The Use of Imaging Mass Spectrometry to Study Peptide Toxin Distribution in Australian Sea Anemones*
Michela L. Mitchell A G , Brett R. Hamilton B C , Bruno Madio D , Rodrigo A. V. Morales A , Gerry Q. Tonkin-Hill E , Anthony T. Papenfuss E , Anthony W. Purcell F , Glenn F. King D , Eivind A. B. Undheim B and Raymond S. Norton AA Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Vic. 3052, Australia.
B Centre for Advanced Imaging, University of Queensland, St Lucia, Qld 4072, Australia.
C Centre for Microscopy and Microanalysis, University of Queensland, St Lucia, Qld 4072, Australia.
D Institute for Molecular Bioscience, University of Queensland, St Lucia, Qld 4072, Australia.
E Bioinformatics Division, Walter & Eliza Hall Institute of Research, Parkville, Vic. 3052, Australia.
F Department of Biochemistry and Molecular Biology and Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Clayton, Vic. 3800, Australia.
G Corresponding author. Email: michela.mitchell@monash.edu

Michela Mitchell holds a Bachelor of Applied Science - Coastal Management (Hons) from the University of New England, Northern Rivers, and a Masters by Research from Southern Cross University (2010), specialising in sea anemone taxonomy. She is currently undertaking a doctorate in medicinal chemistry at Monash University (Monash Institute Pharmaceutical Sciences) with Professor Ray Norton, investigating peptide diversity in sea anemone tissues. |
Australian Journal of Chemistry 70(11) 1235-1237 https://doi.org/10.1071/CH17228
Submitted: 26 April 2017 Accepted: 4 August 2017 Published: 28 August 2017
Cnidarians (e.g. jellyfish, coral, and sea anemones) are one of the oldest known venomous lineages, and have a unique envenomation system to deliver venoms. Their toxins are encapsulated in microscopic organelles (cnidae) that are embedded throughout the ectodermal tissue of the animal. Each type of cnidae performs a specific biological function, ranging from adherence to delivery of toxin for prey capture. Discrete morphological regions of sea anemones contain specific complements of cnidae to deliver toxins for distinct biological functions, e.g. cnidae in tentacles are used for prey capture.[1]
There is a dearth of knowledge regarding the diversity of sea anemone toxins and their distribution in relation to specific morphological regions of the animals. Peptide toxins (Mr <10 kDa) contained in venoms are of particular interest as a source of drug leads because of their typically high potency and target selectivity.[2] One such peptide, ShK, from the sea anemone Stichodactyla helianthus, forms the basis of the first-in-class drug Dalazatide, currently in clinical trials to treat psoriasis.[3] To date, Australian sea anemones have been largely overlooked in the search for novel peptide toxins, and excluded from toxin evolution studies within a global context.
To mine toxins from venomous animals for the purpose of drug discovery, a venomics strategy (proteomics and transcriptomics) is employed. This strategy has been applied successfully to venomous animals with a centralised venom delivery system (venom sac or gland), such as cone snails, spiders, scorpions, and centipedes.[4–7] It has been used less frequently with cnidarians, which lack a centralised delivery system.[8] For cnidarians, traditional toxin isolation techniques for proteomic analysis, such as electric stimulation, fail to capture a comprehensive venom inventory owing to the external and internal distribution of cnidae. Moreover, the use of transcriptomics alone will only identify putative toxins with homology to known peptides,[9] thereby overlooking novel scaffolds of potential interest as drug leads.
We have examined the utility of incorporating matrix-assisted laser desorption/ionisation imaging mass spectrometry (MALDI-IMS) into a venomics strategy to discern the tissue distribution of peptides and infer biological functions. MALDI-IMS is commonly used in clinical applications to identify protein changes within cancers, detect biomarkers within tissues, and as a tool for drug discovery.[10–12] Recently, the application of MALDI-IMS has been extended to examine peptidome complexity in centipede venom glands.[13,14]
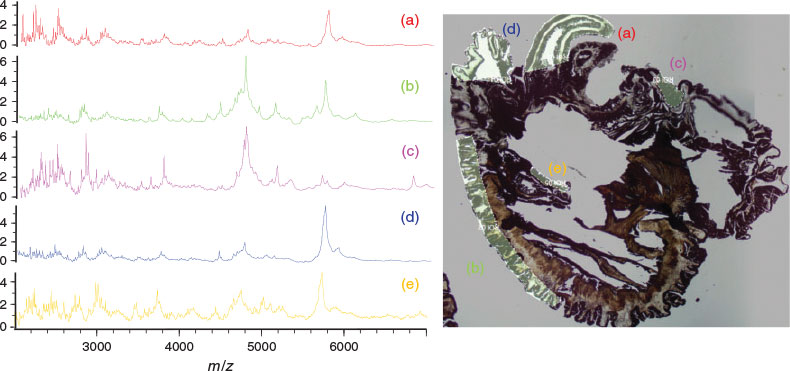
We conducted a pilot MALDI-IMS study using transverse sections of the sea anemone, Oulactis muscosa, a species found along the eastern Australian coastline. Regions of interest (ROI) were selected based on biological functions and associated cnidae profile (Fig. 1a–e). External ROI included tentacles (prey capture and immobilisation), acrorhagi and frill (defence), and column (external defence). Internal ROI included the actinopharynx (throat) and mesenterial filaments, both used in prey immobilisation and digestion. Fig. 1 displays the unique individual spectra produced for each ROI, reflecting mass diversity within each tissue region (individual masses for ROI spectra and comparison provided in Supplementary Material, Fig. S1).
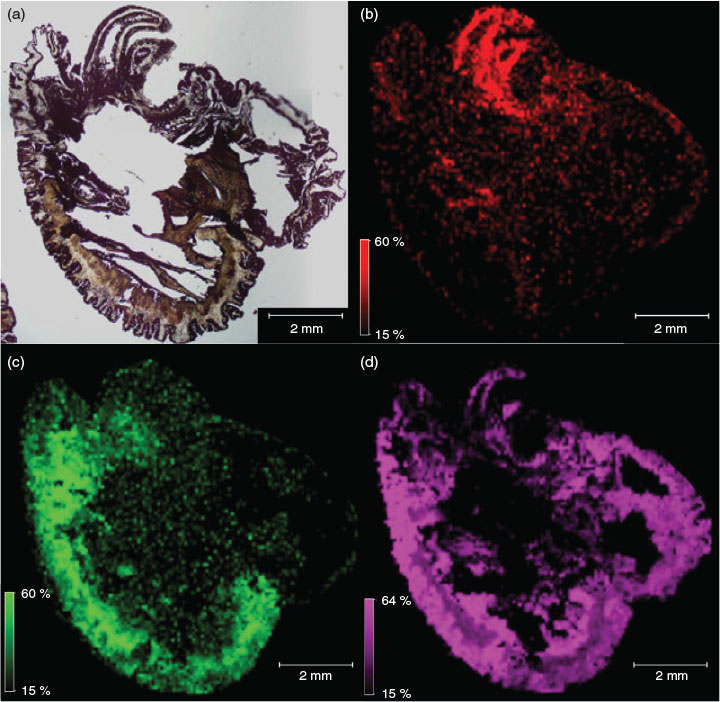
Fig. 2 exemplifies the capability of MALDI-IMS, displaying the distribution of three individual peptide masses, from which we can potentially draw inferences linking biological function to morphology. Fig. 2b highlights a putative peptide toxin, as it occurs solely in the ectoderm of the tentacles, where cnidae are densely packed. Fig. 2c shows a peptide with a distribution restricted to muscular tissue, implying a physiological role, and Fig. 2d shows a ubiquitously distributed peptide.
By utilising a venomics strategy that combines transcriptomics and proteomics with MALDI-IMS, we can potentially identify and correlate peptides according to tissue-specific regions. These results will aid our understanding of the functional evolution of sea anemone peptide toxins, while providing a library of novel peptides and scaffolds that may be useful as pharmacological tools or drug leads.
Supplementary Material
Individual masses for ROI spectra and comparison (Fig. S1) are available on the Journal’s website.
Conflicts of Interest
The authors declare no conflicts of interest
Acknowledgements
This project is funded in part by Australian Research Council linkage grants LP150100621 and LP140100832, an Australian Government Research Training Program Scholarship and a Monash University–Museum Victoria Scholarship top-up.
References
[1] G. Kass-Simon, A. A. Scappaticci, Can. J. Zool. 2002, 80, 1772.| Crossref | GoogleScholarGoogle Scholar |
[2] R. S. Norton, Expert Opin. Drug Discov. 2017, 12, 611.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXmtlOms7o%3D&md5=72d3730248bc6614d48c6b030752bb2dCAS |
[3] K. G. Chandy, R. S. Norton, Curr. Opin. Chem. Biol. 2017, 38, 97.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXmtFOhtL8%3D&md5=5df63395628a7432bfdc7b879f5fad9bCAS |
[4] S. D. Robinson, H. Safavi-Hemami, S. Raghuraman, J. S. Imperial, A. T. Papenfuss, R. W. Teichert, A. W. Purcell, B. M. Olivera, R. S. Norton, J. Proteomics 2015, 114, 38.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvF2mtrjF&md5=377085d78b38ce292722dbea9b461280CAS |
[5] C. Santibáñez-López, J. Cid-Uribe, C. Batista, E. Ortiz, L. Possani, Toxins 2016, 8, 367.
| Crossref | GoogleScholarGoogle Scholar |
[6] Z.-C. Liu, R. Zhang, F. Zhao, Z.-M. Chen, H.-W. Liu, Y.-J. Wang, P. Jiang, Y. Zhang, Y. Wu, J.-P. Ding, W.-H. Lee, Y. Zhang, J. Proteome Res. 2012, 11, 6197.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1Kntb%2FN&md5=cb31c02ff148ddb0737a5241ffb02acdCAS |
[7] S. S. Pineda, E. A. B. Undheim, D. B. Rupasinghe, M. P. Ikonomopoulou, G. F. King, Future Med. Chem. 1699, 2014, 15.
| Crossref | GoogleScholarGoogle Scholar |
[8] D. Ponce, L. D. Brinkman, J. Potriquet, J. Mulvenna, Toxins 2016, 8, 102.
| Crossref | GoogleScholarGoogle Scholar |
[9] J. Macrander, M. Broe, M. Daly, Genome Biol. Evol. 2016, 8, 2358.
| Crossref | GoogleScholarGoogle Scholar |
[10] J. O. R. Gustafsson, M. K. Oehler, A. Ruszkiewicz, S. R. McColl, P. Hoffmann, Int. J. Mol. Sci. 2011, 12, 773.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhvFOltbg%3D&md5=37127d7e4f50979254a2acbe2ec73a76CAS |
[11] Y. Sugihara, K. Watanabe, Á. Végvári, Bioanalysis 2016, 8, 575.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xjs1WnsLk%3D&md5=723dce539c038cae6f4ca1f5bf6d589cCAS |
[12] L. H. Cazares, D. A. Troyer, B. Wang, R. R. Drake, O. J. Semmes, Anal. Bioanal. Chem. 2011, 401, 17.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXlsFSgtLw%3D&md5=e84f1dce803cb5f8fa3382d32693c395CAS |
[13] E. A. B. Undheim, K. Sunagar, B. R. Hamilton, A. Jones, D. J. Venter, B. G. Fry, G. F. King, J. Proteomics 2014, 102, 1.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXntVequ78%3D&md5=085c4ff253b8881c5d9ae79bacaa99efCAS |
[14] E. A. B. Undheim, B. R. Hamilton, N. D. Kurniawan, G. Bowlay, B. W. Cribb, D. J. Merritt, B. G. Fry, G. F. King, D. J. Venter, Proc. Natl. Acad. Sci. USA 2015, 112, 4026.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXksF2itLo%3D&md5=2eb6a89c50721d91d50ff6732bc61b81CAS |
* Michela Mitchell received the Royal Australian Chemical Institute – Peptide User Group – Winter Symposium 2016 Early Career Investigator Award and best student oration.