Rumen bacteria and feed efficiency of beef cattle fed diets with different protein content
M. C. Parra A , D.F. Costa
A , D.F. Costa  A , S. J. Meale B and L. F. P. Silva
A , S. J. Meale B and L. F. P. Silva  A *
A *
A Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Gatton, Qld 4343, Australia.
B School of Agriculture and Food Science, The University of Queensland, Gatton, Qld 4343, Australia.
Animal Production Science - https://doi.org/10.1071/AN21508
Submitted: 1 October 2021 Accepted: 24 February 2022 Published online: 13 April 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: Beef cattle feed efficiency is challenged in northern Australian production systems due to the limited dietary protein, leading to changes in rumen bacterial populations and fermentation outcomes.
Aims: Two types of diets with different dietary protein contents were used to evaluate changes in rumen bacterial composition and diversity, aiming to correlate rumen bacterial populations with feed and rumen efficiency parameters.
Methods: In total, 90 Brahman steers (341 ± 45 kg BW) were selected for this trial, but rumen fluid was collected from 85 Brahman steers, at 0 and 4 h after feeding, during a feed-efficiency trial. The steers were fed with a low-protein diet, including 70% rumen-degradable protein and 8.8% crude protein (CP) for 60 days, followed by a high-protein diet for the same period (13.5% CP). Liveweight and dry-matter intake measurements, as well as urine, faeces and rumen fluid samples, were collected to determine feed and rumen efficiency, and ruminal bacteria composition. Steers were clustered into groups using principal component analysis and Ward’s hierarchical method, and differences in feed-efficiency parameters among clusters were compared.
Key results: Rumen bacterial composition differed between diets (P < 0.01) and diversity changes were more related to bacterial richness (P < 0.01). In a low-protein diet, there were four distinct clusters of steers, on the basis of rumen bacteria, in which the most efficient steers, with a better residual feed intake (P = 0.06) and lower rumen ammonia concentration (P < 0.01) before feeding, had the highest relative abundance of Prevotella (P < 0.01). While in a high-protein diet, no differences were observed on feed or rumen fermentation parameters among steer clusters.
Conclusion: In a low-protein diet, rumen bacterial shifting might contribute to upregulate nitrogen recycling, favouring feed efficiency.
Implications: Identifying ruminal bacterial populations involved in nitrogen recycling upregulation might be useful to select the most efficient cattle fed low-protein diets.
Keywords: Bos indicus, feed efficiency, low protein diet, nitrogen recycling, Prevotella, rumen ammonia, rumen bacteria composition, rumen maturation, rumen microbiome.
Introduction
Maintaining a balanced energy:protein ratio in the diet is crucial for maximising rumen fermentation efficiency, supply of microbial protein, and growth efficiency of cattle (Poppi and McLennan 1995). Nevertheless, in the dry-tropic beef cattle-production systems dietary protein content is a limiting nutritional factor, challenging the rumen and feed efficiency and restraining growth rates of high genotypic-value animals. Ruminants have an extraordinary capacity to subsist consuming low-protein diets by altering host and rumen microbiology features. However, low dietary protein content tends to shift rumen bacterial populations, favouring the growth of non-ammonia-dependant bacteria (Marini and Van Amburgh 2003; Belanche et al. 2012). This microbial shift is accompanied by the upregulation of nitrogen (N) recycling from the liver into the rumen, with the main purpose of providing enough N for microbial growth with limited ruminal ammonia (NH3-N) concentration (Reynolds and Kristensen 2008). These rumen bacterial changes, in addition to modifications of rumen cell-wall characteristics and urea transporters, facilitate N flow into the rumen. The hypothesis was that rumen bacteria modulated feed efficiency by sustaining microbial growth and favouring N recycling.
Studies have shown a correlation between N recycling and feed efficiency. For instance, Carmona et al. (2020) demonstrated that in a low-protein diet, efficient beef cattle, in terms of residual gain (RG), had lower N excretion, meaning greater N preservation and N utilisation efficiency. The association of ruminal bacterial populations and activities with cattle feed efficiency also has been identified by Shabat et al. (2016). Overall, depending on bacterial composition, diversity, activities and relevant metabolite outputs, the ability to meet energy and amino acid requirements of the host might be positively or negatively influenced, altering feed efficiency. However, there is limited knowledge about the role of rumen bacteria in relation to feed efficiency of beef cattle fed protein-limiting diets. Thus, the objectives of the current study were to evaluate the role of rumen microbiota in modulating feed efficiency in tropically adapted cattle receiving two diets with different protein contents.
Material and methods
All experimental procedures were performed at the Queensland Animal Science Precinct (Gatton, Queensland, Australia) and approved by the University of Queensland Animal Ethics Committee.
Animals and experimental design
In total, 90 Brahman steers [341 ± 45 kg initial body weight (iBW); 19.2 ± 3.4 months] were selected on the basis of their genotypic value, using Illumina BovineSNP50 BeadChip (Neogen, Gatton) to increase genomic parentage. Due to limitations of only 30 individual pens, steers were distributed into three blocks (30 steers each), with uniformity on their liveweight (LW) within blocks. The experimental period for the first block occurred from late-May to early October 2018, second block between mid-January to late June 2019 and last block from late September 2019 to mid-February 2020. Differences in LW were expected as growth stage and genomic variability were different.
Steers were housed in individual pens and adapted over 10 days to a low-protein diet (LP) fed ad libitum for 60 days, followed by the same duration on a high-protein diet (HP). At the end of each subperiod, steers were transferred in groups of 10 animals into metabolism crates for 7 days, using the first 2 days for adaptation and 5 days for collections. Subperiods lasted 77 days, with a total experimental period duration of 154 days. Feed offered was adjusted daily with previous day feed intake, targeting 5% refusals to minimise forage selection. Due to the measurement of parameters related to N recycling mechanism, it was not possible to evaluate both diets at same time period. In this sense, authors are aware about the influence of steer age in the response of rumen bacteria to these diets and their potential capacity to modulate feed efficiency.
The experimental diet consisted of Rhodes grass hay (Chloris gayana) chopped to approximately 5.5 cm, and a 3-mm pelleted concentrate, formulated to provide 70% of rumen-degradable protein (RDP) requirements in the LP [8.8% crude protein (CP)] diet and 100% with HP diet (13.5% CP). Diet composition was formulated following feeding standard NRC (2000) and presented in Carmona et al. (2020). Offered hay and concentrate samples, in addition to individual daily refusal consisting of a mixture of hay and concentrate, were collected, weighed and bulked weekly. These bulked samples were dried in a forced-air oven at 60°C for 72 h for initial dry-matter (DM) estimation.
Feed-efficiency parameters
The average daily gain (ADG) was determined using a linear regression of LW over time. During each experimental period, LW of unfasted steers was measured on two consecutive days at the beginning of experiment, a procedure repeated every 30 days, with additional single measurements in 1 day only conducted every fortnight. Feed conversion ratio (FCR) was calculated as DM intake (DMI) per unit of ADG, while gain to feed ratio (GF) was estimated on the inverse relationship. Expected ADG, as well as expected DMI, were obtained from linear regression of DMI and ADG over LW. Residual gain (RG) was estimated using expected and actual ADG (Crowley et al. 2010), while residual feed intake (RFI) was determined by calculating the difference between actual and expected DMI (Archer et al. 1997). The digestible organic matter (OM) intake was calculated using the digestibility value obtained in the metabolism crates, using total faecal output and intake for that period. The calculated digestibility was then extrapolated for the whole feeding period.
Rumen fluid, faeces and urine collection
At end of each subperiod within experimental-block periods, rumen fluid was collected via oesophageal tubing at 0 and 4 h after feeding on the same day. Rumen fluid was filtered through four layers of cheesecloth, and pH was measured immediately (Edge Benchtop HI2002, Hanna Instruments, Melbourne, Vic., Australia). Initial rumen fluid was not discarded unless saliva was visually observed. In addition, pH measurements were assessed to monitor whether samples were within optimum levels, ruling out saliva contamination. Subsamples were transferred into tubes for ammonia (NH3-N) estimation (6 mL of rumen fluid + 2 mL 0.5 M H2SO4) and stored at −20°C. Further subsamples (1 mL) were immediately flash-frozen in liquid N and stored at −80°C for DNA extraction. Volatile fatty acids (VFA) were not measured in the current study as the differences on CP between diets were not expected to generate fluctuations in the synthesis of individual VFA and their proportion total VFA.
Within metabolism crates, after 2 days for adaptation, samples of daily faeces were collected from each crate, weighed, and stored (10% of total output) at 4°C until the end of the collection period, resulting in five storage days for the samples taken the first day, 4 days for the samples taken the second day, and so on. At the end of the 5 days, a representative subsample for each steer was collected from the mixture of daily samples and dried in a forced-air oven at 60°C for 72 h and stored to estimate N and OM content. Daily samples of urine were collected and mixed with 5% of H2SO4 to maintain the pH level under 4 and inhibit microbial growth. The amount of sulfuric acid inclusion was corrected daily, based on individual urine weights from the previous day. Subsamples were collected, representing 10% of total urine output per steer, and stored at 4°C until the last sampling collection day. These subsamples were mixed, and representative samples were collected to estimate total N, NH3-N and purine derivatives (PD), using the latest for calculation of microbial CP production (MCP).
Laboratory analyses
Dry-matter content of feed refusals and faeces samples were determined at 105°C (AOAC 2005, Method 934.01) and OM content was calculated discounting the ash content determined at 550°C for 8 h (AOAC 2005, Method 942.05). Ash-free NDF content was estimated following the procedure described in Mertens (2002). Nitrogen content was measured following Dumas combustion by the method described in Sweeney (1989), using LECO CN928 carbon/nitrogen combustion analyser (LECO Corporation; St Joseph, MI, USA). Crude protein was calculated by multiplying N content by 6.25.
Rumen ammonia concentration was estimated using the distillation and titration method (Buchi 321 distillation unit, Flawil, St Gallen, Switzerland) described in Preston (1995). Purine derivative concentration in urine was estimated following the methods of Czauderna and Kowalczyk (1997) and George et al. (2006), using high-performance liquid chromatography with Prodigy 250 × 46 mm, 5 μm, ODS C18 reverse-phase column (Phenomenex, Torrence, CA, USA). Microbial protein production (MCP) was estimated with the formula of Chen and Gomes (1992), with the value for excretion of endogenous PD for Bos indicus cattle from Bowen et al. (2006). The efficiency of MCP synthesis (EMPS) was calculated as g of MCP/kg digestible OMI (DOMI). Retained N was calculated subtracting total N excretion in urine and faeces from total N intake. Nitrogen use efficiency (NUE) was determined by dividing retained N (g/day) over apparently digested N (g/day).
The genomic DNA in rumen samples was extracted using bead-beating, followed by a column purification procedure (Popova et al. 2010). Genomic DNA was amplified, sequenced and analysed following the procedure described by Popova et al. (2010), using V3-4 region of bacterial 16S rRNA gene and 515f (5ʹ-GTGCCAGCMGCCGCGGTAA-3ʹ) and 806r (5ʹ-GACTACHVGGGTWTCTAAT-3ʹ) primers. Chimeras were removed and the remaining sequences were assigned taxonomy at 97% similarity using Green Genes database (version 13.8; DeSantis et al. 2006).
Statistical analyses
From the cohort of 90 steers, five steers were removed from the study due to aggressive behaviour (two steers) and illness, leading to very low intake (three steers). Therefore, 85 Brahman steers were considered for statistical analysis. Data were analysed as a completely randomised block design, using the MIXED procedure of SAS, (SAS Institute Inc. 2019, version 9.4). Experimental period was treated as a random factor within the model.
Differences in ruminal microbial abundance between high- and low-protein diets were determined using DESeq2 package in R (Love et al. 2014). Alpha diversity indexes were analysed with Phyloseq package (version 1.26.0) on R software, using the number of operational taxonomy units (OTUs) per sample (McMurdie and Holmes 2013). Bray–Curtis dissimilarities were estimated with vegan package 2.5-3 (Oksanen et al. 2015) in R. Differences between low- and high-protein diets were calculated with permutational multivariate analysis of variance (PERMANOVA).
Ruminal microbial populations were associated with feed-efficiency parameters by using two alternative analyses. The abundance of individual bacterial genera was correlated with these parameters by using PROC CORR procedure on SAS software. Further, principal-component analysis (PROC PRINCOMP) was performed for each diet, considering bacterial genera population to identify patterns between steers. Steers were grouped on the basis of the four principal components, using PROC CLUSTER, explaining approximately 62% and 53.5% (R-square) of total bacterial variance composition across steers, for LP and HP respectively. Ward’s hierarchical method was used for clustering analysis, and the MIXED procedure to calculate differences in feed-efficiency parameters among steer clusters based on ruminal bacteria populations.
Cluster means were compared using the LSMEANS option of the MIXED procedure, considering experimental period as a random effect, and cluster as a fixed effect. Differences were considered significant at P ≤ 0.05 and tendencies were declared when P ≤ 0.10. Shapiro–Wilk test was performed to estimate normality of residuals, and homogeneity of variances was calculated using the Levene test.
Results
Bray–Curtis dissimilarity index demonstrated that dietary protein content caused differentiation in OTU distribution (P < 0.001; Fig. 1a). Similarly, ruminal microbial diversity changed between diets. Shannon index demonstrated that the LP diet led to greater diversity, as indicated by bacterial richness (P < 0.01; Fig. 1b), and not evenness (data not shown), than did the HP diet across both sampling times. Within the most abundant bacterial communities, Bacteroidetes, Firmicutes, Euryarchaeota (P < 0.01), Proteobacteria (P = 0.01), Spirochaeta, Cyanobacteria and Actinobacteria (P < 0.01) were greater in LP steers (Fig. 1c), while Verrucomicrobia, SR1, TM7, LD1 and Lentisphaerae (P < 0.01) presented a higher abundance in the HP diet.
To further understand the effect of diet, ruminal bacterial genera were correlated with feed-efficiency parameters independently for each diet. In the LP diet, Succiniclasticum, Streptococcus and Trepomena were correlated with RFI. In addition, Trepomena, Anaeroplasma and Streptococcus presented a correlation with NUE (Fig. 2). In the HP diet, Succinivibrio, Megasphaera and two unclassified genera belonging to the Victivallaceae and Lachnospiraceae families were correlated with feed-efficiency parameters (Fig. 3). Interestingly, there were multiple bacterial genera related to feed efficiency that exhibited shifting correlations relative to the time of sampling.
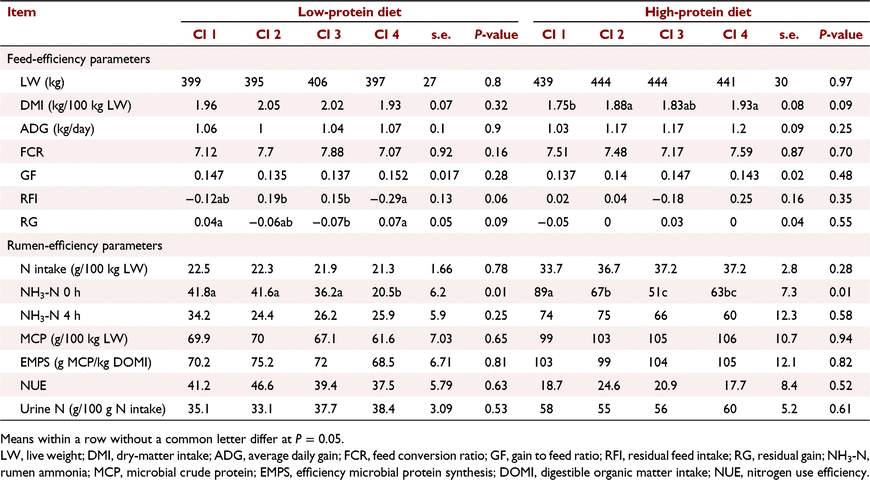
To estimate the correlation between rumen bacteria and feed efficiency, steers were classified into four clusters depending on ruminal bacterial profiles and diet, explaining 62% and 53.5% of the total variation in LP and HP diets respectively (R-square). With a LP diet, steers were clustered on the basis of the abundance of Prevotella, Ruminobacter and unclassified genera belonging to Succinivibrionacea and Bacteroidales families. For instance, steers in Cluster 3 had a high abundance of Ruminobacter at 4 h after feeding sampling and were the least feed-efficient steers. While steers in Cluster 4 were the most feed-efficient steers, with the greatest abundance of Prevotella in both sampling times (Fig. 4). Interestingly, Cluster 4 steers had lower ammonia concentrations at 0 h after feeding than did Cluster 3 steers (20.5 vs 36.2; P < 0.01). Tendencies for high RFI (−0.29 vs 0.15; P = 0.06) and RG (0.07 vs −0.07; P = 0.09) were observed in Cluster 4 steers (Table 1).
|
|
|
In contrast, no differences were observed in feed-efficiency parameters among steers clusters fed an HP diet (Table 1). Cluster 1 had a high abundance of unclassified Bacteroidales at 4 h after feeding samples and a low abundance of Prevotella in both sampling times (Fig. 5). For Cluster 2, Prevotella was dominant at both sampling times, while Cluster 3 steers had a greater population of Fibrobacter at 0 h after feeding, unclassified Succinivibrionaceae at 4 h after feeding, and Succinivibrio at both sampling times. Cluster 4 presented high Ruminobacter and low Prevotella populations, at both sampling times (data not shown). It was observed that Cluster 1 showed the greatest NH3-N concentration compared with the other clusters, especially Cluster 3, which had the lowest concentration (80 vs 51 mg/L; P < 0.01).
|
|
Discussion
Our initial hypothesis was that rumen bacteria can modulate cattle feed efficiency. The present study demonstrated that this modulation occurred when the cattle were fed a LP diet, but not when fed an HP diet. This might be related to the vast difference in the ruminal bacterial composition between the diets and the greater bacterial diversity, related only to bacterial richness and not evenness. Although changes in bacterial composition were expected, the high bacterial diversity in the LP diet confirmed a great variability of bacterial communities among the individual steers (Fig. 1b), suggesting alternative mechanisms to sustain microbial growth and host adaptation to the protein-limiting diet.
The existence of alternative adaptation mechanisms to LP diets is supported by observing the differences in bacterial populations between the diets. For instance, Firmicutes and Bacteroidetes, known as the most dominant and diverse phyla in the rumen, had a great abundance in the LP diet (P < 0.05). The low rumen ammonia concentration in LP diets generated a shift on bacterial populations, including specific cellulolytic species, allowing non-dependant NH3-N bacterial communities to become more competitive and dominant (Hristov et al. 2004). Moreover, there are certain bacterial communities, including Prevotella, which have a low ammonia saturation constant, allowing to maintain growth even in LP diets (Belanche et al. 2012). In the present study, the greater bacterial diversity among steers when in the LP diet suggests alternative ruminal bacterial communities shifting among steers, leading to diverse methods of adaptation to the protein-limiting environment.
This variation in bacterial communities among steers and between sampling times can also be illustrated with the correlation results between individual bacterial genera and feed-efficiency parameters. In the current study, diet and sampling collection times modified the intensity and direction of correlations. A clear example is Fibrobacter, which showed a positive correlation with FCR and no correlation with RFI when sampled before feeding, but no correlation with FCR and negative correlation with RFI when sampled 4 h after feeding in the LP diet. Similarly, Treponema presented a negative correlation with RFI at 4 h after feeding in the LP diet and no correlation with RFI in the HP diet. Variation of rumen conditions (i.e. temperature, pH, redox potential and oxygen) generates differences in rumen bacterial growth, diversity, activities, distribution among the rumen and interaction among communities, explaining the differences in bacterial abundances and possibly the association with feed-efficiency parameters among sample collection times (Li et al. 2009; de Assis Lage et al. 2020). Therefore, the correlation between rumen microbiota and feed-efficiency parameters cannot be explained by analysing bacterial abundance independently. In this sense, a more complete analysis of the bacteria population profile must be applied.
To have a better picture of how changes in the rumen bacterial population might be influencing feed efficiency, steers were clustered on the basis of the overall rumen bacterial similarity. The Prevotella abundance, before and after feeding, was an important factor separating the cluster of steers in the LP diet. The high Prevotella abundance in more efficient steers is opposite to the results of Carberry et al. (2012) but similar to those of Myer et al. (2015). The Prevotella genus is characterised by the capacity to degrade multiple substrates (i.e. peptide, starch, hemicellulose and pectin), which can be interpreted as less energy efficient because of the high energy demand in the multiple metabolite synthesis and a lower contribution to meeting host energy requirements (Shabat et al. 2016). However, these multiple fermentation pathways allow other non-cellulolytic bacterial communities to maintain microbial growth, possibly contributing to feed efficiency. Nonetheless, the correlation of Prevotella with feed efficiency depends on the diet and Prevotella species (Lopes et al. 2021).
It is likely that in the current study, in the LP diet, Prevotella would be scavenging N from different substrates (peptides and urea), allowing this genus to become more competitive and abundant within the rumen of more efficient steers (Cluster 4) at both sampling times. Greater Prevotella abundance is controversial in efficient steers as it is involved in multiple roles in rumen fermentation, which might cause more energy used for metabolic pathways instead for host utilisation. However, a recent study demonstrated that this genus has an important role in the synthesis of amino acids, and carbohydrate metabolites, which influence animal metabolism (Xue et al. 2020). For further studies, it is recommended to identify metabolites linked with Prevotella and its effects on protein metabolism and microbiota growth, especially in a LP diet.
Apart from a greater Prevotella abundance at both sampling times, the cluster with the more efficient steers in a LP diet also had a lower rumen NH3-N concentration 0 h before feeding. Evidence suggests that rumen ammonia concentration influences the transfer of blood urea into the rumen (Lapierre and Lobley 2001). Therefore, in a LP diet, bacterial growth is more reliant on alternative N sources and upregulation of the N recycling mechanism, rather than ammonia from dietary CP (Li et al. 2017). For instance, when beef cattle are fed LP diets, the amount of N recycled into the gastrointestinal tract can be doubled, from 43% to 85% of N intake (Huntington 1989; Silva et al. 2019).
The upregulation in N recycling is accompanied by a shift in ruminal microbial population, promoting ureolytic bacteria species (Lapierre and Lobley 2001). These ureolytic bacteria species promote urea-N transport through the rumen by hydrolysing urea, which maintains rumen wall gradient for urea diffusion, or by releasing toxin-like compounds that can be considered as urea transporters and promote rumen urea permeability (Lapierre and Lobley 2001; Kristensen et al. 2010). Nevertheless, there are other factors that modulate N recycling, including rumen pH, VFA and fermentable carbohydrate concentrations (Silva et al. 2019). In this sense, low dietary CP content challenges bacteria populations to obtain N from different sources, shifting bacterial compositions and resulting in a greater transference of urea into the rumen. However, N recycling efficiency also depends on the host and changes in rumen-wall characteristics such as ruminal epithelial urea permeability and urea transporters (Marini and Van Amburgh 2003; Kristensen et al. 2010).
Feed efficiency among steer clusters was not affected by the HP diet. Given that the high-protein diet was formulated to meet the requirements for rumen-degradable protein, differences in feed efficiency could be explained by phenotypic expression, rather than bacterial modulation and upregulation of N-recycling in the rumen (Carmona et al. 2020). On the contrary, rumen bacterial communities stabilise with age and maturation of the host, which may have limited feed efficiency variability among clusters fed the HP diet (Liu et al. 2017; Costa-Roura et al. 2020). However, if steer age were an explanation for the absence of an interaction between microbial profiles and feed-efficiency parameters, this effect would also be observed in the LP diet, as samples were collected at the end of each diet period. Moreover, Costa-Roura et al. (2020) explained that over time, rumen bacterial diversity and abundance fluctuate in steers fed a LP diet initially, but feeding time, microbial adaptation and rumen maturation may decrease or eliminate these fluctuations entirely. In this sense, diet and animal age are likely contributors to the outcomes observed in steers fed the HP diet. However, LP diets challenged ruminal bacterial profiles, as indicated by species richness, likely modifying mechanisms that guarantee microbial growth and host adaptation, likely improving feed efficiency.
Conclusion
In conclusion, the results corroborate the hypothesis that rumen bacteria can modulate feed efficiency, but only in protein-limiting diets. Therefore, the identification of ruminal bacterial populations involved in N recycling upregulation might be useful to select for more efficient cattle.
Data availability
Data are available from the authors by request.
Conflicts of interest
Luis Prada e Silva is an Associate Editor of Animal Production Science but was blinded from the peer-review process for this paper.
Declaration of funding
This trial was funded by Meat and Livestock Australia (MLA). The funders did not participate in the elaboration of the manuscript and further submission for publication.
Acknowledgements
We acknowledge the contribution of A. Connolly, A. S. V. Palma, J. Reid, L. O. Lima, M. Reis, S. Kennedy, P. Carmona and T. Breed for the tremendous help in the field and P. Isherwood and K. Raymond for the important guidance with laboratorial analyses throughout this study.
References
AOAC (2005) ‘Official methods of analysis.’ 18th edn. (AOAC International: Gaithersburg, MD, USA)Archer JA, Arthur PF, Herd RM, Parnell PF, Pitchford WS (1997) Optimum postweaning test for measurement of growth rate, feed intake, and feed efficiency in British breed cattle. Journal of Animal Science 75, 2024–2032.
| Optimum postweaning test for measurement of growth rate, feed intake, and feed efficiency in British breed cattle.Crossref | GoogleScholarGoogle Scholar | 9263047PubMed |
Belanche A, Doreau M, Edwards JE, Moorby JM, Pinloche E, Newbold CJ (2012) Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. The Journal of Nutrition 142, 1684–1692.
| Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation.Crossref | GoogleScholarGoogle Scholar | 22833657PubMed |
Bowen MK, Poppi DP, McLennan SR, Doogan VJ (2006) A comparison of the excretion rate of endogenous purine derivatives in the urine of Bos indicus and Bos taurus steers. Australian Journal of Agricultural Research 57, 173–177.
| A comparison of the excretion rate of endogenous purine derivatives in the urine of Bos indicus and Bos taurus steers.Crossref | GoogleScholarGoogle Scholar |
Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM (2012) Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Applied and Environmental Microbiology 78, 4949–4958.
| Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle.Crossref | GoogleScholarGoogle Scholar | 22562991PubMed |
Carmona P, Costa DFA, Silva LFP (2020) Feed efficiency and nitrogen use rankings of Bos indicus steers differ on low and high protein diets. Animal Feed Science and Technology 263, 114493
| Feed efficiency and nitrogen use rankings of Bos indicus steers differ on low and high protein diets.Crossref | GoogleScholarGoogle Scholar |
Chen XB, Gomes MJ (1992) ‘Estimation of microbial protein supply to sheep and cattle based on urinary excretion of purine derivatives: an overview of technical details.’ (International Feed Resources Unit, Rowett Research Institute: Aberdeen, UK)
Costa-Roura S, Balcells J, de la Fuente G, Mora-Gil J, Llanes N, Villalba D (2020) Effects of protein restriction on performance, ruminal fermentation and microbial community in Holstein bulls fed high-concentrate diets. Animal Feed Science and Technology 264, 114479
| Effects of protein restriction on performance, ruminal fermentation and microbial community in Holstein bulls fed high-concentrate diets.Crossref | GoogleScholarGoogle Scholar |
Crowley JJ, McGee M, Kenny DA, Crews DH, Evans RD, Berry DP (2010) Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance-tested beef bulls. Journal of Animal Science 88, 885–894.
| Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance-tested beef bulls.Crossref | GoogleScholarGoogle Scholar | 19966161PubMed |
Czauderna M, Kowalczyk J (1997) Simultaneous measurement of allantoin, uric acid, xanthine and hypoxanthine in blood by high-performance liquid chromatography. Journal of Chromatography B: Biomedical Sciences and Applications 704, 89–98.
| Simultaneous measurement of allantoin, uric acid, xanthine and hypoxanthine in blood by high-performance liquid chromatography.Crossref | GoogleScholarGoogle Scholar | 9518182PubMed |
de Assis Lage CF, Räisänen SE, Melgar A, Nedelkov K, Chen X, Oh J, Fetter ME, Indugu N, Bender JS, Vecchiarelli B, Hennessy ML, Pitta D, Hristov AN (2020) Comparison of two sampling techniques for evaluating ruminal fermentation and microbiota in the planktonic phase of rumen digesta in dairy cows. Frontiers in Microbiology 11, 1447
| Comparison of two sampling techniques for evaluating ruminal fermentation and microbiota in the planktonic phase of rumen digesta in dairy cows.Crossref | GoogleScholarGoogle Scholar |
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, achimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology 72, 5069–2.
| Greengenes, achimera-checked 16S rRNA gene database and workbench compatible with ARB.Crossref | GoogleScholarGoogle Scholar | 16820507PubMed |
George SK, Dipu MT, Mehra UR, Singh P, Verma AK, Ramgaokar JS (2006) Improved HPLC method for the simultaneous determination of allantoin, uric acid and creatinine in cattle urine. Journal of Chromatography B 832, 134–137.
| Improved HPLC method for the simultaneous determination of allantoin, uric acid and creatinine in cattle urine.Crossref | GoogleScholarGoogle Scholar |
Hristov AN, Etter RP, Ropp JK, Grandeen KL (2004) Effect of dietary crude protein level and degradability on ruminal fermentation and nitrogen utilization in lactating dairy cows. Journal of Animal Science 82, 3219–3229.
| Effect of dietary crude protein level and degradability on ruminal fermentation and nitrogen utilization in lactating dairy cows.Crossref | GoogleScholarGoogle Scholar | 15542468PubMed |
Huntington GB (1989) Hepatic urea synthesis and site and rate of urea removal from blood of beef steers fed alfalfa hay or a high concentrate diet. Canadian Journal of Animal Science 69, 215–223.
| Hepatic urea synthesis and site and rate of urea removal from blood of beef steers fed alfalfa hay or a high concentrate diet.Crossref | GoogleScholarGoogle Scholar |
Kristensen NB, Storm AC, Larsen M (2010) Effect of dietary nitrogen content and intravenous urea infusion on ruminal and portal-drained visceral extraction of arterial urea in lactating Holstein cows. Journal of Dairy Science 93, 2670–2683.
| Effect of dietary nitrogen content and intravenous urea infusion on ruminal and portal-drained visceral extraction of arterial urea in lactating Holstein cows.Crossref | GoogleScholarGoogle Scholar | 20494176PubMed |
Lapierre H, Lobley GE (2001) Nitrogen recycling in the ruminant: a review. Journal of Dairy Science 84, E223–E236.
| Nitrogen recycling in the ruminant: a review.Crossref | GoogleScholarGoogle Scholar |
Li M, Penner GB, Hernandez-Sanabria E, Oba M, Guan LL (2009) Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. Journal of Applied Microbiology 107, 1924–1934.
| Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen.Crossref | GoogleScholarGoogle Scholar | 19508296PubMed |
Li F, Guan LL, McBain AJ (2017) Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Applied and Environmental Microbiology 83, e00061-17
| Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle.Crossref | GoogleScholarGoogle Scholar | 28235871PubMed |
Liu C, Meng Q, Chen Y, Xu M, Shen M, Gao R, Gan S (2017) Role of age-related shifts in rumen bacteria and methanogens in methane production in cattle. Frontiers in Microbiology 8, 1563
| Role of age-related shifts in rumen bacteria and methanogens in methane production in cattle.Crossref | GoogleScholarGoogle Scholar | 28855896PubMed |
Lopes DRG, de Souza Duarte M, La Reau AJ, Chaves IZ, de Oliveira Mendes TA, Detmann E, Bento CBP, Mercadante MEZ, Bonilha SFM, Suen G, Mantovani HC (2021) Assessing the relationship between the rumen microbiota and feed efficiency in Nellore steers. Journal of Animal Science and Biotechnology 12, 2853
| Assessing the relationship between the rumen microbiota and feed efficiency in Nellore steers.Crossref | GoogleScholarGoogle Scholar |
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550
| Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.Crossref | GoogleScholarGoogle Scholar | 25516281PubMed |
Marini JC, Van Amburgh ME (2003) Nitrogen metabolism and recycling in Holstein heifers. Journal of Animal Science 81, 545–552.
| Nitrogen metabolism and recycling in Holstein heifers.Crossref | GoogleScholarGoogle Scholar | 12643500PubMed |
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217
| Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data.Crossref | GoogleScholarGoogle Scholar | 23630581PubMed |
Mertens D (2002) Gravimetric determination of amylase-treated neutral detergent fiber in feeds with refluxing in beakers or crucibles: collaborative study. Journal of AOAC International 85, 1217–1240.
Myer PR, Smith TPL, Wells JE, Kuehn LA, Freetly HC (2015) Rumen microbiome from steers differing in feed efficiency. PLoS ONE 10, e0129174
| Rumen microbiome from steers differing in feed efficiency.Crossref | GoogleScholarGoogle Scholar | 26030887PubMed |
NRC (2000) ‘Nutrient requirements of beef cattle.’ 7th rev. edn. (National Academies Press: Washington, DC, USA)
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara B, Simpson G, Solymos P, Stevens H, Wagner H (2015) Vegan: community ecology package. PR Package Version 2.2-1 2, 1–2. Available at https://CRAN.R-project.org/package=vergan
Popova M, Martin C, Morgavi DP (2010) Improved protocol for high-quality co-extraction of DNA and RNA from rumen digesta. Folia Microbiologica 55, 368–372.
| Improved protocol for high-quality co-extraction of DNA and RNA from rumen digesta.Crossref | GoogleScholarGoogle Scholar | 20680573PubMed |
Poppi DP, McLennan SR (1995) Protein and energy utilization by ruminants at pasture. Journal of Animal Science 73, 278–290.
| Protein and energy utilization by ruminants at pasture.Crossref | GoogleScholarGoogle Scholar | 7601744PubMed |
Preston TR (1995) Biological and chemical analytical methods. In ‘Tropical animal feeding: a manual for research workers’. (Ed. TR Preston) pp. 191–264. (FAO: Rome, Italy)
Reynolds CK, Kristensen NB (2008) Nitrogen recycling through the gut and the nitrogen economy of ruminants: an asynchronous symbiosis. Journal of Animal Science 86, E293–E305.
| Nitrogen recycling through the gut and the nitrogen economy of ruminants: an asynchronous symbiosis.Crossref | GoogleScholarGoogle Scholar | 17940161PubMed |
Shabat SKB, Sasson G, Doron-Faigenboim A, Durman T, Yaacoby S, Berg Miller ME, White BA, Shterzer N, Mizrahi I (2016) Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. The ISME Journal 10, 2958–2972.
| Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants.Crossref | GoogleScholarGoogle Scholar |
Silva LFP, Dixon RM, Costa DFA (2019) Nitrogen recycling and feed efficiency of cattle fed protein-restricted diets. Animal Production Science 59, 2093–2107.
| Nitrogen recycling and feed efficiency of cattle fed protein-restricted diets.Crossref | GoogleScholarGoogle Scholar |
Sweeney RA (1989) Generic combustion method for determination of crude protein in feeds: collaborative study. Journal of AOAC International 72, 770–774.
| Generic combustion method for determination of crude protein in feeds: collaborative study.Crossref | GoogleScholarGoogle Scholar |
Xue M-Y, Sun H-Z, Wu X-H, Liu J-X, Guan LL (2020) Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8, 64
| Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance.Crossref | GoogleScholarGoogle Scholar | 32398126PubMed |


