Genetic diversity in the U.S. hard red winter wheat cultivars as revealed by microsatellite markers
B. Prasad A , M. A. Babar B F , X. Y. Xu C , G. H. Bai D and A. R. Klatt EA Rice Research and Extension Center, University of Arkansas, Stuttgart, AR 72160, USA.
B Department of Agronomy, Kansas State University, Manhattan, KS 66506, USA.
C Department of Crop and Soil Sciences, University of Georgia, Athens, GA 30602, USA.
D USDA-ARS, Kansas State University, Manhattan, KS 66506, USA.
E Department of Plant and Soil Sciences, Oklahoma State University, Stillwater, OK 74078, USA.
F Corresponding author. Email: mababar@ksu.edu, mdalibabar@yahoo.com
Crop and Pasture Science 60(1) 16-24 https://doi.org/10.1071/CP08052
Submitted: 13 February 2008 Accepted: 17 October 2008 Published: 5 January 2009
Abstract
Knowledge of the genetic diversity existing in previously released hard red winter wheat (HRWW, Triticum aestivum L.) cultivars in the Great Plains region, United States, is essential for effective utilisation of these genetic resources in the various HRWW breeding programs. To ascertain a measure of the genetic diversity of the existing US HRWW, 60 cultivars were analysed with 62 microsatellite markers distributed throughout the wheat genome. Marker data were subjected to distance-based analysis and analysis of molecular variances. In total, 341 polymorphic alleles were scored with a range of 2–12 alleles per locus. Genetic diversity gradually increased in cultivars released after the 1970s. Cultivars released in the 1990s had the highest allelic richness (4.79), gene diversity (0.60), and polymorphic information content (0.56). Levels of genetic diversity were similar between the major HRWW breeding programs. Cluster analysis resulted in eight clusters. Cluster grouping gave close matches with pedigrees and with regional distribution of the cultivars. Using decadal information, cultivars released from 1900–1969 were grouped into one cluster, cultivars from 1990–2005 were grouped into a separate cluster, whereas cultivars from the 1980s did not group with any other decades. Analysis of molecular variance revealed a significant variation among the clusters, signifying that a true genetic variation existed among the clusters. The higher proportion of genetic variation explained by cultivars within clusters compared with among clusters indicates greater genetic diversity among cultivars within clusters. Our results indicate that genetic diversity of Great Plains HRWW cultivars has increased in the past century, and the trend is continuing.
Additional keywords: molecular markers, cluster analysis.
Introduction
Genetic diversity is the foundation for crop genetic improvement, and thus the most important consideration in any plant breeding program. Information related to genetic diversity among adapted lines helps breeders in selecting parents for hybrid production with maximum heterosis and combining useful genes in an adapted genetic background. In contrast to wide genetic diversity, a narrow genetic base is a big limitation to breeding for adaptation to different biotic and abiotic stresses. A major concern for many modern plant breeding programs is the narrow genetic base of their germplasm (Velle 1993; Clunies-Ross 1995). Several authors have argued that the narrowness in genetic diversity could lead to an increased vulnerability to diseases and pests, as well as the ability of plants to respond to changing environmental conditions (Clunies-Ross 1995; Tripp 1996). Thus, quantifying genetic diversity among existing germplasm helps to address this concern. Also, the knowledge of genetic relationships among different genetic materials allows breeders to eliminate unwanted duplication in germplasm and increase the efficiency of breeding programs.
The Great Plains region is the largest winter wheat (Triticum aestivum L.) growing area in the USA, and hard red winter wheat (HRWW) is the major market class of wheat produced. Each of the states in the Great Plains region has at least one major winter wheat breeding program, plus the two private breeding programs, AgriPro and Pioneer, have been active in the region. The western part of the southern and central Great Plains is characterised as having low or relatively low rainfall, low incidence of fungal diseases, moderate incidence of viral diseases, and moderate to high incidence of aphids, including Greenbugs and the Russian Wheat Aphid. The breeding programs at the University of Nebraska (UNL), Colorado State University (CSU), Kansas State University (KSU) at Hays, Oklahoma State University (OSU), Texas A & M University (TAMU) at Amarillo, and the two private breeding programs (Pioneer and AgriPro) have developed cultivars with good drought tolerance, good quality, and resistance to these biotic stresses. The eastern and central parts of the Great Plains have higher rainfall and much higher incidence of fungal diseases, especially leaf rust and stripe rust. High yield potential, good industrial quality, and high levels of resistance to biotic stresses are the priority breeding objectives. The programs at KSU (Manhattan), OSU, TAMU (Vernon), UNL, and the two private breeding programs have developed successful cultivars for these regions. Collectively, these programs have released most of the cultivars that were included in this study. The first widely cultivated genotype in the region was ‘Turkey’, a landrace from southern Russia, and this cultivar was dominant until the end of the 1920s. Since then, considerable progress has been achieved by conventional plant breeding programs. From 1930 to 1950, the dominated cultivars were Blackhull, Pawnee, Wichita, Triumph, and Comanche, and all were derivatives of Turkey (Cox et al. 1986). Genetic diversity was first introduced to incorporate better rust resistance, primarily from US spring and winter wheat materials. Subsequently, in the late 1960s and 1970s, CIMMYT germplasm was used as a source of the semi-dwarf character and for better stem rust resistance (A. R. Klatt, unpublished 2007). The first widely grown HRWW semi-dwarf variety was ‘Newton’ and it was obtained by crossing a CIMMYT spring wheat semi-dwarf advanced line to Scout. In the 1970s, breeding programs focussed primarily on incorporating shorter straw and stem rust resistant genes into adapted, high-yielding lines. During the 1980s and 1990s, the main breeding objectives were to incorporate resistance to other diseases, especially leaf rust, and to improve industrial quality traits. Thus, during the last four decades, breeding programs have focussed on a few major objectives. Therefore, it is important to know the extent of genetic diversity that has been achieved and that currently exists among released cultivars in the Great Plains. Twenty years ago, Cox et al. (1986) made a landmark study on the genetic diversity in the commercially cultivated HRWW and soft red winter wheat (SRWW) varieties grown between 1919 and 1984. That study revealed that genetic diversity had increased in HRWW, but decreased in SRWW. According to their results, up to the 1950s there was a high degree of genetic uniformity, most probably due to the widespread use of Turkey and its derivatives, but uniformity had decreased somewhat from 1919 to 1949. After 1950, diversity increased, maybe due to the introduction of different disease resistance and quality genes from a variety of germplasm. In the past two decades, many new cultivars have been released. Also, in the last 2–3 decades there has been increased collaboration between breeding programs, many use similar parents in their crossing program, and much sharing of germplasm (gene pool) has occurred between different breeding programs. There has not been a recent study to provide information on how genetic diversity has changed in modern HRWW cultivars except Fufa et al. (2005), who reported a comparison between phenotypic and molecular marker-based approaches of estimating genetic diversity, using a set of 30 HRWW cultivars only from the Northern Great Plains of the USA. As a result, there is a need to quantify the genetic diversity existing among the different parts of the Great Plains HRWW cultivars released in the last several decades.
The use of molecular markers for the evaluation of genetic diversity of wheat has recently received a great deal of attention from molecular geneticists and wheat breeders. With the help of molecular markers, geneticists can monitor subtle changes in DNA sequences in genotypes released at different times (Ortiz 2001). This approach would help breeders to assess the allelic combinations selected generation after generation, and thus provide a genetic ideotype for future marker-assisted selection (Christiansen et al. 2002). This information would help breeders to incorporate useful genetic variation into adopted gene pools by selecting for marker alleles linked to loci controlling important agronomic or quality traits (Tanksley and McCouch 1997). Several authors have studied genetic diversity in wheat by using different molecular markers, such as microsatellite (Devos et al. 1995; Bohn et al. 1999; Donini et al. 2000; Manifesto et al. 2001; Christiansen et al. 2002; Huang et al. 2002; Dreisigacker et al. 2004; Fufa et al. 2005), amplified DNA fragment polymorphism markers (Schut et al. 1997; Barrett et al. 1998; Manifesto et al. 2001), and random amplified polymorphic DNA markers (Joshi and Nguyen 1993). Microsatellite markers, also known as simple sequence repeats (SSR), have proved to be the most suitable molecular markers for studying genetic diversity in wheat because of its multi-allelic nature, chromosome specificity, high polymorphism, and distribution throughout the genomes (Röder et al. 1998a, 1998b). Most of the genetic diversity studies in wheat have been conducted by using genetic materials from different geographic locations or from different mega-environments or from different market classes. So far, genetic diversity in the HRWW cultivars released in different eras in the Great Plains has not been well documented. The objective of this study was to assess the extent and trend for genetic diversity among HRWW cultivars released from different USA Great Plains breeding programs in different eras.
Materials and methods
Genetic materials and DNA extraction
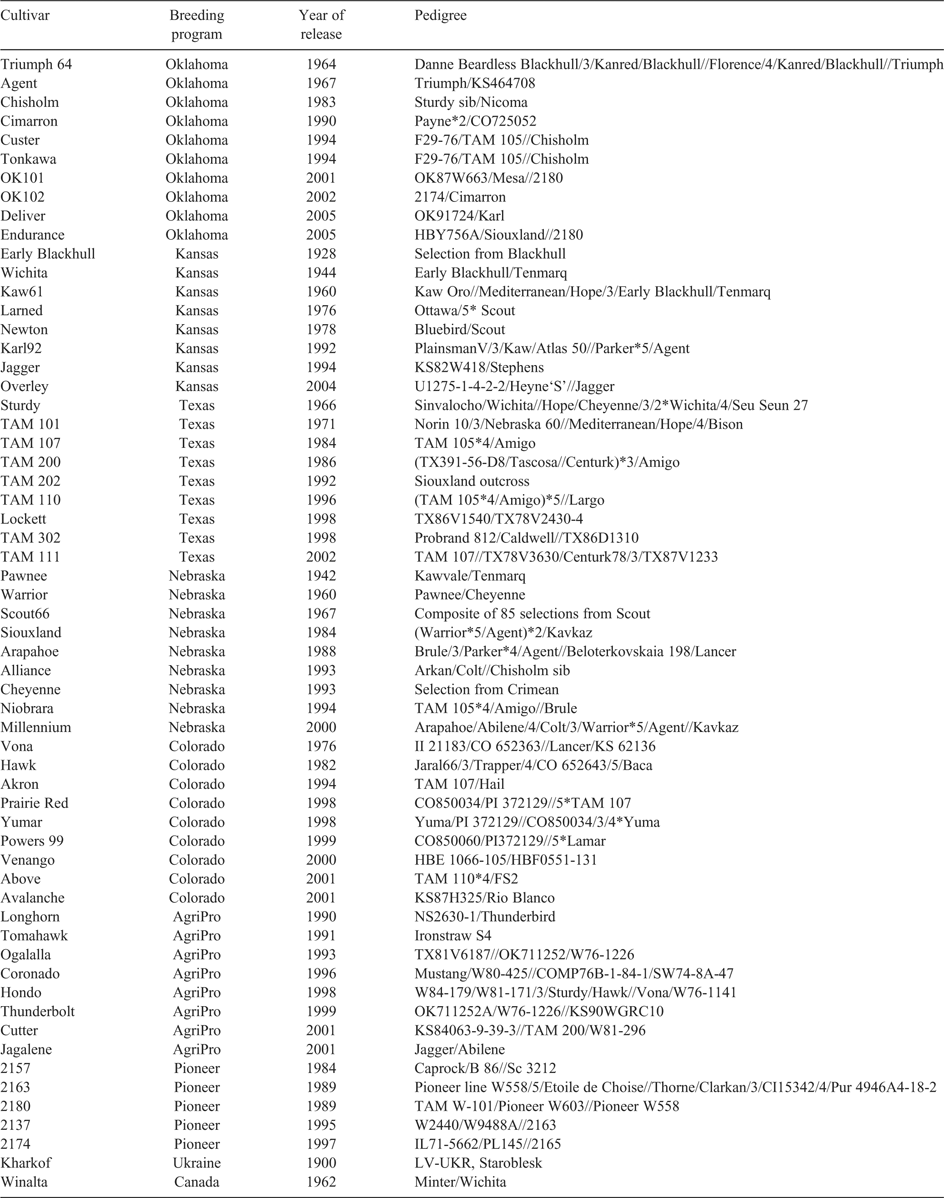
In total, 60 wheat cultivars representing the Great Plains HRWW regions of the USA, released from 1900 to 2005, were used in this study (Table 1). The cultivars were grouped into six decadal periods based on their year of release, namely <1960 (1900–59), 1960s (1960–69), 1970s (1970–79), 1980s (1980–89), 1990s (1990–99), and 2000s (2000–05). Comparatively higher numbers of cultivars were selected from the 1990s because a significant number of cultivars were released during the 1990s in the Great Plains. Also, many of these cultivars were still under commercial cultivation and were sown on a significant proportion of the total wheat area in the region at the time of this study. The cultivars were also grouped into 8 different classes based on the breeding programs, namely Oklahoma, Kansas, Texas, Nebraska, Colorado, AgriPro, Pioneer, and other (Table 1). In each breeding group, the cultivars were selected from different decades of release, and listed along with their known pedigrees. Pure seeds of the cultivars were collected from the National Plant Germplasm System (NPGS, USDA-ARS), and from the wheat breeders of Oklahoma, Kansas, Texas, Colorado, and Nebraska. Leaves were collected from 6–10 seedlings of each cultivar at the 2-leaf stage for DNA isolation using Qiagen DNeasy Plant Mini Kit (Qiagen Inc.; Valencia, CA, USA).
|
SSR analysis
The experiment was conducted in the USDA-ARS Small Grain Genotyping Laboratory in Kansas State University, Manhattan, KS, and Wheat Genetic Laboratory in Oklahoma State University, Stillwater, OK. Sixty-two pairs of SSR primers comprising 19 XGWM (Röder et al. 1998a), 20 BARC (Song et al. 2005), and 23 WMC (Gupta et al. 2002) primers were used in the study. The primers were selected from all 21 chromosomes of hexaploid wheat. The PCR was performed in a volume of 10 µL containing 2 µL of DNA (20 ng/µL) and 8 µL of the master mix that consisted of 3.2 µL of ddH20, 1 µL of 10 × PCR buffer, 1.1 µL of 25 mm MgCl2, 0.5 µL of 5 mm dNTPs, 0.5 µL of each forward and reverse primer (1 p.m./µL), 1 µL of tailed fluorescence-labelled (IRD-700 or IRD-800) M13 primer (Oetting et al. 1995), and 0.2 µL of Taq polymerase (5 U/µL). The PCR was performed by using a touchdown universal program consisting of 5 cycles of denaturing at 95°C for 45 s, annealing at 68°C for 5 min decreasing by 2°C in each subsequent cycle, and extension at 72°C for 1 min. In the next 5 cycles, the annealing temperature was started at 58°C for 2 min and decreased by 2°C in each subsequent cycle. An additional 25 cycles consisted of 45 s of denaturing at 95°C, 1 min of annealing at 50°C, and 1 min of extension at 72°C with a final extension cycle at 72°C for 5 min. The PCR product was denatured at 95°C for 5 min before it was separated in a 6.5% polyacrylamide gel on Li-Cor IR-4200 or IR-4300 DNA sequencer (Li-Cor Inc.; Lincoln, NE, USA).
Data analyses
Marker data were scored as presence or absence of an allele corresponding to each SSR locus using the software GeneImageIR ver. 4.05 (BD Biosciences, USA). Nei’s (1972) genetic distance algorithm was used to calculate the pair-wise genetic distances among the cultivars, and clusters were shown in a neighbour joining tree. This is a hierarchical algorithm for clustering genotypes into different groups. Another neighbour joining tree was created using the decadal information (among the six defined decades). PowerMarker software (Liu and Muse 2005) was used to calculate the genetic distance among the 60 cultivars as well as among the decadal groups. Frequency and number of alleles, gene diversity, and polymorphic information content (PIC) were calculated for each SSR locus, chromosome, and genome for the entire set of cultivars, six decadal groups, and the regions/breeding programs using the PowereMarker software. Gene diversity was defined by Weir (1996) as the probability of two randomly chosen alleles from a population being different, and PIC value was defined by Botstein et al. (1980) as the value of a marker for detecting polymorphism within a population which depends on the number of detectable alleles and their frequency distribution. Tree view program MEGA ver.4.0 (Tamura et al. 2007) was used to construct the cluster trees. Levels of genetic variation within and among clusters as identified from the cluster analysis were estimated from the allelic frequencies using the analysis of molecular variance (AMOVA) (Weir and Cockerham 1984), and were implemented by using the software Arlequin ver. 3.0 (Excoffier et al. 2005). The AMOVA analyses were also tested using the groups designated by the decades of cultivar release and breeding programs. These analyses separate the total molecular variance into components of inter and intra groups and test the level of significance among the groups. Wright’s fixation index (FST) was used to estimate the variance within the groups, and statistical significance of the index was evaluated using a 1000-times permutation of the genotypes among and within the groups. Genetic diversity among the clusters was evaluated based on the pair-wise FST comparison that measures the fixation of alleles in the population.
Results and discussion
Diversity of the SSR markers
Allele frequency, number of alleles/locus, gene diversity, and PIC values for each SSR marker were calculated, and a total of 341 polymorphic alleles were scored with a mean of 5.5 alleles per primer pair (data not shown). Number of alleles for each primer pair varied greatly, ranging from 2 (BARC 1097; GWM 210) to 12 (GWM 261). Studies conducted earlier suggested that around 350 alleles were sufficient to assess the relationship between wheat accessions (Zhang et al. 2002). Major allelic frequency ranged from 0.23 (XGWM 604) to 0.98 (WMC 477), with a mean of 0.51. Gene diversity ranged from 0.03 (WMC 477) to 0.86 (WMC 707), with an average of 0.62 (data not shown). Among the detected alleles, 97 (28.4%) alleles were considered as rare alleles that occurred at a frequency of less than 5%. Estimated gene diversity in our study was 0.62, which was a little higher than the estimate of 0.57 from 68 advanced CIMMYT lines (Dreisigacker et al. 2004) and the estimate of 0.55 from 134 durum wheat accessions (Maccaferri et al. 2005), but lower than the estimates of 0.66 in 43 bread wheat cultivars from 7 USA market classes (Chao et al. 2007) and 0.66 in 559 French wheat accessions (Roussel et al. 2004), and 0.77 in 998 accessions from 68 countries (Huang et al. 2002). Studies reported earlier with higher gene diversity can be attributed to the use of genetic materials from well defined market classes or landraces, or genotypes from gene banks, or cultivars from different geographic locations (Huang et al. 2002; Roussel et al. 2004; Chao et al. 2007). Although we have used only one market class of wheat, the moderately high gene diversity indicates that the breeders in the Great Plains wisely used genetic materials for different sources of genes. The PIC values ranged from 0.03 (WMC 477) to 0.85 (BARC 184), with an average of 0.58 per primer pair (data not shown). Allelic richness (6.1), gene diversity (0.69), and PIC values (0.65) were the highest for chromosome 4, which suggests that most genetic variations were incorporated into chromosome 4 in the HRWW cultivars studied. Among the three genomes, gene diversity (0.63) and PIC value (0.63) were comparatively higher for the B genome, which means that more genetic diversity among the cultivars used in the study may reside in the B genome (data not shown). Hai et al. (2007) and Huang et al. (2002) also reported the highest gene diversity and PIC values in the B genome.
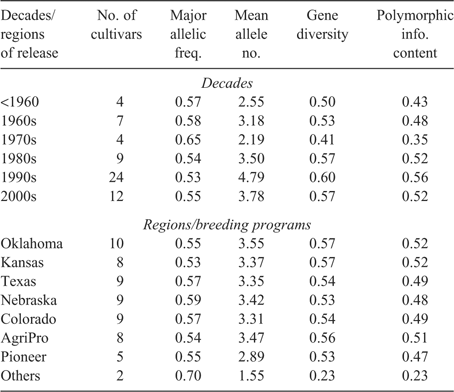
Major allelic frequency, alleles detected, gene diversity, and PIC values of the cultivars were estimated for the decade of cultivar release, and for the regions/breeding programs where the cultivars were released (Table 2). Mean allele number (4.79), gene diversity (0.60), and PIC (0.56) were the highest for the cultivars released during the 1990s. Cultivars from the 1980s and 2000s had similar gene diversity and PIC values. Cultivars from the 1970s showed a decrease in mean allele number, gene diversity, and PIC values compared with the other decadal groups (Table 2). In general, diversity measurements were higher in the cultivars released after the 1980s than in those released before the 1980s. However, cultivars released from 2000 to 2005 showed a decrease in allelic richness compared with those released in the 1990s. This might be due to the fact that, during the 1990s, a significant number of cultivars were released from different breeding programs, and no single cultivar dominated the commercial cultivation as in some of the previous decades. As a result, a comparatively larger number of cultivars from the 1990s have been used in this study. Also during the 1990s, the two private breeding programs released many cultivars that might have represented different gene pools or new diversity. The cultivars released during the 2000s represent only a 5-year period instead of a 10-year period, and thus may result in decreased diversity measurements. However, the reduction of allelic richness in the 1970s and 2000s could be due to the elimination of deleterious genes rather than the erosion of useful genetic resources in those years. No substantial differences were observed among the major HRWW breeding programs of the Great Plains in terms of allelic richness, gene diversity, and PIC values, except a comparatively low allelic richness in the Pioneer genetic materials (Table 2). These observations indicate that the major HRWW breeding programs of the Great Plains are consistent in maintaining genetic diversity in their released cultivars for the last several decades.
|
The increase in gene diversity and allelic richness after the 1970s reflects the use of diverse sets of germplasm and the introduction of new genetic variation. The breeders in the southern Great Plains introduced considerable spring wheat into the HRWW gene pool in the late 1960s and early 1970s. Mexican semi-dwarf wheat was the major source of spring wheat diversity added to the HRWW germplasm pool. ‘Newton’, released in 1978, had a CIMMYT spring wheat background, which created a truly new diversity in the wheat germplasm from the southern Great Plains (A. K. Fritz, pers. comm., 2007). Moreover, integration and selection for several important yield and adaptive traits also led to an increase in average gene diversity and allelic richness.
Genetic relationships among the HRWW cultivars
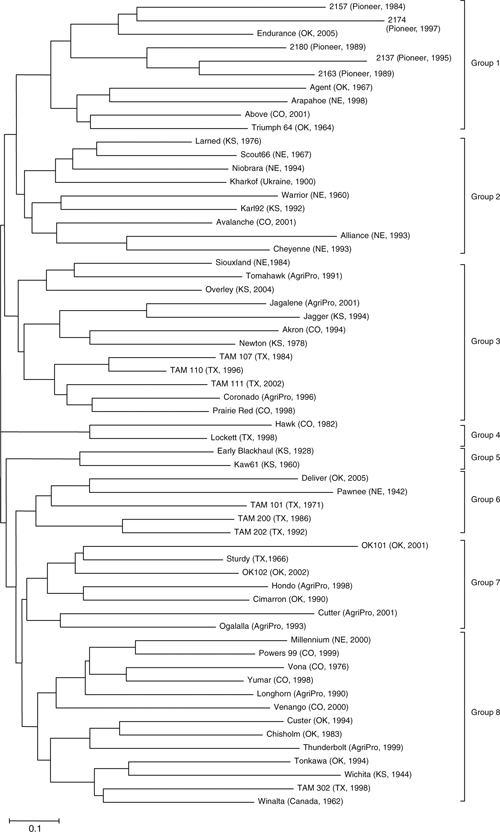
Cluster analysis using the Nei’s genetic distance algorithm separated the 60 HRWW cultivars into 8 clusters (Fig. 1). A wide range of genetic distances, from 0.33 to 1.79, was observed among the cultivars. The furthest genetic distance (1.79) was between cv. TAM 200 and 2174, and the shortest distance (0.33) was between TAM 107 and TAM 110. The cv. TAM 110 was developed using TAM 107 as a backcross parent (Table 1). The clusters (Fig. 1) generally agreed with the pedigree or genetic background of the cultivars. The cultivars did not cluster according to the defined decades of release, but in several cases cultivars released from a specific breeding program were grouped together. For example, cultivars released in different states from Pioneer lines, such as 2137, 2157, 2163, 2174, and 2180, were grouped together. Several cultivars released from Nebraska, such as Scout66, Niobrara, Warrior, Alliance, and Cheyenne, grouped together. Two historical cultivars, Early Blackhull and Kaw61 from Kansas, formed a unique cluster. The Texas-released cvv. TAM 101, TAM 200, and TAM 202 grouped together. Jagger, the most widely grown cultivar from the Great Plains in 2000–05, grouped with Jagalene and Overley, which was as expected because Jagger is one of the parents of Jagalene and Overley. All clusters contained cultivars from multiple states, except the smallest cluster with only two Kansas cultivars (Early Blackhull and Kaw61), suggesting that frequent exchange and sharing of breeding materials occurred across HRWW breeding programs from different states in different time periods. The fact that the predominant cluster was based on regional adaptation (Fig. 1) within different parts of the Great Plains indicates the likeliness of widespread sharing of related ancestral lines within the various breeding programs.
|
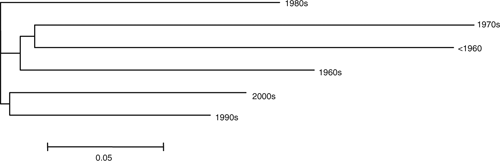
To evaluate the temporal variation and relationship among different decades, marker data were used to group different decades. The analysis identified 3 major decadal groups, i.e. 1980s decadal group, pre-1980 decadal group, and post-1980s decadal group (Fig. 2). Breeding objectives before 1980 mainly focussed on incorporating semi-dwarfing genes, especially Rht1, and stem rust resistance genes. In the 1980s, breeding objectives shifted mainly to leaf rust resistance, which continues to be a very important objective in all the HRWW breeding programs along with end-use quality. Cox et al. (1986) reported a low genetic diversity among the cultivars released before 1950 in the Great Plains HRWW region due to the widespread and continuous use of Turkey, a land race, and its derivatives Pawnee, Wichita, Triumph, and Comanche. They also reported an increase in genetic diversity among the cultivars from 1950 to the 1980s, due to the extensive use of semi-dwarfing genes from soft red spring wheat in the 1960s and 1970s (Cox et al. 1986). Our results indicate that although the breeding objectives for HRWW were relatively narrow across decades and a lot of cultivars shared a common ancestry, genetic diversity was maintained among the commercial cultivars to date by the introduction of new genes from different gene pools. Breeders always selected for cultivars that were well adapted to the variable regional environments while introducing genetic diversity from several germplasm pools, especially from the Mexican spring wheat and European germplasm.
|
Genetic diversity in the HRWW cultivars
The distribution of molecular variation among and within the identified clusters of 60 HRWW cultivars as indicated in Fig. 1 was estimated by AMOVA procedure (Table 3). The AMOVA revealed that around 10% of the total variation resided among the clusters, whereas around 90% of the total molecular variation existed among the cultivars within the clusters. The higher proportion of genetic variation explained by the cultivars within the clusters indicated a large variation among the cultivars within a cluster as a result of intensive breeding activities within each breeding program, which made use of diverse genetic materials while selecting for local adaptation. Roussel et al. (2005) also reported similar trends of larger within-group genetic variation in wheat cultivars from different regions of Europe. Our results suggest that a significant amount of genetic diversity was incorporated into the HRWW cultivars in this study.
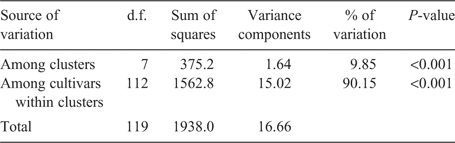
|
To investigate the diversity changes based on the decades and breeding programs, the AMOVA procedure was also carried out using the groups created by decades of cultivar release and sources of cultivar release (breeding programs) as indicated in Table 2. The AMOVA revealed a significant proportion of genetic variation as explained among the decades as well as among the cultivars within decades (Table 4). The higher proportion of genetic variation was attributed to among cultivars within decades variance (94.68%) compared with the among decades variance (5.32%). When the different breeding programs were subjected to an AMOVA test (Table 5), the proportion of genetic variation was greater for among cultivars within breeding program variation (86.49%) compared with the among breeding programs variation (13.51%), although both of them were highly significant. When the three AMOVA procedures were compared for among groups variation, the genetic variation was best explained by the among breeding programs variation, which means true genetic variations exist between the different breeding programs in the Great Plains.
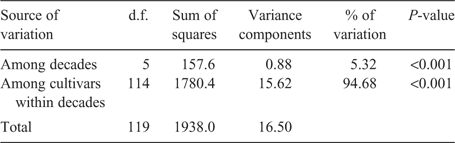
|
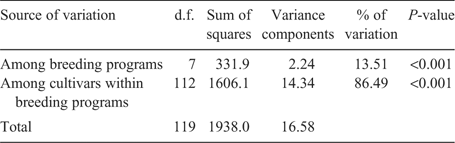
|
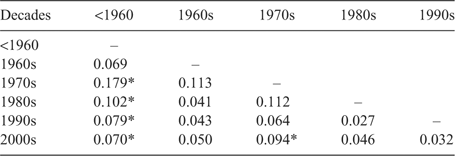
Determination of FST value of the polymorphic loci for all accessions revealed that the FST values ranged from 0.017 (WMC 477) to 0.427 (BARC 184), with an average of 0.288 (data not shown). These results suggest that 28.8% of the total variation in allele frequency was due to genetic variation among clusters. It has been reported that markers with higher FST values have greater power to produce genetic distance estimates (Watkins et al. 2003). Genetic variation among the clusters was tested by computing pair-wise FST values between the clusters (Tables 6–8). Most of the pair-wise FST values were statistically significant (P < 0.05) for groups created by cluster analysis (Table 6) and breeding programs (Table 8). Higher numbers of pair-wise comparisons based on the decadal groups resulted in a non-significant difference among the pairs of decades compared with the other two groups (cluster analysis and breeding programs). This indicates that the enhancement of genetic diversity may not be well classified as true decadal groups compared with the breeding programs or the groups identified in the cluster analysis. The significant FST values among the clusters (Tables 6 and 8) suggest that a real difference between the clusters exists and a significant amount of genetic diversity is still present among the HRWW cultivars used in this study from different breeding programs of the Great Plains.
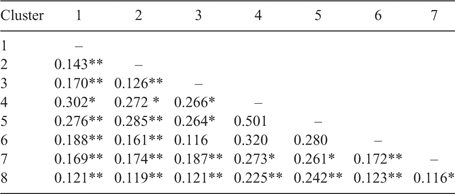
|
|

|
These results suggest that there has been no real loss of genetic diversity in the studied HRWW cultivars. In fact, a gradual increase of allelic richness was observed from 1980 to 1999, indicating the incorporation of important alleles for better agronomic, disease resistance, and quality traits until 1999. After 2000, a decrease in allelic richness was due to the qualitative changes in allelic composition. Replacement with alleles favourable for adaptation is a key feature for today’s breeding approach, which has led to the apparent loss of allelic richness (Landjeva et al. 2007), but the maintenance of gene diversity and high PIC values in the 2000s releases indicate positive changes in the most recent cultivars. Genetic diversity may not be well assessed using a particular decade because the pattern of changes may not happen in a defined number of years. But the overall results of the study indicate that the wheat breeders in the Great Plains adopted a very effective breeding strategy to introduce new variation in the existing gene pool when needed, and maintained that variation over a long period of time.
Acknowledgments
We thank NPGS (USDA-ARS) and the wheat breeders of Oklahoma, Kansas, Texas, Colorado, and Nebraska for providing the germplasm. Our sincere thanks go to Dr Allan Fritz for his comments and information regarding the pedigree of some genotypes.
Barrett BA,
Kidwell KK, Fox PN
(1998) Comparison of AFLP and pedigree-based genetic diversity assessment methods using wheat cultivars from the Pacific Northwest. Crop Science 38, 1271–1278.
|
CAS |

Bohn M,
Utz HF, Melchinger AE
(1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Science 39, 228–237.
|
CAS |

Botstein D,
White RL,
Skolnick M, Davis RW
(1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32, 314–331.
|
CAS |
PubMed |

Chao S,
Zhang W,
Dubcovsky J, Sorrells M
(2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Science 47, 1018–1030.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Christiansen MJ,
Andersen SB, Oritiz R
(2002) Diversity changes in an intensively bred wheat germplasm during the 20th century. Molecular Breeding 9, 1–11.
| Crossref | GoogleScholarGoogle Scholar |

Clunies-Ross T
(1995) Mangolds, manure and mixtures: The importance of crop diversity on British farms. The Ecologist 25, 181–187.

Cox TS,
Murphy JP, Rodgers DM
(1986) Changes in genetic diversity in the red winter wheat regions of the United States. Proceedings of the National Academy of Sciences of the United States of America 83, 5583–5586.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Devos KM,
Bryan GJ,
Collins AJ,
Stephenson P, Gale MD
(1995) Application of two microsatellite sequences in wheat storage proteins as molecular markers. Theoretical and Applied Genetics 90, 247–252.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Donini P,
Law JR,
Koebner RMD,
Reeves JC, Cooke RJ
(2000) Temporal trend in the diversity of UK wheat. Theoretical and Applied Genetics 100, 912–917.
| Crossref | GoogleScholarGoogle Scholar |

Dreisigacker S,
Zhang P,
Warburton ML,
van Ginkel M,
Hoisington D, Melchinger AE
(2004) SSR and pedigree analyses of genetic diversity among CIMMYT wheat lines targeted to different mega-environments. Crop Science 44, 381–388.
|
CAS |

Excoffier L,
Laval G, Schneider S
(2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1, 47–50.
|
CAS |

Fufa H,
Baenziger PS,
Beecher I,
Dweikat V,
Graybosch RA, Eskridge KM
(2005) Comparison of phenotypic and molecular marker-based classifications of hard red winter wheat cultivars. Euphytica 145, 133–146.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Gupta K,
Balyan S,
Edwards J,
Isaac P,
Korzun V,
Röder M,
Gautier MF,
Joudrier P,
Schlatter R,
Dubcovsky J,
De La Pena C,
Khairallah M,
Penner G,
Hayden J,
Sharp P,
Keller B,
Wang C,
Hardouin P,
Jack P, Leroy P
(2002) Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theoretical and Applied Genetics 105, 413–422.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hai L,
Wagner C, Friedt W
(2007) Quantitative structure analysis of genetic diversity among spring bread wheats (Triticum aestivum L.) from different geographical regions. Genetica 130, 213–225.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Huang XQ,
Börner A,
Röder MS, Ganal MW
(2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theoretical and Applied Genetics 105, 699–707.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Joshi CP, Nguyen HT
(1993) RAPD (random amplified polymorphic DNA) analysis based intervarietal genetic relationships among hexaploid wheat. Plant Science 93, 95–103.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Landjeva M,
Korzum V, Borner A
(2007) Molecular markers: actual and potential contributions to wheat genome characterization and breeding. Euphytica 156, 271–296.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Liu K, Muse SV
(2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Maccaferri M,
Sanguineti MC,
Noli E, Tuberosa R
(2005) Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Molecular Breeding 15, 271–289.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Manifesto MM,
Schlatter AR,
Hopp HP,
Suarez EY, Dubcovsky J
(2001) Quantitative evaluation of genetic diversity in wheat germplasm using molecular markers. Crop Science 41, 682–690.
|
CAS |

Nei M
(1972) Genetic distance between populations. American Naturalist 106, 283–292.
| Crossref | GoogleScholarGoogle Scholar |

Oetting WS,
Lee HK,
Flanders DJ,
Wiesner GL,
Seller TA, King RA
(1995) Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30, 450–458.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Röder MS,
Korzun V,
Gill BS, Ganal MW
(1998b) The physical mapping of microsatellite marker in wheat. Genome 41, 278–283.
| Crossref | GoogleScholarGoogle Scholar |

Röder MS,
Korsun V,
Wendehake K,
Plaschke J,
Tixier MH,
Leroy P, Ganal MW
(1998a) A microsatellite map of wheat. Genetics 149, 2007–2023.
| PubMed |

Roussel V,
Koenig J,
Beckert M, Balfourier F
(2004) Molecular diversity in French bread wheat accessions related to temporal trends and breeding programs. Theoretical and Applied Genetics 108, 920–930.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Roussel V,
Leisova L,
Exbrayat F, Stehno Z
(2005) SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theoretical and Applied Genetics 111, 162–170.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Schut JW,
Qi X, Stam P
(1997) Association between relationship measures based on AFLP markers, pedigree data and morphological traits in Barley. Theoretical and Applied Genetics 95, 1161–1168.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Song QJ,
Shi JR,
Singh S,
Fickus EW,
Costa JM,
Lewis J,
Gill BS,
Ward R, Cregan PB
(2005) Development and mapping of microsatellite (SSR) markers in wheat. Theoretical and Applied Genetics 110, 550–560.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software ver. 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tanksley SD, McCouch SR
(1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tripp R
(1996) Biodiversity and modern crop varieties: sharpening the debate. Agriculture and Human Values 13, 48–63.
| Crossref | GoogleScholarGoogle Scholar |

Velle R
(1993) The decline of diversity of European agriculture. The Ecologist 23, 64–69.

Watkins WS,
Rogers AR,
Ostler CT,
Wooding S,
Bamshad MJ,
Brassington A-M E,
Carroll ML,
Nguyen SV,
Walker JA,
Prasad BVR,
Reddy PG,
Das PK,
Batzer MA, Jordel LB
(2003) Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Research 13, 1607–1618.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Weir BS, Cockerham CC
(1984) Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370.
| Crossref | GoogleScholarGoogle Scholar |

Zhang XY,
Li CW,
Wang LF,
Wang HM,
You GX, Dong YS
(2002) An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties. I. Information from large-scale planted varieties and cornerstone breeding parents in Chinese wheat improvement and production. Theoretical and Applied Genetics 106, 112–117.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



