Industrial perspective: capturing the benefits of genomics to Irish cattle breeding
B. W. Wickham A D , P. R. Amer B , D. P. Berry C , M. Burke A , S. Coughlan A , A. Cromie A , J. F. Kearney A , N. Mc Hugh C , S. McParland C and K. O’Connell AA Irish Cattle Breeding Federation, Highfield House, Shinagh, Bandon, County Cork, Ireland.
B AbacusBio Limited, PO Box 5585, Dunedin, New Zealand.
C Teagasc, Moorepark Research Centre, Fermoy, County Cork, Ireland.
D Corresponding author. Email: bwickham@icbf.com
Animal Production Science 52(3) 172-179 https://doi.org/10.1071/AN11166
Submitted: 5 August 2011 Accepted: 7 February 2012 Published: 6 March 2012
Journal Compilation © CSIRO Publishing 2012 Open Access CC BY-NC-ND
Abstract
Genomics is a technology for increasing the accuracy with which the genetic merit of young potential breeding animals can be determined. It enables earlier selection decisions, thus reducing generation intervals and gives rise to more rapid annual rates of genetic gain. Recently, the cost of genomics has reduced to the point where it enables breeding-program costs to be reduced substantially. Ireland has been a rapid adopter of genomics technology in its dairy-cattle breeding program, with 40% of dairy-cow artificial inseminations in 2010 being from bulls evaluated using genomic information. This rapid adoption has been facilitated by a comprehensive database of phenotypes and genotypes, strong public funding support for applied genomics research, an international network of collaborators, a short path between research and implementation, an overall selection index which farmers use in making breeding decisions, and a motivated and informed breeding industry. The shorter generation interval possible with genomic selection strategies also allows exploitation of the already accelerating rate of genetic progress in Ireland, because elite young dairy bulls are considerably superior to the small numbers of bulls that entered progeny test 6 years ago. In addition, genomics is having a dramatic impact on the artificial-insemination industry by substantially reducing the cost of entry, the cost of operation, and shifting the focus of breeding from bulls to cows. We believe that the current industry structures must evolve substantially if Irish cattle farmers are to realise the full benefits of genomics and be protected from related risks. Our model for future dairy breeding envisages a small number of ‘next generation research herds’, 1000 ‘bull breeder herds’ and an artificial-insemination sector using 30 new genomically selected bulls per year to breed the bulk of replacements in commercial milk-producing herds. Accurate imputation from a low-density to a higher-density chip is a key element of our strategy to enable dairy farmers to afford access to genomics. This model is capable of delivering high rates of genetic gain, realising cost savings, and protecting against the risks of increased inbreeding and suboptimal breeding goals. Our strategy for exploiting genomic selection for beef breeding is currently focussed on genotyping, using a high-density chip, a training population of greater than 2000 progeny-tested bulls representing all the main beef breeds in Ireland. We recognise the need for a larger training population and are seeking collaboration with organisations in other countries and populations.
Introduction
Over the past 13 years, Ireland has established a new infrastructure to facilitate the genetic improvement of both dairy and beef cattle. Prime responsibility for leading the development rests with the Irish Cattle Breeding Federation Society Ltd (ICBF), Shinagh, Bandon, County Cork, Ireland, established in 1997 with the objective of achieving the greatest possible genetic improvement in the national cattle herd for the benefit of Irish farmers, the dairy and beef industries, and members. This development has been funded by a unique partnership involving farmers, breeders, service providers and government.
In the present paper, we outline our strategy for ensuring the Irish dairy and beef cattle-breeding industry is able to benefit to the greatest possible extent from developing genomic technologies. The present paper deals with our research strategy, our implementation strategy, progress made so far, and how our industry needs to change in the future if it is to fully benefit from current and future developments in genomic technologies.
Irish cattle industry
The Irish cattle industry is based on 2 million calvings per year, with 1.1 million in dairy herds, from predominately Holstein–Friesian cows, and 0.9 million in beef suckler herds. The industry involves a large amount of cross-breeding, with 38% of dairy-cow calvings to beef-sire breeds, mainly Angus and Hereford, and 61% of calvings of suckler cows being to a beef breed different from the breed of the cow. The five main beef breeds are Charolais, Limousin, Simmental, Angus and Hereford.
Dairy and beef production systems in Ireland are seasonal (Berry et al. 2006a; Mc Hugh et al. 2010). Some 80% of the milk and meat products of the Irish cattle industry are exported into a wide range of international markets.
Industry structure – the formation of ICBF
ICBF was established in 1997 and commenced operations in 1998, with its current structure being finalised in 2000. Its main activities are those associated with developing the cattle-breeding infrastructure in Ireland, operating the cattle breeding database, providing genetic evaluation services, and providing information useful for cattle-breeding decisions.
ICBF is owned by the cattle industry, with 18% of shares held by each of the artificial-insemination (AI), milk-recording (MR) and herd-book (HB) sectors and the remaining 46% held by the organisations representing farmers. The ICBF board of 16 comprises 15 persons appointed by the shareholders and one appointed by the Irish Department of Agriculture Fisheries and Food.
Since its inception, much of ICBF’s work has been focussed on improving the quantity and quality of data available for cattle breeding. New technologies have been adopted (Berry et al. 2006b; Pabiou et al. 2011) and business arrangements established with both shareholders and industry stakeholders. The overall goal has been to ensure that Irish farmers have access to high-quality information for use in breeding more profitable cattle.
Since 2008, ICBF and its research partners have made a major commitment to researching and implementing genomic selection in dairy-cattle breeding and are currently extending this effort to beef-cattle breeding.
Breeding and selection criteria
A widespread industry consultation supported by extensive research resulted in an agreed breeding objective and selection criteria for dairy cattle (Veerkamp et al. 2000) and beef cattle (Amer et al. 2001) in Ireland. The objective in both cases is farm-level profitability by accounting for the most significant sources of income and cost. An overall index, the economic breeding index (EBI), and suckler-beef value, both expressed in economic terms (€), are the selection criteria for dairy and beef, respectively. These overall indexes are built up from several sub-indexes, also expressed in economic terms. Examples are readily available on the ICBF website (www.icbf.com, verified 1 May 2011). A recent well powered study (Ramsbottom et al. 2011) relating herd EBI to overall herd profitability across 1131 herd-years on Irish dairy farms clearly showed that profit per lactation was within expectations based on differences in EBI.
ICBF provides the genetic evaluation system for both dairy- and beef-cattle breeding in Ireland. This genetic evaluation system operates in close association with the ICBF database. ICBF is a full participant in the activities of Interbull, the international dairy genetic evaluation organisation and is currently providing leadership for the establishment of Interbeef.
The genetic evaluation system used by ICBF is an across-breed system with a single base for each set of traits. It covers all breeds and crosses of cattle in Ireland and the full range of traits. For some traits (e.g. calving and carcass), the evaluation system uses data from dairy and suckler herds, and the resulting evaluations, in these cases, are comparable across dairy and beef breeds. Strong emphasis is placed on obtaining peer review of the procedures and statistical models used (Olori et al. 2002; Crowley et al. 2010; Berry et al. 2011; Mc Hugh et al. 2011; Pabiou et al. 2012).
Phenotypes – the ICBF cattle-breeding database
At the time ICBF was formed, there were a large number of separate computer systems. Each had its own data-collection system and supported the information needs of one or other aspects of the cattle-breeding industry. For example, each pedigree breed society HB, of which there were 18 at that time, had its own system, and each MR organisation, of which there were eight in 1998, had its own system. At that time, the Irish Department of Agriculture Fisheries and Food also operated separate systems for genetic evaluations and the official calf-registration and cattle-movement monitoring. These systems used several different animal identifications and held limited cross-references.
ICBF established its cattle-breeding database using a software system from a Dutch cattle-breeding organisation. Creating the database involved an enormous effort to negotiate agreements for the sharing of data, to establish shared-data collection systems and to consolidate the existing computer files into a single shared database. The key principles underpinning the agreement among organisations to share data are summarised in Table 1.

|
ICBF established a team of information-technology developers, supported by several contractors, to customise the Dutch system to meet the needs of the Irish breeding industry. This customisation has now reached the point where the ICBF database requires no support from suppliers of the Dutch system.
The ICBF cattle-breeding database supports, through the use of a range of new technologies (Cromie et al. 2008) the information needs of MR, HB, AI organisations and cattle farmers. Farmers are able to access their own data in the database through the web, or on paper, via a service known as HerdPlus (www.icbf.com, verified 15 February 2012).
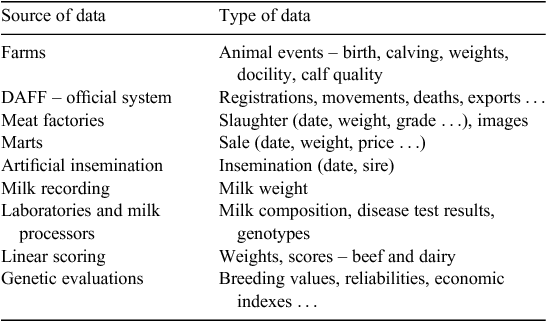
This database has and is playing a fundamental role in facilitating the research and implementation of genomic selection in Irish cattle breeding. In all, 90% of all dairy and beef cattle in Ireland are recorded in this database. Table 2 provides a summary of data and data sources included in this system. The database now also stores 13 739 genotyped animals (Table 3), consisting of >2 billion single-nucleotide polymorphisms.
|

|
Potential contribution of genomics
Ideally, to locate the most profitable cattle for use in Ireland, we would like to rank all potential selection candidates on the merit of future progeny, irrespective of breed. That is, to rank them on the Irish selection criteria. For an Irish breeder, the candidates for selection are currently bulls worldwide available through AI, natural mating bulls in Ireland, and cows and heifers mostly in the breeder’s own herd. The availability of candidates is complicated by disease regulations and disease risks.
Genomics has the potential to help the Irish breeder, in the short term, through a combination of one or more of the following three factors:
-
To increase the accuracy of the genetic evaluation of selection candidates, especially natural mating bulls and replacement females.
-
To reduce the age at which accurate evaluations are available on selection candidates, especially for bulls available through AI.
-
To increase the number of potentially available selection candidates, especially for bulls available through AI.
In the longer term, and providing the technology is used appropriately, we expect that genomics will give rise to more rapid annual rates of genetic change (Mc Hugh et al. 2011). Providing this change is for a combination of traits giving rise to more profitable commercial production of milk and meat, there will be substantial added benefits to farmers in Ireland.
Implementation
The implementation of genomic selection in Ireland has followed closely on the availability of research results, industry acceptance, and a decision to proceed. Developments since this initial decision to proceed have included the availability of a larger training population, accurate imputation from lower-density (3K) to higher-density (50K) chips (Berry and Kearney 2011), extending genomic evaluations to the traditional Friesian, and the inclusion of body-conformation traits. This has been achieved in a well coordinated and integrated manner, through a combination of research, systems, operations, and communications.
Research
Ireland’s cattle genomics research program is part of a longer-term strategy to build a more profitable cattle-farming sector. The research was undertaken by both the ICBF and Teagasc, Fermoy, County Cork, Ireland. The research strategy comprises several elements and resources, as summarised in Table 4. Genomic selection in Ireland is undertaken using genomic best linear unbiased prediction (Van Raden 2008) and are described in detail by Berry et al. (2009). When originally launched in 2009, the input variables for the genomic predictions were de-regressed proofs from the domestic proof run on 985 Holstein–Friesian AI bulls. The Illumina Bovine50 beadchip (www.illumina.com, verified 15 February 2012) was the genotyping platform used. Following considerable sharing of genotypes with international collaborators, research showed a benefit of replacing the de-regressed domestic proofs with de-regressed multiple-across-country-evaluation proofs from Interbull on a larger number of animals. This resulted in a considerable increase in the training population size. The dependent variables are weighted by their respective reliability less parental contribution. A restriction is imposed that the reliability of the training animals must be >40%. This, therefore leads to different-sized training populations for the different traits, depending on (1) the respective heritability and number of effective daughters, and (2) whether or not the trait is included in the suite of traits evaluated by Interbull. The number of animals currently included in the training population for the milk-production traits is 4550, whereas the number of animals currently included in training population for calving interval is 2315. Direct genomic values are blended, using selection index methodology, with traditional breeding values of the ungenotyped back-pedigree. In 2011, the average reliability of genomic estimated breeding values of young animals was 50%. Research is currently underway to expand genomic selection in non-Holstein animals, by acquiring genotypes on cross-bred and alternative-breed cows with phenotypes and also the limited number of bulls proven in Ireland.

|
Systems
The key systems that were developed to support the implementation of genomic selection for dairy cattle in early 2009 were those associated with genomic data storage and genetic evaluations. Initially, in 2009, flat file systems were used to store the genotypes but these have been replaced by a genotype storage system integrated into the ICBF database. ICBF’s genetic evaluation systems, web screens and active bull lists were upgraded to incorporate genomic data into the existing trait-based evaluations system and progress has been reported elsewhere (Kearney et al. 2009, 2010).
Operations
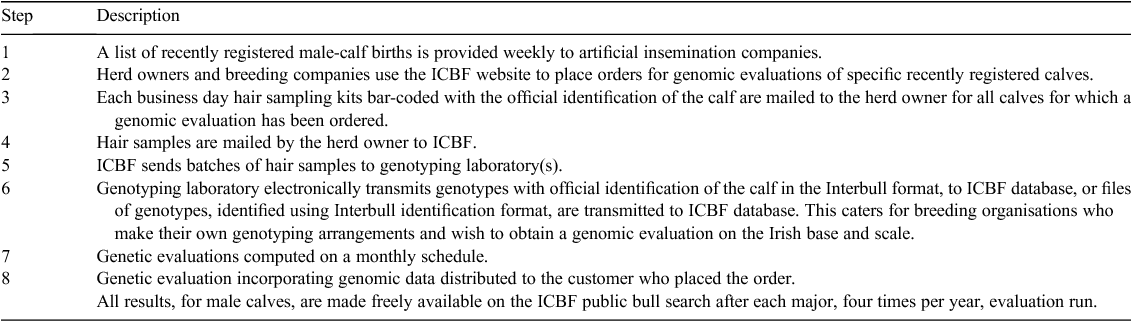
A genomic evaluation service to farmers and breeding organisations has recently been launched in Ireland. This user-pays-service, €50, including tax per animal for Illumina 3K-based evaluation, is focussed on providing genomic evaluations of new-born dairy calves, males and females. This service is available to farmers and breeding companies (Irish or international) alike. It is a fully integrated service that makes extensive use of the ICBF database and associated systems to save costs, minimise errors and minimise turn-around times. The service involves eight main steps (Table 5).
|
Communications
Communications with the Irish cattle-breeding industry, that is AI companies (including semen importers), breed associations, MR organisations and advisory services, are primarily focussed on three to four open public meetings annually – the cattle breeding consultation meetings. Copies of all presentations to these meetings are routinely placed in the publications section of the ICBF website (http://www.icbf.com, verified 1 May 2011), with meeting summaries and developments also reported in a weekly update, which is also available in the publications section of the ICBF website. The cattle-breeding consultation meetings operate on an annual cycle, with the meetings earlier in the year focussed on research plans and priorities, and those later in the year focusing on implementation plans. Most changes to evaluations are implemented later in the year, so as to ensure complete and stable information for the main spring breeding season in Ireland. Between these major meetings, the breeding industry has ready access to scientific staff, receives the results of periodic test runs and interacts with ICBF’s operational team. The subject of genomics has been a routine part of this communication system since 2008.
Communication with farmers for the implementation of genomic selection in 2009 primarily focussed on a relatively small set of key messages, through a range of well established communication channels, as summarised in Table 6.

|
The communication channels include farmer discussion groups, national farming press (for a period of 16 weeks there was a full page feature titled ‘Best Practice in Cattle Breeding’ in the weekly Irish Farmers Journal), farm-walks, and a Discussion Group competition. The latter focuses on rewarding groups of farmers who have excelled in making sustainable genetic gain.
Progress – so far
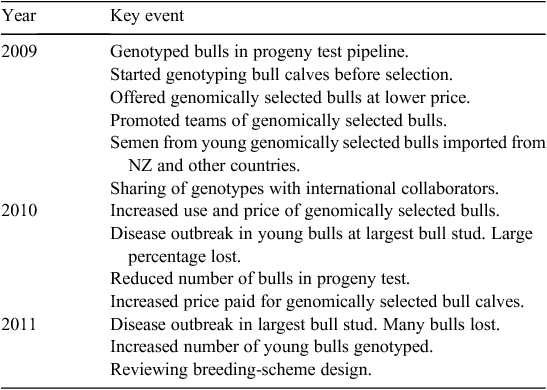
The use of genomically selected (GS) dairy bulls in AI is now in its third year in Ireland. The response of the Irish AI industry to the availability of genomic technology is summarised in Table 7 and that of farmers in Table 8. The AI industry has moved rapidly to the provision of younger GS dairy bulls, with higher EBIs selected using genomic information. These are known as GS bulls. In the past 2 years, the situation has been complicated by repeated outbreaks of infectious bovine rhinotracheitis in the bull stud of the largest AI company in Ireland, which will lead to an even greater use of young bulls in the future. Farmers have responded by using the higher-index GS bulls and have taken the advice to use teams of bulls to compensate for the lower reliability of individual bulls.
|
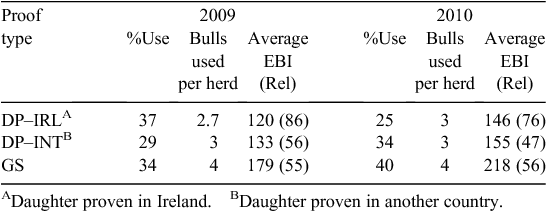
|
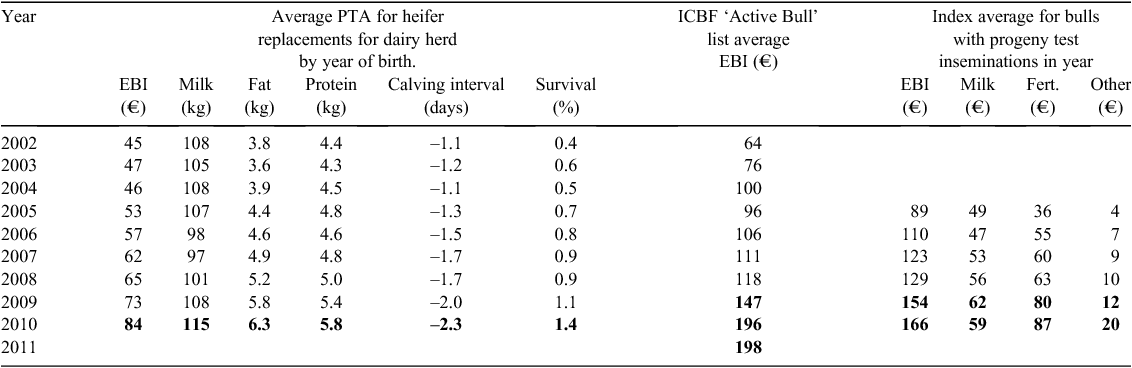
The impact of genomics on genetic trends is shown in Table 9. Genomic technology was first introduced early in 2009 and its impact can be immediately seen in the average EBI of the bulls available in AI – the ‘Active Bull’ list – and the bulls used in progeny testing. Further gains in these two indicators also occurred in 2010. The flow through to the birth of replacements for the dairy herd is seen in the dairy replacements born in 2010. The contribution of fertility to the bulls being selected on the basis of genomic information for progeny testing was observed in 2009 and 2010, where a large part of the EBI increase is explained by increases in the fertility sub-index of the EBI.
Future breeding-scheme design
Breeding-scheme design was a priority from the establishment of ICBF in 1997, and before adopting genomic selection, the implications of the breeding-scheme design were considered. An initial analysis conducted in 2008 (Sonesson et al. 2008) about the impact of genomics identified the potential for more rapid genetic gain and lower costs and was a key element in the decision to pursue the technology vigorously. The study of Sonesson et al. (2008) identified the risk of increased inbreeding if the use of genomic selection was not well implemented. It was on the basis of this finding, and the very large impact of one bull on the Active Bull List, that the practicality of guiding the mating of elite cows in the Irish dairy herd was tested (McParland et al. 2009). Further research included extensive simulation studies that identified the important potential role of females and the need to maintain some form of progeny test (Mc Hugh et al. 2011). An extensive industry consultation was conducted during 2010 and culminated with the completion of a cost–benefit evaluation of a range of future options (Amer 2010). The benefits and costs of these options are summarised in Table 10.

|
On the basis of the present results, the current focus for the future breeding scheme for Irish dairy cattle comprises three types of herds – commercial, bull breeder and next generation. Table 11 contains a summary of the key attributes and roles of these herds.
Summary
In the past 13 years, the Irish cattle-breeding industry has undergone a complete redevelopment of its data-gathering and genetic-evaluation infrastructure. The comparatively recent availability of genomic technologies has been quickly assimilated by this new infrastructure, through a combination of the following:
-
research based on Irish phenotypes, Interbull evaluations and genotypes from Irish and international collaborations;
-
rapid implementation of research findings into the Irish genetic evaluations systems for dairy cattle;
-
deployment of an operational system for efficiently providing genomic evaluations to anybody for new-born male and female selection candidates;
-
effective communications with both the breeding industry and farmers;
-
research on breeding-scheme design and related cost–benefit studies; and
-
plans for ensuring the Irish cattle-breeding industry is structured to enable the rapidly evolving genomics technology to be fully exploited for the benefit of Irish cattle farmers.
Irish farmers, research scientists, HBs and AI companies have responded by making good use of the information now available on the role that genomics can play in cattle breeding. As a result, Irish farmers are now able to better exploit the potential of genetics as a tool for improving the profitability of their enterprises.
References
Amer PR (2010) Implications of alternative breeding program structures for dairy cattle in Ireland. Available at http://www.icbf.com/publications/files/cost_benefit_analysis_final.pdf [Verified 13 February 2012]Amer PR, Simm G, Keane MG, Diskin MG, Wickham BW (2001) Breeding objectives for beef cattle in Ireland. Livestock Production Science 67, 223–239.
| Breeding objectives for beef cattle in Ireland.Crossref | GoogleScholarGoogle Scholar |
Berry DP, Kearney JF (2011) Imputation of genotypes from low- to high-density genotyping platforms and implications for genomic selection. Animal 5, 1162–1169.
| Imputation of genotypes from low- to high-density genotyping platforms and implications for genomic selection.Crossref | GoogleScholarGoogle Scholar |
Berry DP, O’Brien B, O’Callaghan EJ, O’Sullivan K, Meaney WJ (2006a) Temporal trends in bulk tank somatic cell count and total bacterial count in Irish dairy herds during the past decade. Journal of Dairy Science 89, 4083–4093.
| Temporal trends in bulk tank somatic cell count and total bacterial count in Irish dairy herds during the past decade.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVWgsb3P&md5=c96e9bf3b8b09590aa026b9571742746CAS |
Berry DP, Burke M, O’Keefe M, O’Connor P (2006b) Do-it-yourself milk recording as a viable alternative to supervised milk recording in Ireland. Irish Journal of Agricultural and Food Research 45, 1–12.
Berry DP, Kearney JF, Harris BL (2009). Genomic selection in Ireland. Interbull Bulletin 39. Uppsala, Sweden, 26–29 January 2009.
Berry DP, Evans RD, McParland S (2011) Evaluation of bull fertility in dairy and beef cattle using cow field data. Theriogenology 75, 172–181.
| Evaluation of bull fertility in dairy and beef cattle using cow field data.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3M%2FitVWiuw%3D%3D&md5=c2470d452b310f6ea4c9702ad96d922bCAS |
Cromie A, Wickham B, Coughlan S, Burke M (2008) The impact of new technologies on performance recording and genetic evaluation of dairy and beef cattle in Ireland. Proceedings ICAR biennial conference, Niagara, June 2008.
Crowley JJ, McGee M, Kenny DA, Crews DH, Evans RD, Berry DP (2010) Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance tested beef bulls. Journal of Animal Science 88, 885–894.
| Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance tested beef bulls.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXktVOqurk%3D&md5=92c906a0415427e4b45ff696b8375241CAS |
Kearney F, Cromie A, Berry D (2009) Implementation and uptake of genomic evaluations in dairy cattle in Ireland. Available at http://www.icbf.com/publications/files/itb_082009_FK.pdf [Verified 13 February 2012]
Kearney F, Cromie A, Berry D (2010) Implementation and uptake of genomic evaluations in dairy cattle in Ireland. Available at http://www.icbf.com/publications/files/wcgalp_fk.pdf [Verified 13 February 2012]
Mc Hugh N, Fahey AG, Evans RD, Berry DP (2010) Factors associated with selling price of cattle at livestock marts. Animal 4, 1378–1389.
| Factors associated with selling price of cattle at livestock marts.Crossref | GoogleScholarGoogle Scholar |
Mc Hugh N, Meuwissen THE, Cromie AR, Sonesson AK (2011) Use of female information in dairy cattle genomic breeding programs. Journal of Dairy Science 94, 4109–4118.
| Use of female information in dairy cattle genomic breeding programs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpsVylurY%3D&md5=6ed4c4c06edcc3fc24a726565a920408CAS |
McParland S, Kearney JF, Lopez-Villalobos N, Berry DP (2009) Optimal system of contract matings for use in a commercial population. Irish Journal of Agricultural and Food Research 48, 43–56.
Olori VE, Meuwissen THE, Veerkamp RF (2002) Calving interval and survival breeding values as measure of cow fertility in a pasture-based production system with seasonal calving. Journal of Dairy Science 85, 689–696.
| Calving interval and survival breeding values as measure of cow fertility in a pasture-based production system with seasonal calving.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xis1Krtbk%3D&md5=b1e26be1f92d7b5468bc772663d687d4CAS |
Pabiou T, Fikse WF, Cromie AR, Keane MG, Nasholm A, Berry DP (2011) Use of digital images to predict carcass cut yields in cattle. Livestock Science 137, 130–140.
| Use of digital images to predict carcass cut yields in cattle.Crossref | GoogleScholarGoogle Scholar |
Pabiou T, Fikse WF, Am PR, Cromie AR, Näsholm A, Berry DP (2012) Genetic relationships between carcass cut weights predicted from video image analysis and other performance traits in cattle. Animal in press.
Ramsbottom G, Cromie AR, Horan B, Berry DP (2011) Relationship between dairy cow genetic merit and profit on commercial spring calving dairy farms. Animal
| Relationship between dairy cow genetic merit and profit on commercial spring calving dairy farms.Crossref | GoogleScholarGoogle Scholar |
Sonesson A, Meuwissen T, Cromie A (2008) Genomic selection in Irish dairy cattle breeding scheme. Available at http://www.icbf.com/publications/files/Genomic_Selection_in_Irish_dairy_cattle_breeding_Report_Dec_2008.pdf [Verified 13 February 2012]
Van Raden PM (2008) Efficient methods to compute genomic predictions. Journal of Dairy Science 91, 4414–4423.
| Efficient methods to compute genomic predictions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlajtLzO&md5=d5c2fddefa3e236291c053ec3e512ae0CAS |
Veerkamp RF, Meuwissen THE, Dillon P, Olori V, Groen AF, van Arendonk JAM, Cromie AR (2000) Dairy breeding objective and programs for Ireland. Available at http://www.icbf.com/publications/files/Final_RBI_report_25_11_2000.pdf [Verified 13 February 2012]