.
| Genome-wide marker-assisted selection combining all pedigree phenotypic information with genotypic data in one step: an example using broiler chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXotl2lsg%3D%3D&md5=c031b0d13581b2262848206e16b79808CAS |
de Roos APW, Hayes BJ, Spelman RJ, Goddard ME (2008) Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics 179, 1503–1512.
| Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1cvmsFeltw%3D%3D&md5=a6c49348245b1efce69bcdfe46d3a613CAS |
de Roos APW, Hayes BJ, Goddard ME (2009) Reliability of genomic predictions across multiple populations. Genetics 183, 1545–1553.
| Reliability of genomic predictions across multiple populations.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1MfhtFSqsg%3D%3D&md5=b9d5bd700e46018d2915502cf78fc7daCAS |
Garrick DJ (2010) The nature, scope and impact of some whole genome analyses in beef cattle. In ‘Proceedings of the 9th world congress on genetics applied to livestock production’. Communication 23.
Goddard ME (2009) Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136, 245–257.
| Genomic selection: prediction of accuracy and maximisation of long term response.Crossref | GoogleScholarGoogle Scholar |
Goddard ME (2012) Uses of genomics in livestock agriculture. Animal Production Science 52, 73–77.
| Uses of genomics in livestock agriculture.Crossref | GoogleScholarGoogle Scholar |
Goddard ME, Hayes BJ (2009) Mapping genes for complex traits in domestic animals and their use in breeding programs. Nature Reviews. Genetics 10, 381–391.
| Mapping genes for complex traits in domestic animals and their use in breeding programs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtVKrsLw%3D&md5=5318e6beaf602f17794e33ee2b300564CAS |
Goddard ME, Hayes BJ, Meuwissen THE (2010a) Genomic selection in farm animal species – lessons learnt and future perspectives. In ‘Proceedings of the 9th world congress on genetics applied to livestock production’. Communication 701.
Goddard ME, Hayes BJ, Meuwissen THE (2010b) Genomic selection in livestock. Genetic Research Cambridge 92, 413–421.
| Genomic selection in livestock.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjs1Clt7k%3D&md5=eeda12b1ce683152888107d3874bb5f0CAS |
Graser H-U, Tier B, Johnston DJ, Barwick SA (2005) Genetic evaluation for the beef industry in Australia. Australian Journal of Experimental Agriculture 45, 913–921.
| Genetic evaluation for the beef industry in Australia.Crossref | GoogleScholarGoogle Scholar |
Habier D, Tetens J, Seefied F-R, Lichtner P, Thaller G (2010) The impact of genetic relationship information on genomic breeding values in German Holsteins. Genetics, Selection, Evolution 42, 5–17.
| The impact of genetic relationship information on genomic breeding values in German Holsteins.Crossref | GoogleScholarGoogle Scholar |
Harris BL, Johnson DL (2010) Genomic predictions for New Zealand dairy bulls and integration with national genetic evaluation. Journal of Dairy Science 93, 1243–1252.
| Genomic predictions for New Zealand dairy bulls and integration with national genetic evaluation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitlOis78%3D&md5=22c05949294bfed382460384b5f73857CAS |
Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME (2009a) Invited review. Genomic selection in dairy: progress and challenges. Journal of Dairy Science 92, 433–443.
| Invited review. Genomic selection in dairy: progress and challenges.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXit1Kju7s%3D&md5=23939241311e2729579adc8fdcbcfdabCAS |
Hayes BJ, Visscher PM, Goddard ME (2009b) Increased accuracy of artificial selection by using the realised relationship matrix. Genetic Research Cambridge 91, 47–60.
| Increased accuracy of artificial selection by using the realised relationship matrix.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXit1aisbc%3D&md5=bd9e1502e7793d6b8df1216149b86bfaCAS |
Johnston DJ, Graser H-U (2010) Estimated gene frequencies and GeneSTAR markers and their size of effects on meat tenderness, marbling, and feed efficiency in temperate and tropical beef cattle breeds across a range of production environments. Journal of Animal Science 88, 1917–1935.
| Estimated gene frequencies and GeneSTAR markers and their size of effects on meat tenderness, marbling, and feed efficiency in temperate and tropical beef cattle breeds across a range of production environments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmvVeqtrk%3D&md5=7de295e469abf66a1b5df8ec99e3b192CAS |
Johnston DJ, Tier B, Graser H-U (2009) Integration of DNA markers into BREEDPLAN EBVs. Proceedings of the Association for the Advancement of Animal Breeding and Genetics 18, 30–33.
Johnston DJ, Jeyaruban GJ, Graser H-U (2010) Evaluation of Pfizer Animal Genetics HD 50K MVP calibration. Available at http://agbu.une.edu.au/pdf/Pfizer_50K_ September%202010.pdf [Verified 1 June 2011]
Kachman SD (2008) Incorporation of marker scores into national genetic evaluations. Available at http://www.beefimprovement.org/PDFs/Kansas%20City%20Missouri%202008.pdf [Verified 21 January 2011]
Kinghorn BP (2012) The use of genomics in the management of livestock. Animal Production Science 52, 78–91.
| The use of genomics in the management of livestock.Crossref | GoogleScholarGoogle Scholar |
Kizilkaya K, Fernando RL, Garrick DJ (2010) Genomic prediction of simulated multibreed and purebred performance using fifty thousand single nucleotide polymorphism genotypes. Journal of Animal Science 88, 544–551.
| Genomic prediction of simulated multibreed and purebred performance using fifty thousand single nucleotide polymorphism genotypes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXktVOqt74%3D&md5=fd9a487bbe918237da46d74065069b64CAS |
Legarra A, Aguilar I, Misztal I (2009) A relationship matrix including full pedigree and genomic information. Journal of Dairy Science 92, 4656–4663.
| A relationship matrix including full pedigree and genomic information.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVKqtr3E&md5=52dad86381f223dab5059bdac03e2e99CAS |
Lewis J, Abas Z, Dadousis C, Lykidis D, Paschou P, Drineas P (2011) Tracing cattle breeds with principal components analysis ancestry informative SNPs. PLoS ONE 6, e18007
| Tracing cattle breeds with principal components analysis ancestry informative SNPs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltVWlsLs%3D&md5=98f67eb3e0e78be0d48388f7cabbab08CAS |
MacNeil MD, Northcutt SL, Schnabel RD, Garrick DJ, Woodward BW, Taylor JF (2010) Genetic correlations between carcass traits and molecular breeding values in Angus cattle. In ‘Proceedings of the 9th world congress on genetics applied to livestock production’. Communication 482, Leipzig.
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Misztal I, Legarra A, Aguilar I (2009) Computing procedures for genetic evaluation including phenotypic records, full pedigree, and genomic information. Journal of Dairy Science 92, 4648–4655.
| Computing procedures for genetic evaluation including phenotypic records, full pedigree, and genomic information.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVKqtr3L&md5=5949d33b867e99a0c757ecbf6e73c3dcCAS |
Northcutt SL (2010) Pulling it all together: genomic-enhanced EPDs. Available at http://www.angus.org/AGI/GenomicEnhancedEPDs.pdf [Verified 13 January 2011]
Northcutt SL (2011) Genomic choices. Available at http://www.angus.org/AGI/GenomicChoiceApril2011.pdf [Verified 10 September 2011]
Pérez-Enciso M, Ferretti L (2010) Massive parallel sequencing in animal genetics: where froms and wheretos. Animal Genetics 41, 561–569.
| Massive parallel sequencing in animal genetics: where froms and wheretos.Crossref | GoogleScholarGoogle Scholar |
Schaeffer LR (2006) Strategy for applying genome-wide selection in dairy cattle. Journal of Animal Breeding and Genetics 123, 218–223.
| Strategy for applying genome-wide selection in dairy cattle.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD28vlvFCitA%3D%3D&md5=f2323721cc79f7ab1ccb3d6057ddefe7CAS |
Swan AA, Brown DJ, Tier B, van der Werf JH (2011) Use of genomic information to estimate breeding values for carcass traits in sheep. Proceedings of the Association for the Advancement of Animal Breeding and Genetics. 19, 331–334.
Swan AA, Johnston DJ, Brown DJ, Tier B, Graser H-U (2012) Integration of genomic information into beef cattle and sheep genetic evaluations in Australia. Animal Production Science 52, 126–132.
| Integration of genomic information into beef cattle and sheep genetic evaluations in Australia.Crossref | GoogleScholarGoogle Scholar |
The Bovine HapMap Consortium (2009) Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532.
| Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds.Crossref | GoogleScholarGoogle Scholar |
Van Eenennaam AL, van der Werf JH, Goddard ME (2011) The value of using DNA markers for beef bull selection in the seedstock sector. Journal of Animal Science 89, 307–320.
| The value of using DNA markers for beef bull selection in the seedstock sector.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhvFSntb0%3D&md5=e5e80496b989f4adbda2021a961016aeCAS |
VanRaden PM, Van Tassel CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor JF, Schenkel FS (2009) Invited review: reliability of genomic predictions for North American Holstein bulls. Journal of Dairy Science 92, 16–24.
| Invited review: reliability of genomic predictions for North American Holstein bulls.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXlsVOrsw%3D%3D&md5=e20b7d5db6872bfe41fe1c7cba312240CAS |
Weber K, Bennett G, Keele J, Snelling W, Thallman RM, Van Eeennaam AL, Kuehn L (2011) Genomic selection in beef cattle: training and validation in multibreed populations. In ‘Proceedings plant and animal genome conference XIX’, San Diego, 2011. Poster P514.
Weigel KA, Van Tassell CP, O’Connell JR, VanRaden PM, Wiggans GR (2010) Prediction of unobserved single nucleotide polymorphisms genotypes of Jersey cattle using reference panels and population-based imputation algorithms. Journal of Dairy Science 93, 2229–2238.
| Prediction of unobserved single nucleotide polymorphisms genotypes of Jersey cattle using reference panels and population-based imputation algorithms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnvVans7Y%3D&md5=175dbe8742bc1ea65d367ddf18da76f9CAS |
1AGBU is a joint venture of NSW DPI and the University of New England.


