Quantitative molecular assays for evaluating changes in broiler gut microbiota linked with diet and performance
V. A. Torok A B C , C. Dyson A , A. McKay A and K. Ophel-Keller AA South Australian Research and Development Institute, Urrbrae, SA 5064, Australia.
B Australian Poultry CRC, University of New England, Armidale, NSW 2351, Australia.
C Corresponding author. Email: valeria.torok@sa.gov.au
Animal Production Science 53(12) 1260-1268 https://doi.org/10.1071/AN12272
Submitted: 3 August 2012 Accepted: 13 March 2013 Published: 21 May 2013
Journal Compilation © CSIRO Publishing 2013 Open Access CC BY-NC-ND
Abstract
Changes in the levels of specific gut bacteria have been linked to improved broiler feed efficiency. Quantitative polymerase chain reaction (qPCR) assays were developed to five potential performance-related bacteria (Lactobacillus salivarius, L. crispatus, L. aviarius, Gallibacterium anatis and Escherichia coli) and generic eubacteria. These were used to screen broiler gut samples from four geographically diverse Australian feeding trials showing significant treatment-related differences in feed efficiency. It was our aim to validate the association of particular bacteria with broiler feed efficiency across a broad range of environmental and dietary conditions, and hence to evaluate their predictive potential for monitoring broiler performance. Across trials L. salivarius, L. crispatus, L. aviarius, E. coli and total eubacterial numbers were significantly altered by diet, environment (litter), and/or sex of birds. Furthermore, changes in the numbers of these gut bacteria were significantly linked to broiler performance. Lactobacilli and total eubacteria were significantly decreased in birds that were more feed efficient. E. coli was not consistently linked with either improved or decreased performance and these discrepancies may be due to differences at the strain level which were not detectable using our assays. G. anatis was detected only in two of the four trials and found not to be significantly linked with broiler performance. These qPCR assays have been useful in either validating or disproving previous reported findings for the association of specific gut bacteria with broiler feed efficiency. This qPCR format can be easily expanded to include other organisms and used as a quantitative screening tool in evaluating dietary additives for improved broiler production.
Additional keywords: feed conversion efficiency, microbial diversity, poultry, poultry nutrition.
Introduction
Gut microbiota play an important role in animal health, performance and nutrition (Gordon and Pesti 1971; Hammons et al. 2010). Although knowledge of the ideal gut microbiota is still incomplete, it is apparent that a variety of diets can equally support optimal broiler performance and maintain a healthy gut microbial balance (Torok et al. 2011b). Several factors can influence the host’s gut microbiota, including age, genetics, rearing environment and stress, although the greatest determinant by far is the host’s diet (Lu et al. 2003; Burkholder et al. 2008; Torok et al. 2009, 2011b; Lumpkins et al. 2010). All these factors also have the potential to influence the animal’s performance as measured by growth rate, feed efficiency or productivity, hence indicating a close relationship between gut microbiota and animal performance (Torok et al. 2011b).
Numerous studies have been undertaken to determine the influence of dietary changes on the structure of the gut microbial community, by using microbiological culturing techniques, culture-independent molecular techniques and indirect measurement of bacterial metabolic products (Bjerrum et al. 2006; Choct et al. 1999; Hubener et al. 2002; Zhu et al. 2002; Yin et al. 2010). However, understanding how diet-induced changes in the composition of the gut bacterial community relate to important metabolic changes, and ultimately to broiler health and performance, is less clear. Diet-related changes in gut microbiota do not always translate into altered broiler performance (Gunal et al. 2006; Pedroso et al. 2006; Geier et al. 2009), so caution needs to be exercised in identifying gut bacteria linked with broiler performance from limited animal trials. As such, tools need to be developed that can quantitatively validate findings across numerous performance trials, as many molecular techniques (denaturing gradient gel electrophoresis, terminal restriction fragment length polymorphism, pyrotag sequencing and phlyogenetic microarrays) used to investigate microbial community structure are at best semiquantitative (Amend et al. 2010; Zhou et al. 2011; Fraher et al. 2012).
We have previously shown that diet-related changes in gut microbiota are linked to broiler performance as measured by feed efficiency, and have identified a common suite of bacterial groups associated with significant changes in performance across various feeding trials (Torok et al. 2011b). Using microbial profiling, we were able to identify eight operational taxonomic units (OTU) related to differences in broiler performance across three Australian feeding trials. Targeted cloning and sequencing of these eight OTUs revealed that they could represent 26 different bacterial species or phylotypes. In the present study, we developed quantitative polymerase chain reaction (qPCR) assays to the following two of these performance-related OTUs: OTU 492, which was associated with improved broiler performance and potentially represented Escherichia coli or Gallibacterium anatis, and OTU 564–566, which was associated with decreased broiler performance and potentially represented Lactobacillus crispatus, L. salivarius or L. aviarius. It was our aim to use these assays, along with a generic eubacterial qPCR assay, to screen banks of chicken gut DNA generated from four broiler feeding trials showing significant performance differences, hence determining the diagnostic potential of these quantitative assays in determining broiler performance.
Materials and methods
Broiler feeding trials
Four independent broiler feeding trials demonstrating significant differences in bird performance as measured by feed efficiency (apparent metabolisable energy; AME or feed conversion ratio; FCR) were investigated.
In Trial I, gut samples were collected from two groups of 24 broiler chickens (12 males and 12 females) that were each fed a barley-based control diet or barley-based diet supplemented with non-starch polysaccharide (NSP)-degrading feed enzyme product (Torok et al. 2008). Previously, AME had been shown to be significantly higher for chickens fed the barley-plus-enzyme diet than for chickens fed the control barley diet. No effect of bird gender on AME, or interaction between diet and sex, was detected (Torok et al. 2008).
In Trial II, gut samples were collected from 48 male broiler chickens that had been fed one of the following four diets: wheat control supplemented with xylanase; sorghum B; commercial sorghum; and commercial sorghum supplemented with phytase (n = 12 birds/treatment) (Torok et al. 2011b). It had been previously shown that liveweight gain was significantly (P < 0.05) decreased for birds on the wheat control diet supplemented with xylanase as compared with those on the sorghum B, commercial sorghum and commercial sorghum supplemented with phytase diets (Perez-Maldonado and Rodrigues 2009). Furthermore, feed conversion efficiency was significantly (P < 0.05) decreased for birds at 0–42 days on the wheat control diet as compared with those on any of the sorghum-based diets (Perez-Maldonado and Rodrigues 2009).
In Trial III, gut samples were collected from 96 broiler chickens that had been raised on one of the following four treatments: paper litter and low-fibre diet; wood litter and low-fibre diet; paper litter and high-fibre diet; and wood litter and high-fibre diet (n = 24 birds/treatment; 12 males and 12 females) (Torok et al. 2011b). It had been shown that litter type in combination with diet composition significantly (P < 0.05) influenced FCR (Torok et al. 2011b). Birds were more feed efficient on a low-fibre diet if housed on wood litter than if housed on paper litter. Sex of birds also significantly (P < 0.001) influenced FCR, with males having a better feed efficiency than females. The bodyweight of male chickens was also significantly (P < 0.001) greater than that of the female chickens. Litter type in combination with sex was found to significantly (P < 0.05) alter bodyweight, with females growing slower on paper litter than on the wood litter (Torok et al. 2011b).
In Trial IV, gut samples were collected from 96 broiler chickens that had been fed one of four dietary treatments A, B, F and G (n = 24 birds/treatment; 12 males and 12 females) (Torok et al. 2011b). Although diet composition varied, Treatments A and B were predominantly sorghum based, while Treatments F and G were predominantly wheat based. Significant differences in FCR among dietary treatments had been previously reported for these birds (Torok et al. 2011b). Birds on the Treatment G and F diets had significantly improved feed efficiency compared with birds on Treatment A and B diets. Birds fed the Treatment G diet had the most significantly improved feed efficiency of birds on any of the diets investigated. Sex of birds also significantly (P < 0.001) influenced FCR, with males having a better feed efficiency than females.
Nucleic acid extraction
Total nucleic acid was extracted from the digesta and associated tissue from the ilea and caeca of individual chickens as previously described (Torok et al. 2011b, 2008). Total nucleic acid was extracted from ~0.5 g of freeze-dried material.
Quantitative PCR
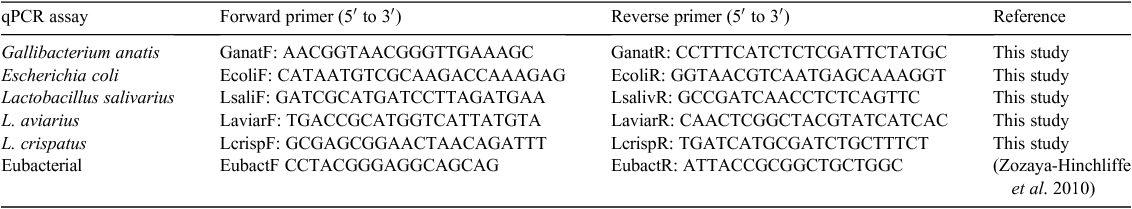
Quantitative PCR primers (Table 1) were designed to target organisms using Primer3Plus (Untergasser et al. 2007) and tested for specificity in silico by using Primer-BLAST (http://blast.ncbi.nlm.nih.gov/, verified 26 April 2013). All qPCR assays were based on SYBR green detection. Sensitivity of qPCR assays was tested against dilutions of known amounts of target (plasmid standards). Sequences of 16S rRNA representative of L. crispatus (JF798181), L. salivarius (JF798161), L. aviarius (JF798223), G. anatis (JF798066), E. coli (JF798201) and eubacteria (JF798192) were cloned into pGEM-T (Promega, Madison, WI, USA), as previously described (Torok et al. 2011a, 2011b). Plasmids were quantified using spectrophotometry (NanoDrop 2000, Thermo Scientific, Wilmington, DE, USA). The number of plasmid copies per microlitre was calculated using the following formula:
|
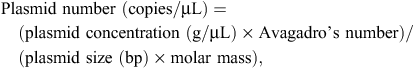
where Avagadro’s number is 6.022 × 1023 molecules/mol and the average molar mass for double-stranded DNA is 660 g/mol.
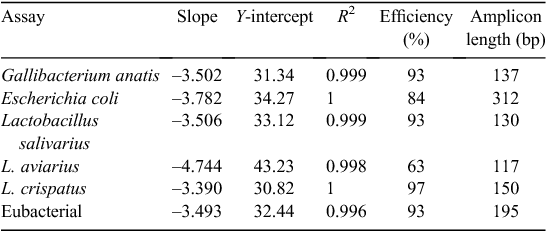
All plasmids were diluted to 2 × 1010 copies/µL. Standard curves were produced from a 10-fold dilution series, using the following range: 106 to 1 copy for specific bacteria and 108 to 102 copies for general eubacterial detection. The efficiency of each assay was determined using the plasmid standards (Table 2). Specificity of qPCR assays was confirmed visually using gel electrophoresis, as well as by determining the nucleotide sequence of resulting PCR amplicons. Purified PCR amplicons were cloned and recombinant plasmid recovered and sequenced, as previously described (Torok et al. 2011a). All qPCR amplicons were generated from total nucleic acid extract obtained from chicken gut digesta and associated gut tissue that was representative of a diverse bacterial template. Sequencing was performed from five independent clones per assay. All sequences were ≥99% identical with the target.
|
All qPCR assays were run in a 384-well format, with master mix and template being dispensed using a Biomek 3000 Laboratory Automation Workstation (Beckman Coulter, Fullerton, CA, USA). qPCR was performed in a reaction volume of 10 µL, containing 2 µL of total nucleic extract, 50 nM each primer (Table 1; Sigma Aldrich, Castle Hill, NSW, Australia) and 2 × Power SYBR Green PCR Master Mix (Applied Biosystems, Mulgrave, Vic, Australia), according to manufacturer’s recommendations. All reactions were run on a 7900HT Sequence Detection System (Applied Biosystems), with the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. To determine the specificity of all SYBR green qPCR assays, a dissociation-curve analysis of amplified DNA fragments was performed following PCR in the range of 60−95°C. All data were analysed using the 7900HT v2.3 SDS software (Applied Biosystems). Raw data were analysed for L. salivarius, E. coli, G. anatis and L. crispatus assays, by using a cycle threshold (Ct) of 0.2 and a baseline of 3–10. Assays for eubacteria used a Ct of 0.2 and a baseline of 2–10 and assays for L. aviarius used a Ct of 0.05 and an automatic baseline. Bacterial copies were calculated using standard curves.
Statistical analyses
Data obtained from individual birds were analysed by ANOVA, with significant (P < 0.05) differences determined between treatment means by the least significant difference (l.s.d.) method in Genstat v14 (VSN International, Hemel Hempstead, UK). The bacterial copy numbers were logged (ln(x)) before analysis by ANOVA. The covariate ln(copy number) eubacteria was used to standardise gross bacterial levels.
Results
Trial I
Quantitative PCR of L. salivarius showed that this microorganism was present in significantly higher numbers within the ileum of the improved performing broilers on the barley plus enzyme diet than within the ileum of birds on the barley control diet (Table 3). However, within the caeca, the numbers of L. salivarius were significantly higher in the birds fed the barley control diet (Table 4). Within the caeca, the numbers of E. coli were also significantly increased in the poorer-performing birds on the barley control diet. No diet-related differences were detected in total eubacterial or L. crispatus numbers in either gut sections. G. anatis and L. aviarius were not detected in any of the birds from this trial. Gender of birds was found to significantly influence total eubacterial numbers within the caeca, with males having increased numbers of eubacteria compared with females (Table 4).
Trial II
The only bacterial species significantly affected by diet was E. coli within the caeca (Tables 3, 4). Better-performing birds on the sorghum-based diets had significantly higher numbers of E. coli than did the poorer-performing birds on the wheat control diet. G. anatis was only infrequently detected within the ilea and caeca of birds within this trial (in 10 of 48 birds), with the same birds testing positive for both gut sections.
Trial III
Total eubacterial, E. coli and L. aviarius numbers within the ilea (Table 3), as well as, L. salivarius numbers within the caeca (Table 4), were significantly increased in birds fed the low-fibre diet. Ileal and caecal eubacterial numbers were also significantly increased in birds raised on wood litter as opposed to paper litter. Male broilers had significantly increased ileal eubacterial and L. aviarius numbers (Table 3) and decreased caecal L. crispatus numbers (Table 4) compared with female birds. Significant diet by sex and litter by sex interactions were also detected for ileal bacterial communities (Table 3). Females on the high-fibre diet had lower numbers of total eubacteria than did females on the low-fibre diet, while males on the high-fibre diet had lower numbers of L. aviarius than did males on the low-fibre diet. Furthermore, males raised on wood litter had significantly lower L. crispatus numbers than did males raised on paper litter (Table 3). Within this trial, L. aviarius was only infrequently detected within the ilea (in 34 of 96 birds) and caeca (in 15 of 96 birds), while no G. anatis was detected in either the ileum or caeca of any of these chickens.
Trial IV
Diet had a significant influence on most of the bacterial species investigated. Total eubacterial numbers were significantly decreased within both the ileum (Table 3) and caeca (Table 4) of the better-performing birds fed Treatment F, and Treatment F or G diets, respectively. Diet also had a significant influence on E. coli numbers. Within the ilea, E. coli was significantly influenced by diet but was not clearly linked to performance, while within the caeca, E. coli was significantly increased in the better-performing birds fed Treatments F and G (Table 4). Gender also had a significant influence on E. coli communities within the caeca, with males having higher numbers than females (Table 4). A significant diet by sex interaction for ileal E. coli was detected, with females fed Treatment A diet having higher numbers of E. coli than those fed Treatment B diet. Furthermore, significant differences between the sexes were noted only for birds fed the Treatment G diet, with males having higher E. coli numbers than females.
Unlike for other trials, G. anatis was detected in the ileum and caeca of most birds in the present trial, but numbers were not significantly influenced by diet nor linked to performance.
Lactobacilli were significantly influenced by both diet and sex. L. aviarius, L. crispatus and L. salivarius were significantly reduced within the ilea of better-performing birds on Treatment F and or G diets (Table 3). L. salivarius and L. crispatus were also significantly increased in the caeca of males (Table 4). Within the ilea, a significant diet by sex interaction was observed for both L. salivarius and L. crispatus. The most feed-efficient male and female birds fed Treatment G diet had significantly lower numbers of L. crispatus than the same-sex birds on the lower-performing diets (Treatments A and B). Furthermore, L. salivarius and L. crispatus differed significantly between the sexes only for the less feed-efficient birds fed Treatment A diet, with males having higher ileal numbers of these species than females (Table 3).
Discussion
Of the 26 bacterial species or phylotypes previously identified as potentially representing eight performance-related OTUs, only six phylotypes could be identified to the species (>97% nucleotide-sequence identity) (Torok et al. 2011b). In the present study, we developed qPCR assays to five of these known phylotypes (L. salivarius, L. crispatus, L. aviarius, E. coli and G. anatis), so as to determine whether they are consistently linked to broiler performance from a variety of feeding trials differing in feed composition, environment, broiler breed and bird age. qPCR results supported previous findings that OTU 564–566 (L. salivarius, L. crispatus, L. aviarius) is associated with decreased broiler performance. However, qPCR results for OTU 492 (E. coli and G. anatis), previously thought to be associated with improved performance, showed discrepancies across trials. These findings are promising in identifying broiler gut bacteria associated with performance, but also highlighted the need to validate research findings across various performance trials with specific quantitative tools such as qPCR.
Across four broiler feeding trials, each being linked to significant changes in performance as determined by feed efficiency, it was found that L. salivarius, L. crispatus, L. aviarius, E. coli and total eubacterial numbers were significantly altered by dietary treatment. Furthermore, changes in the numbers of these bacteria could be linked to broiler performance. Although diet-related changes in the composition of bacterial communities varied among feeding trials, we found that across trials, the numbers of lactobacilli and total eubacteria were significantly increased in less feed-efficient birds. Some lactobacilli, including L. salivarius, L. aviarius and L. crispatus, have been previously reported to have bile-deconjugating activity (Knarreborg et al. 2002; Majidzadeh Heravi et al. 2011). This bacterial activity lowers the detergent properties of bile acids in the emulsification of fat and leads to growth depression in chickens (Knarreborg et al. 2002; Harrow et al. 2007). Furthermore, increase in the total number of eubacteria in the less feed-efficient chickens may be a sign of bacterial overgrowth, which can lead to impaired gut health (Teirlynck et al. 2011) and, hence, impaired performance.
Conflicting results for the association of E. coli with performance were obtained across feeding trials. Increased numbers of caecal E. coli were linked to improved performance in feeding Trials II and IV, but in feeding Trial I, E. coli was linked to poorer performance. Furthermore, within the ilea, significant diet-related changes in E. coli numbers were not linked to broiler performance. These discrepancies may be due to differences at the strain level and, hence, functional level of the bacteria. Although, E. coli is considered a pathogen of humans and livestock, not all strains of E. coli have been shown to be detrimental, with some strains (E. coli Nissle) being used as a probiotic in both humans and livestock (Waidmann et al. 2003; Barth et al. 2009). Any strain differences present within trials could not be differentiated by using our species-specific qPCR approach.
Lactobacillus salivarius, L. crispatus and E. coli were detected in all trials, while L. aviarius and G. anatis were only infrequently or sporadically detected. Interestingly, in two trials, we were unable to detect G. anatis, while in one trial, most birds harboured this organism without exhibiting any detrimental effects on performance. This pattern of colonisation is consistent with previous observations where birds within a flock were either all colonised with G. anatis or were free of this organism (Bojesen et al. 2003a). This ‘all or none’ colonisation has been shown to be linked to the biosecurity level on the farm. The importance of Gallibacterium species as pathogens in poultry is unclear. Gallibacterium is part of the commensal microbiota of the upper respiratory and the lower genital tracts of healthy chickens (Bojesen et al. 2003b). However, Gallibacterium isolates have also been recovered in pure culture from a range of pathological lesions in chickens, including septicemia, oophoritis, follicle degeneration, salpingitis with or without peritonitis, peritonitis, enteritis and diseases of the respiratory tract (Bojesen et al. 2003b). Our finding that this organism can be widespread in healthy broilers is an indication that it may not be the absence or presence of the organism that is important but rather its metabolic function, which again may be linked to differences among strains. Indeed, several G. anatis biovars have been detected with clonal differences identified between tracheal and cloacal isolates from within an individual bird (Bojesen et al. 2003b). Detection of G. anatis in healthy chicken may have greater implications for detrimental performance within the layer and breeder industries due to the longer life span of those birds. Our studies on broilers were all performed at or <6 weeks of age, whereas previous investigations were conducted in layers and breeder at 13–63 weeks of age (Bojesen et al. 2003a).
We found that bird gender also had a significant influence on L. salivarius, L. crispatus, L. aviarius, E. coli and total eubacterial numbers within the gut. This is supportive of previous findings in which male and female chickens were found to differ in their ileal bacterial communities (Lumpkins et al. 2008). The fact that gender does influence gut microbiota is not surprising, considering that gender also influences performance (feed efficiency and growth rate) (Lumpkins et al. 2008). Better understanding of these gender-related differences in gut microbiota may, in future, lead to the development of gender-specific broiler diets to optimise gut microbial balance and performance.
Advances in high-resolution methods for determining the structure of microbial community, such as pyrotag sequencing and gut bacterial phylogenetic microarrays, will help identify suites of bacteria potentially linked to broiler performance. Several studies have already used pyrotag sequencing to investigate the community structure of the chicken gut (Callaway et al. 2009; Yin et al. 2010; Hume et al. 2011; Stanley et al. 2012). Although this methodology gives a detailed phylogenetic description of the complexity of the microbial community, quantitative interpretation of results should be performed with some caution (Amend et al. 2010; Zhou et al. 2011). Furthermore, direct comparison of pyrotag sequencing and phylogentic microarray results have indicated discrepancies in the phylogenetic groups identified due to the different classification schemes used by the two techniques (Claesson et al. 2009). If we are to extrapolate research findings from individual broiler feeding or performance trials, tools are needed for rapid and specific identification of organisms across trials. qPCR is ideal and can be developed into a diagnostic format which can be easily adopted by both industry and research organisations because the infrastructure for this technology is widespread, specific, quantitative, affordable and of high throughput, compared with other platforms.
The qPCR assays developed for L. salivarius, L. crispatus, L. aviarius, E. coli and total bacteria have been used to show that dietary and environmental treatments alter these bacterial communities and that they are linked to broiler feed efficiency. This qPCR approach has been shown to be a valid approach in screening large numbers of samples for the quantitative determination of organism numbers. Our qPCR results have also validated or disproved earlier findings for the association of specific bacteria with broiler performance. The qPCR assays developed could be used to screen future broiler feeding and performance trials, as well as expanded to include further gut bacteria thought to be linked to broiler performance, poultry health (pathogens) or consumer product safety (zoonotic agents). Understanding the dynamics of the chicken gut microbial community is necessary if we wish to develop further strategies for improved feed efficiency and growth rate, avoid intestinal diseases and proliferation of food-borne pathogens, and identify better feed additives and nutrient levels that influence beneficial microbial communities.
Acknowledgements
We thank Ms Ina Dumitrescu for technical assistance. This research was conducted within the Poultry CRC, established and supported under the Australian Government’s Cooperative Research Centres Program.
References
Amend AS, Seifert KA, Bruns TD (2010) Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Molecular Ecology 19, 5555–5565.| Quantifying microbial communities with 454 pyrosequencing: does read abundance count?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlagsb0%3D&md5=d7587fefc4f59d2077510f353d001a8fCAS | 21050295PubMed |
Barth S, Duncker S, Hempe J, Breves G, Baljer G, Bauerfeind R (2009) Escherichia coli Nissle 1917 for probiotic use in piglets: evidence for intestinal colonization. Journal of Applied Microbiology 107, 1697–1710.
| Escherichia coli Nissle 1917 for probiotic use in piglets: evidence for intestinal colonization.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsVKhu7nE&md5=24c9756d7c9925755bf32c610c050275CAS | 19457029PubMed |
Bjerrum L, Engberg RM, Leser TD, Jensen BB, Finster K, Pedersen K (2006) Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and cellular-based techniques. Poultry Science 85, 1151–1164.
Bojesen AM, Nielsen SS, Bisgaard M (2003a) Prevalence and transmission of haemolytic Gallibacterium species in chicken production systems with different biosecurity levels. Avian Pathology 32, 503–510.
| Prevalence and transmission of haemolytic Gallibacterium species in chicken production systems with different biosecurity levels.Crossref | GoogleScholarGoogle Scholar | 14522706PubMed |
Bojesen AM, Torpdahl M, Christensen H, Olsen JE, Bisgaard M (2003b) Genetic diversity of Gallibacterium anatis isolates from different chicken flocks. Journal of Clinical Microbiology 41, 2737–2740.
| Genetic diversity of Gallibacterium anatis isolates from different chicken flocks.Crossref | GoogleScholarGoogle Scholar | 12791918PubMed |
Burkholder KM, Thompson KL, Einstein ME, Applegate TJ, Patterson JA (2008) Influence of stressors on normal intestinal microbiota, intestinal morphology, and susceptibility to Salmonella enteritidis colonization in broilers. Poultry Science 87, 1734–1741.
| Influence of stressors on normal intestinal microbiota, intestinal morphology, and susceptibility to Salmonella enteritidis colonization in broilers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFWqu7nE&md5=3f5c7e6fe40cbf60d56c6374e7dca69bCAS | 18753440PubMed |
Callaway TR, Dowd SE, Wolcott RD, Sun Y, McReynolds JL, Edrington TS, Byrd JA, Anderson RC, Krueger N, Nisbet DJ (2009) Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poultry Science 88, 298–302.
| Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1M%2FotFWrtA%3D%3D&md5=4cd409a8691519962f7182b88670782bCAS | 19151343PubMed |
Choct M, Hughes RJ, Bedford MR (1999) Effects of a xylanase on individual bird variation, starch digestion throughout the intestine, and ileal and caecal volatile fatty acid production in chickens fed wheat. British Poultry Science 40, 419–422.
| Effects of a xylanase on individual bird variation, starch digestion throughout the intestine, and ileal and caecal volatile fatty acid production in chickens fed wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXlvVaisLk%3D&md5=d5f89cabddb963c0de39807e40c67390CAS | 10475642PubMed |
Claesson MJ, O’Sullivan O, Wang Q, Nikkila J, Marchesi JR, Hauke S, De Vos WM, Ross RP, O’Toole PW (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4, e6669
| Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine.Crossref | GoogleScholarGoogle Scholar | 19693277PubMed |
Fraher MH, O’Toole PW, Quigley EMM (2012) Techniques used to characterize the gut microbiota: a guide for the clinician. Nature Reviews in Gastroenterology and Hepatology. 9, 312–322.
| Techniques used to characterize the gut microbiota: a guide for the clinician.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XotVOhsbY%3D&md5=2ae37a4c3558dd4636483100cb4feb20CAS |
Geier MS, Torok VA, Allison GE, Ophel-Keller K, Hughes RJ (2009) Indigestible carbohydrates alter the intestinal microbiota but do not influence the performance of broiler chickens. Journal of Applied Microbiology 106, 1540–1548.
| Indigestible carbohydrates alter the intestinal microbiota but do not influence the performance of broiler chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtVOisrg%3D&md5=5b506823098028ec9714315f2a5b3519CAS | 19187131PubMed |
Gordon HA, Pesti L (1971) The gnotobiotic animal as a tool in the study of host–microbial relationships. Bacteriological Reviews 35, 390–429.
Gunal M, Yayli G, Kaya O, Karahan N, Sulak O (2006) The effects of antibiotic growth promoter, probiotic or organic acid supplementation on performance, intestinal microflora and tissue of broilers. International Journal of Poultry Science 5, 149–155.
| The effects of antibiotic growth promoter, probiotic or organic acid supplementation on performance, intestinal microflora and tissue of broilers.Crossref | GoogleScholarGoogle Scholar |
Hammons S, Oh PL, Martinez I, Clark K, Schlegel VL, Sitorius E, Scheideler SE, Walter J (2010) A small variation in diet influences the Lactobacillus strain composition in the crop of broiler chickens. Systematic and Applied Microbiology 33, 275–281.
| A small variation in diet influences the Lactobacillus strain composition in the crop of broiler chickens.Crossref | GoogleScholarGoogle Scholar | 20554146PubMed |
Harrow S, Ravindran V, Butler R, Marshall J, Tannock G (2007) Real-time quantitative PCR measurement of ileal Lactobacillus salivarius populations from broiler chickens to determine the influence of farming practices. Applied and Environmental Microbiology 73, 7123–7127.
| Real-time quantitative PCR measurement of ileal Lactobacillus salivarius populations from broiler chickens to determine the influence of farming practices.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnsFOqtA%3D%3D&md5=43f997ba39c4f2b55f63748773034799CAS | 17890342PubMed |
Hubener K, Jahjen W, Simon O (2002) Bacterial responses to differrent dietary cereal types and xylanase supplementation in the intestine of broiler chicken. Archives of Animal Nutrition 56, 167–187.
Hume ME, Barbosa NA, Dowd SE, Sakomura NK, Nalian AG, Martynova-Van Kley A, Oviedo-Rondon EO (2011) Use of pyrosequencing and denaturing gradient gel electrophoresis to examine the effects of probiotics and essential oil blends on digestive microflora in broilers under mixed Eimeria infection. Foodborne Pathogens and Disease 8, 1159–1167.
| Use of pyrosequencing and denaturing gradient gel electrophoresis to examine the effects of probiotics and essential oil blends on digestive microflora in broilers under mixed Eimeria infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlKkt7jF&md5=be74c9b39292d43b1b8c1210f86b567cCAS | 21793655PubMed |
Knarreborg A, Engberg RM, Jensen SK, Jensen BB (2002) Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Applied and Environmental Microbiology 68, 6425–6428.
| Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xptlaru7g%3D&md5=7610d3dbe70c4d4c408323f63819d347CAS | 12450872PubMed |
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial communities of the maturing boiler chicken. Applied and Environmental Microbiology 69, 6816–6824.
| Diversity and succession of the intestinal bacterial communities of the maturing boiler chicken.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXptVyktbs%3D&md5=edc0ffc8175725ab2f8de8ca3c81c055CAS | 14602645PubMed |
Lumpkins BS, Batal AB, Lee M (2008) The effect of gender on the bacterial community in the gastrointestinal tract of broilers. Poultry Science 87, 964–967.
| The effect of gender on the bacterial community in the gastrointestinal tract of broilers.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1c3mtVWitA%3D%3D&md5=cb2550d07e33044829cc4e42ad7a8851CAS | 18420988PubMed |
Lumpkins BS, Batal AB, Lee MD (2010) Evaluation of the bacterial community and intestinal development of different genetic lines of chickens. Poultry Science 89, 1614–1621.
| Evaluation of the bacterial community and intestinal development of different genetic lines of chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtVOntr%2FN&md5=3b68945de1a9e65684489ec72a9d64daCAS | 20634515PubMed |
Majidzadeh Heravi R, Kermanshahi H, Sankian M, Nassiri MR, Heravi Moussavi A, Roozbeh Nasiraii L, Varasteh AR (2011) Screening of lactobacilli bacteria isolated from gastrointestinal tract of broiler chickens for their use as probiotic. African Journal of Microbiology Research 5, 1858–1868.
| Screening of lactobacilli bacteria isolated from gastrointestinal tract of broiler chickens for their use as probiotic.Crossref | GoogleScholarGoogle Scholar |
Pedroso AA, Menten FFM, Lambais MR, Racanicci AMC, Longo FA, Sorbara JOB (2006) Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poultry Science 85, 747–752.
Perez-Maldonado RA, Rodrigues HD (2009) Nutritional characteristics of sorghums from Queensland and New South Wales for chicken meat production. RIRDC Publication No. 09/170. Rural Industries Research and Development Corporation, Kingston, ACT.
Stanley D, Denman SE, Hughes RJ, Geier MS, Crowley TM, Chen H, Haring VR, Moore RJ (2012) Intestinal microbiota associated with differential feed conversion efficiency in chickens. Applied Microbiology and Biotechnology 96, 1361–1369.
| Intestinal microbiota associated with differential feed conversion efficiency in chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1ektL7P&md5=7922fe153cb425bb8500d4e3eb85ed82CAS | 22249719PubMed |
Teirlynck E, Gussem MD, Dewulf J, Haesebrouck F, Ducatelle R, Van Immerseel F (2011) Morphometric evaluation of ‘dysbacteriosis’ in broilers. Avian Pathology 40, 139–144.
| Morphometric evaluation of ‘dysbacteriosis’ in broilers.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MvktlWisA%3D%3D&md5=fb1cbef814211568c96428df01a7955cCAS | 21500033PubMed |
Torok VA, Ophel-Keller K, Loo M, Hughes RJ (2008) Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Applied and Environmental Microbiology 74, 783–791.
| Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhvVWjsr0%3D&md5=9ad23b085c83cf2927e07355256cfe35CAS | 18065621PubMed |
Torok VA, Hughes RJ, Ophel-Keller K, Ali M, MacAlpine R (2009) Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poultry Science 88, 2474–2481.
| Influence of different litter materials on cecal microbiota colonization in broiler chickens.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1Mjltlaktw%3D%3D&md5=2b95d796f8bf1eb2e3f749d17389e48cCAS | 19903943PubMed |
Torok VA, Allison GE, Percy NJ, Ophel-Keller K, Hughes RJ (2011a) Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance. Applied and Environmental Microbiology 77, 3380–3390.
| Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXns1SltL0%3D&md5=f8c425001776b8e1834224072df097e8CAS | 21441326PubMed |
Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K (2011b) Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Applied and Environmental Microbiology 77, 5868–5878.
| Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1OjurvO&md5=897de70efec2698e2aa15b171ed9b1f6CAS | 21742925PubMed |
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research 35, W71–W74.
| Primer3Plus, an enhanced web interface to Primer3.Crossref | GoogleScholarGoogle Scholar | 17485472PubMed |
Waidmann M, Bechtold O, Frick JS, Lehr HA, Schubert S, Dobrindt U, Loffler J, Bohn E, Autenrieth IB (2003) Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125, 162–177.
| Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice.Crossref | GoogleScholarGoogle Scholar | 12851881PubMed |
Yin Y, Lei F, Zhu L, Li S, Wu Z, Zhang R, Gao GF, Zhu B, Wang X (2010) Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. The ISME Journal 4, 367–376.
| Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitVWgtb8%3D&md5=b08c2d61c5fa5ae9d713f2d86453e13fCAS | 19956274PubMed |
Zhou Z, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, Xie J, van Nostrand JD, He Z, Yang Y (2011) Reproducibility and quantitation of amplicon sequencing-based detection. The ISME Journal 5, 1303–1313.
| Reproducibility and quantitation of amplicon sequencing-based detection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXptVynuro%3D&md5=49a13e355a819744f4954f16552c0ddfCAS |
Zhu XY, Zhong TZ, Pandya Y, Joerger RD (2002) 16S rRNA-based analysis of microbiota from the cecum of boiler chickens. Applied and Environmental Microbiology 68, 124–137.
| 16S rRNA-based analysis of microbiota from the cecum of boiler chickens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xjt1WgsQ%3D%3D&md5=de30cbe182ee3c160f9384e034b719f0CAS | 11772618PubMed |
Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ (2010) Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. Journal of Clinical Microbiology 48, 1812–1819.
| Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnt1Cnsrw%3D&md5=f1b1253e14737b3383d1c84ab75c63d0CAS | 20305015PubMed |




