Hypotheses from mitochondrial DNA: congruence and conflict between DNA sequences and morphology in Dolichopodinae systematics (Diptera : Dolichopodidae)
Marc Pollet A B C , Christoph Germann D , Samuel Tanner D E and Marco Valerio Bernasconi D FA Department of Entomology, Royal Belgian Institute of Natural Sciences (RBINS), Vautierstraat 29, B-1000 Brussels, Belgium.
B Research Group Terrestrial Ecology (TEREC), Department of Biology, Ghent University (UGent), K.L.Ledeganckstraat 35, B-9000 Ghent, Belgium.
C Information and Data Center, Research Institute for Nature and Forest (INBO), Kliniekstraat 25, B-1070 Brussels, Belgium.
D Zoological Museum, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.
E Present Address: Space Biology Group, BIOTESC, ETH Zurich, Technopark, Technoparkstr. 1, CH-8005 Zurich, Switzerland.
F Corresponding author. Email: marco.bernasconi@access.uzh.ch
Invertebrate Systematics 24(1) 32-50 https://doi.org/10.1071/IS09040
Submitted: 15 October 2009 Accepted: 8 February 2010 Published: 17 May 2010
Abstract
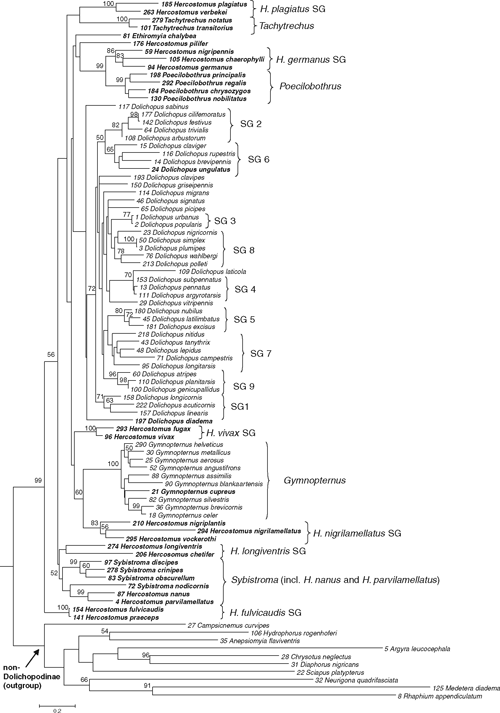
The molecular phylogeny of the subfamily Dolichopodinae (Diptera : Dolichopodidae) is reconstructed based on 79 species of 7 dolichopodine genera as ingroup, and 10 non-dolichopodine species from different genera as outgroup. A Bayesian analysis based on a mitochondrial DNA dataset consisting of 1702 characters (COI : 810; 12S : 366; 16S : 526) was carried out. Genital and non-genital morphological characters from a hitherto unpublished data matrix (based on 57 Dolichopodidae species) were used to explain and support the lineages hypothesised by our molecular phylogenetic analysis. The monophyly of the subfamily Dolichopodinae, and of the genera Dolichopus and Gymnopternus, was confirmed. The molecular analysis yielded nine species groups in Dolichopus that were proposed in previous studies using COI and Cyt-b. No evidence was found to support a clade including Dolichopus, Ethiromyia, and Gymnopternus. The genus Hercostomus proved polyphyletic with respect to Poecilobothrus, Sybistroma, and Gymnopternus. The following lineages were represented by strongly supported clades: Hercostomus germanus species group, H. vivax species group, H. nigrilamellatus species group, H. plagiatus species group, H. longiventris species group, H. fulvicaudis species group, and Poecilobothrus, Gymnopternus, Tachytrechus and Sybistroma (including Hercostomus nanus and H. parvilamellatus). Two clades that were previously established on the basis of morphology were confirmed in our phylogenetic analysis: (i) Poecilobothrus and the flower-feeding Hercostomus germanus species group, and (ii) the H. longiventris lineage and Sybistroma. In most cases, the groups identified in the molecular analysis could be supported and explained by morphological characters. Species of the Hercostomus germanus species group, Poecilobothrus, the Hercostomus longiventris species group, and a Sybistroma subclade have a similar microhabitat affinity.
Additional keywords: molecular phylogeny, Dolichopodidae, Dolichopus, Ethiromyia, Gymnopternus, Hercostomus, Ortochile, Poecilobothrus, Sybistroma, Europe, Cytochrome oxidase I (COI), 12S rDNA, 16S rDNA.
Acknowledgements
We are greatly indebted to Scott Brooks (Agriculture and Agri-Food Canada, Ottawa, Canada) who kindly scored the morphological characters in 18 species not included in his 2005 analysis. We also like to thank Scott for the interesting discussions during the start of this study. Thanks are also due to Valeria Rispoli (Zoological Museum, Zurich, Switzerland) for her contribution with laboratory work. We thank Lucia Pollini (Natural History Museum, Lugano, Switzerland) for providing us with additional D. ungulatus and G. helveticus samples, and Mihail Kechev (University of Plovdiv, Plovdiv, Bulgaria) for samples of P. regalis. We are grateful to Christine Lambkin (Queensland Museum, South Brisbane, Australia) for helpful methodological advice. Many thanks are due to two anonymous referees and to Rudolf Meier (National University of Singapore, Singapore) for providing helpful comments on an earlier draft of this paper. This work has been supported by a grant of the Swiss National Science Foundation to M. V. Bernasconi (Grant 3100A0–115981). This paper is situated within the frame of the PhD thesis of one of the authors (Christoph Germann) and is a contribution of the Zoological Museum of the University of Zurich, Switzerland. Finally, we kindly dedicated it to the memory of our dear friend and colleague Paul I. Ward (director of the Zoological Museum, Zurich), who passed away prematurely on April 19, 2008 and can only be described as a source of unconditional support and permanent motivation for our research project.
Aldrich J. M.
(1905) A catalogue of North American Diptera. Smithsonian Miscellaneous Collections 46(2), 1–680.
[Accessed on 29 February 2008.]
Pollet M., Grootaert P.
(1996) An estimation of the natural value of dune habitats using Empidoidea (Diptera). Biodiversity and Conservation 5(7), 859–880.
| Crossref | GoogleScholarGoogle Scholar |
is indicated between square brackets

|


