Verification of the cryptic species Penaeus pulchricaudatus in the commercially important kuruma shrimp P. japonicus (Decapoda : Penaeidae) using molecular taxonomy
K. H. Tsoi A , K. Y. Ma B , T. H. Wu C , S. T. Fennessy D , K. H. Chu C and T. Y. Chan E FA Department of Science and Environmental Studies, The Hong Kong Institute of Education, Tai Po, Hong Kong.
B Molecular Ecology and Evolution Laboratory, School of Marine and Tropical Biology, James Cook University, Townsville, Qld 4811, Australia.
C Simon F. S. Li Marine Science Laboratory, School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong.
D Oceanographic Research Institute, PO Box 10712, Marine Parade, Durban 4056, South Africa.
E Institute of Marine Biology and Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung 20224, Taiwan.
F Corresponding author. Email: tychan@mail.ntou.edu.tw
Invertebrate Systematics 28(5) 476-490 https://doi.org/10.1071/IS14001
Submitted: 3 January 2014 Accepted: 5 June 2014 Published: 13 November 2014
Journal Compilation © CSIRO Publishing 2014 Open Access CC BY-NC-ND
Abstract
The kuruma shrimp Penaeus japonicus Bate, 1888 (Decapoda : Penaeidae) is economically important in the global shrimp market. It was regarded as the only species in the subgenus Marsupenaeus. However, our previous molecular analyses revealed two cryptic species (Forms I and II) in this species complex. In this study, we confirm the phylogenetic relatedness between the two cryptic species; revise their taxonomic status; and review their range distribution. The name Penaeus pulchricaudatus Stebbing, 1914 (with type-locality off the eastern coast of South Africa), previously considered as a junior synonym of P. japonicus, is fixed for Form II through a neotype selection. P. japonicus (Form I) is only confined to the East China Sea (including Japan, its type-locality) and the northern South China Sea. P. pulchricaudatus is widely distributed in the South China Sea, Australia, the Red Sea, the Mediterranean, and the western Indian Ocean. Phylogenetic analysis shows that P. japonicus is genetically homogeneous yet P. pulchricaudatus exhibits a strong phylogeographical structure. The Mediterranean stock of P. pulchricaudatus originated from the Red Sea population, supporting the Lessepsian migration hypothesis. The presence of two closely related cryptic species in the P. japonicus species complex provides important insights into fishery management and aquaculture development.
Introduction
The kuruma shrimp Penaeus japonicus Bate, 1888 (Decapoda : Penaeidae) is widely distributed in Korea, Japan, the South China Sea, the Malay Archipelagoes, the northern coast of Queensland in Australia, the Red Sea and the western Indian Ocean to eastern South Africa (Pérez Farfante and Kensley 1997). It was first recorded in the Mediterranean in 1924 (Balss 1927) and this ‘Lessepsian migrant’ was believed to invade and spread across the Levantine Coast of the Mediterranean Sea via the Suez Canal (Balss 1927; Galil et al. 2002). This shrimp has subsequently been recorded on the Italian coast (Lumare and Casolino 1986), Aegean Sea (Kevrekidis and Kevrekidis 1996) and English Channel (Clark 1990). Two specimens were captured in 2007 from a depth of ~60 m in the Celtic Sea (Quigley et al. 2013) but the occurrence of a self-sustaining population at its northernmost limit is yet to be supported by current evidence (Quigley 2010). However its successful adaptation in the Mediterranean environment (Kevrekidis and Kevrekidis 1996) and rapid propagation (Gilberto and Héctor 2001) threaten the indigenous species (Galil 2006). For example, the native P. kerathurus (Forskal, 1775) has been eliminated by this invasive alien species from the easternmost areas of the Mediterranean (Galil and Zenetos 2002).
Penaeus japonicus is of economic importance in the global shrimp market and highly prized as the ‘King of Seafood’ in Japan (Shigueno 2001). As this shrimp tolerates handling and long-distance transportation (ASEAN 1978), the market price of living individuals could reach US$127 per kg (Tokyo Metropolitan Central Fish Market, 17 May 2014). Shrimp farms are the main market supplier of the shrimp. Global wild-capture of the shrimp is 2165 tonnes yet the aquaculture production has reached 51 178 tonnes with the net value of 303 million US dollars in 2012 (FAO 2014). It was the first penaeid species successfully cultured on a commercial scale (Hudinaga 1935, 1942). Shrimp farming techniques developed in Japan have been well practiced in many areas ranging from the West Pacific (Liao and Chien 1994) to the Mediterranean (Galil and Zenetos 2002) and French Atlantic coast (Quero and Vayne 1998). However the industry has long been challenged by low egg quality and survival rate of the shrimp larvae (Okauchi et al. 1995; Chuntapa et al. 2003). In order to control production cost and to obtain high quality seeds for better survival and growth performance, most hatcheries still rely on the breeders collected from the wild stocks in nearby waters (Benzie 1998; Delmendo 1989; Treece 1999), including those in China (e. g. Wang 1997), Japan (Mock 2009), Australia (DPI 2013) and the Mediterranean (Scordella and Zecca 2002).
Penaeus japonicus is featured with attractive brownish red bands and named as the kuruma shrimp because of its characteristic wheel-like banding pattern. Yet such body colouration pattern is similar to that of P. canaliculatus (Olivier, 1811) and P. kerathurus, and thus misidentifications have been occasionally reported. Morphological traits including the diagnostic pouch-like thelycum and characteristic features (such as its movable spines in the telson) are effective characters to identify the species. This species is currently described by two scientific names P. japonicus and Marsupenaeus japonicus due to the controversy of the status of the genus Penaeus Fabricius, 1798 (see Pérez Farfante and Kensley 1997; Lavery et al. 2004; Flegel 2007, 2008; McLaughlin et al. 2008; Ma et al. 2011). These names are independently used by various organizations and databases, e.g. FAO, World Register Marine Species (WoRM) and Delivering Alien Invasive Species Inventories for Europe (DAISIE). The present work follows the most updated results (i. e. Ma et al. 2011) for the definition of Penaeus and hence uses Penaeus japonicus. For convenient discussion, the subgenera of Penaeus described in Holthuis (1980) are also used.
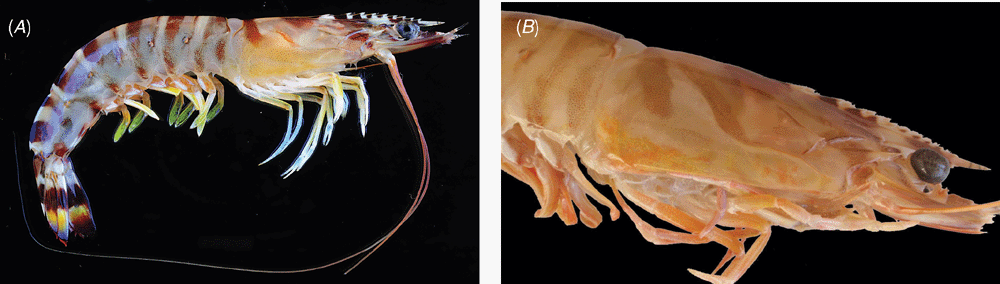
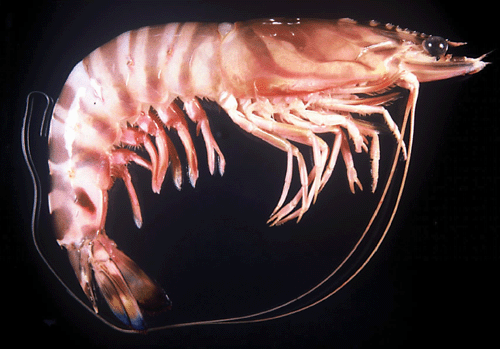
Two morphologically similar colour forms of Penaeus japonicus have been identified in the South China Sea (Tsoi et al. 2005). These two forms, namely I and II, were characterised by diagnostic colour banding patterns on the carapace (see Figs 1, 2), which was generally regarded as a colour variant in previous taxonomic works (Yu and Chan 1986; Chan 1998). These two forms do not differ in other morphological traits or morphometric parameters (Tsoi et al. 2005), yet phylogenetic analyses using mitochondrial (mt) DNA and nuclear DNA markers concordantly reveal that the two forms are closely related (versus other Penaeus spp.) but genetically distinct (Tsoi et al. 2005). No shared mtDNA haplotype was observed among the specimens of these two forms, particularly in their sympatric zone. Hybridization was not detected from AFLP analysis, indicating that the sister taxa are reproductively isolated. All evidence supports the occurrence of cryptic species in the kuruma shrimp. Since the type locality of P. japonicus is Japan, Form I is recognised as the typical form. However, no proper scientific name has yet been given to Form II.
|
In a phylogeographical study of the species beyond the South China Sea, Tsoi et al. (2007) showed that Form I occurs only in Japan and China, including Taiwan, and dominates the populations in the northern South China Sea. This finding was also supported by Tzeng et al. (2004) and Shih et al. (2011), who showed that both forms were found along the Taiwan coast with Form I being dominant. Form II has a much wider range including the South-east Asia, Australia, the western Indian Ocean, the Red Sea and the Mediterranean (Tsoi et al. 2007). Nevertheless, genetic information of specimens from the Red Sea and the western Indian Ocean is lacking in previous studies. The present study thus aims to (1) affirm the identity of Form II and the taxonomic status of the cryptic species in the species complex; (2) ascertain the comprehensive distribution pattern of the species complex and (3) elucidate the phylogenetic relationship of the stocks across its geographical range by extending our sampling effort.
Materials and methods
Sample collection and DNA extraction
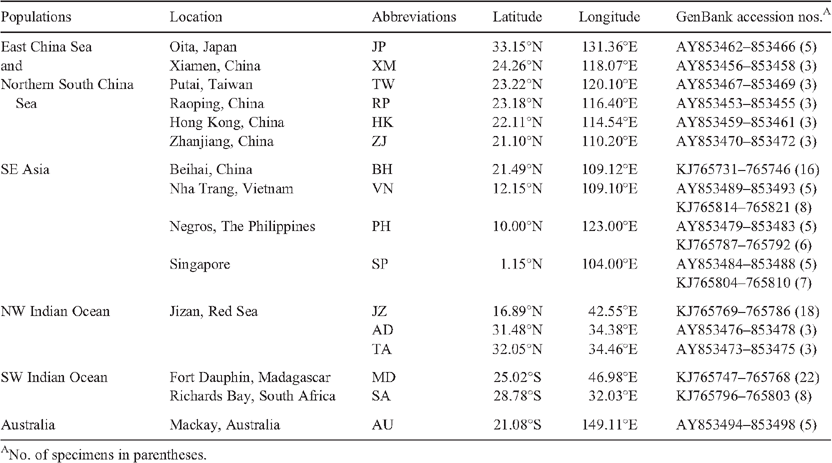
A total of 131 individuals were collected from 16 localities (Fig. 3) including Oita of Japan (JP), Putai of Taiwan (TW), Xiamen (XM), Raoping (RP), Hong Kong (HK), Zhanjiang (ZJ) and Beihai (BH) in China, Negros of the Philippines (PH), Singapore (SP), Nha Trang of Vietnam (VN), Mackay of Australia (AU), Fort Dauphin of Madagascar (MD) and Richards Bay of South Africa (SA), Jizan (JZ) in the Red Sea, and Tel Aviv (TA) and Ashdod (AD) in the Mediterranean (Table 1). Specimens from BH, MD, JZ and SA were collected for the present study while those from the other localities were from our previous study (Tsoi et al. 2007). Specimens were collected from shrimp trawlers operating in the relevant areas, or from nearby fish markets. The shrimps were confirmed to be caught in coastal waters of the corresponding areas and were not imported or obtained from shrimp farms. The specimens were either preserved in 95% ethanol or kept frozen at –70°C before DNA extraction.

|
|
DNA extraction, PCR and sequencing
The total genomic DNA of each shrimp specimen was extracted from the muscle (10–15 mg) of its pleopod IV or V using QIAamp Tissue Kit (QIAGEN). The extracted DNA was kept at −20°C until analyses. The primers PCR−1R and 12S−2 (Chu et al. 2003) were adopted to amplify a 5ʹ segment of the mitochondrial control region (CR) (563 bp). PCR amplification was conducted in a reaction mixture containing 10 ng template DNA, 10 μL Mg2+ free PCR buffer, 2.5 mm MgCl2, 0.12 μM of each primer, 400 μM of dNTPs, 1 unit of Taq polymerase (Promega, Madison, Wisconsin), and ddH2O added up to 50 μL. The PCR thermal cycling profile was as follows: 93°C (3 min), 35 cycles of 93°C (30 s)/48°C (50 s)/72°C (50 s), and 72°C (5 min). The size and quality of amplified DNA fragments were assessed in 1% agarose gel electrophoresis. Prior to DNA sequencing, PCR products were purified using QIAquick gel purification kits (QIAGEN). Bi-directional sequencing was conducted using the corresponding primer sets and ABI PRISM dye-terminator sequencing kits (Applied Biosystems, Foster City, California) with an ABI 3100 automated DNA Sequencer.
Phylogenetic and population genetic analyses
Sequences were aligned using MUSCLE (Edgar 2004) implemented in MEGA v5. 0 (Tamura et al. 2011) with manual adjustments. Phylogenetic relationships among the individuals were analysed using Bayesian inference (BI) carried out in a BEAST v1. 7. 4 (Drummond et al. 2012) substitution model selected by Modeltest2 based on BIC (Darriba et al. 2012). A run of 50 million MCMC generations, sampled every 5000 generations, was conducted using the Yule process tree prior and strict clock model. Tracer v1.5 was used to evaluate convergence of runs (Rambaut and Drummond 2007) and the trees were annotated by using Tree Annotator v1.7.4 (Drummond et al. 2012). Analysis of molecular variance (AMOVA) in Arlequin 3. 11 (Excoffier et al. 2005) was adopted to assess the geographic division among the populations and the significance was determined by 1000 random permutations of sequences. Minimum spanning networks (MSNs) (Rohlf 1973) were constructed. The MSNs of haplotypes were computed in Arlequin 3. 1 (Excoffier et al. 2005). Using the same package, pairwise population FST values (Nei 1977) with the significance of genetic distances tested by 10 000 permutations were calculated. A neutrality test was performed to calculate the Tajima’s D, and Fu & Li’s D values for detecting the occurrence of bottlenecks (or population expansion) and allelic deficiency.
Results
Genetic analyses
Nucleotide composition The aligned partial sequences of CR were deposited with GenBank (Table 1). On average 508 bp of CR fragments were unambiguously determined. The aligned partial CR segment revealed 238 variable and 206 phylogenetically informative sites. The base frequencies were A: 37. 1; %, T: 44. 3%, C: 10.6% and G: 7. 9%. Sequences of CR showed strong AT bias (81. 4%).
Phylogenetic analyses
The best-fit model for the CR was GTR+I+G. The P. japonicus haplotypes are separated into two lineages corresponding to Form I and Form II with very strong support (Fig. 4). Form I was found only in the East China Sea and the northern South China Sea, including specimens from Japan (JP), Xiamen (XM), Taiwan (TW), Hong Kong (HK), Raoping (RP), and Zhanjiang (ZJ). They were collectively defined as the East China Sea clade. Four clades were defined in Form II. The South-east (SE) Asian clade included specimens from Beihai (BH), Vietnam (VN), the Philippines (PH) and Singapore (SP). Yet four specimens from the SE Asia (2 PH, 1 BH and 1 SP) were found to be more closely related to the North-west (NW) Indian Ocean clade, which contained all specimens from the Mediterranean Sea and the Red Sea with the two being reciprocally monophyletic. The South-west (SW) Indian Ocean clade (including the Madagascar and South African populations) exhibited a close relationship with another genetically distinct Australian clade (only comprising the Mackay specimens).

|
Genetic distances and population differentiation
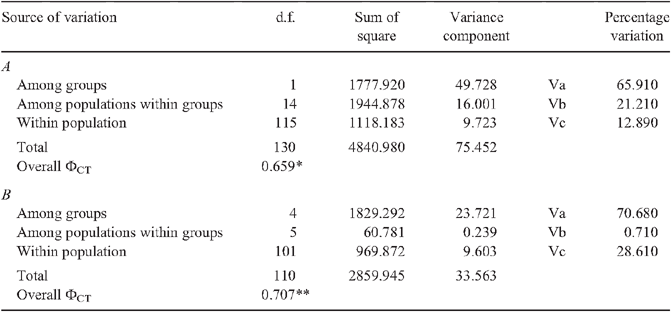
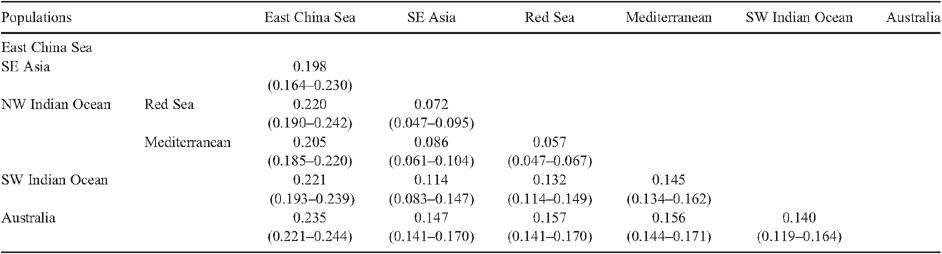
The relative frequency distribution of the clades in each sampling site based on BI analysis of the CR sequence data is presented in Fig. 3. The Kimura 2-parameter distance matrices (K2P) of CR sequence data are shown in Table 2. Mean genetic distance of the CR ranged from 0.057 to 0.235. High genetic divergence supported the genetic differentiation of the two forms (ranging between 0.164 and 0.244), as well as the clades within the Form II (0.047, 0.171). The high and significant FST (0.793, 0.906, P < 0.00001) (Table 3), AMOVA and ΦCT (0.659, P < 0.05) (Table 4) also confirmed that the East China Sea clade (Form I) was genetically distinct from the populations of other regions. This clade was genetically homogeneous with low and insignificant FST (<0.139, P > 0.05), while genetic differentiation was detected among Form II populations, as revealed in FST and AMOVA (Tables 3, 4). The Australian clade was most genetically diverged from the others (FST = 0.707–0.923, P < 0.05). Other genetically distinct clades exhibited relatively lower FST values (FST = 0.488–0.881, P < 0.001). The phylogenetic results revealed a close genetic relationship between the populations of the Red Sea and the Mediterranean in the NW Indian Ocean clade but indicated no recent gene flow between the two regions (FST = 0.784, P < 0.001). The SW Indian Ocean clade was genetically homogeneous with an insignificant FST value (P > 0.05). No genetic drift or bottleneck was detected in any of the populations as supported by insignificant values of Tajima’s D, and Fu & Li’s D (Table 5). Yet a significant and negative value in the SW Indian Ocean population revealed the occurrence of population expansion (Fu & Li’s D = –2.616, P < 0.05).
|

|

|
Minimum spanning networks
CR haplotypes were nested according to the forms to which they belonged and the geographical localities. Haplotypes 01–20 and 21–128 were categorized into Forms I and II, respectively (Fig. 5). These two major assemblages were discriminated by 74 mutational events. Form II was further separated into four clades – the Australian, NW and SW Indian Ocean clades appeared to have originated from the South-east Asian clade. The Australian clade was the most distantly related to the other Form II clades, as separated by more than 53 mutational steps. The NW Indian Ocean clade was separated from the South-east Asian clade by 22 mutational steps. Within the NW Indian Ocean clade, the Mediterranean subclade was split from the Red Sea subclade by 22 steps as well. No haplotype was shared among the Mediterranean and Red Sea subclades.

|
Discussion
Cryptic species differentiation
Cryptic diversity
Our previous studies have identified two genetically distinct colour forms of P. japonicus and revealed their different geographical distributions (Tsoi et al. 2005, 2007). The present study extends the analysis of the CR dataset to new populations, further supporting the occurrence of cryptic species. Significant FST and the high genetic divergence (74 mutational events) support that these two genetically distinctive lineages are phylogenetic species. In addition, Tsoi et al. (2005, 2007) did not perceive any shared composite haplotypes between the two forms in all mtDNA markers used. Using AFLP, hybridization is also not detected across the forms collected from the sympatric zone in AFLP diploid nuclear loci, supporting our contention that the two reproductively isolated forms also likely represent biological species.
Studies from other research groups also support the occurrence of two genetically distinct taxa in the South China Sea (Shih et al. 2011; Tan et al. 2009). However, Tan et al. (2009) reported a new clade from Sanya of China. This new clade was claimed to be genetically diverged from Forms I and II at the level of 8% in the 16S rRNA marker. Our previous studies show that genetic divergences of the two forms are only 1–1.5% for 16S rRNA and 6–7% for a more variable marker COI (Tsoi et al. 2005, 2007). Such a high level (8%) of divergence in the conserved 16S marker is rarely observed in cryptic species delineation of penaeid shrimps. The highest value of the sequence divergence in 16S rRNA data among various Penaeus spp. in Melicertus Rafinesque-Schmaltz, 1814 is 7.3% (Lavery et al. 2004). The 16S rRNA divergence between P. vannamei Boone, 1931 and P. subtilis (Pérez Farfante, 1967) attains 8. 2% yet these species belong to two different subgenera Farfantepenaeus Burukovsky, 1972 and Litopenaeus Pérez Farfante, 1969, respectively (De Francisco and Galetti 2005). Our BLAST search of the shrimp sequences (EU056320–22 and EU056324) of the ‘new clade’ reported by Tan et al. (2009) shows that it probably represents P. semisulcatus De Haan, 1844 as its nucleotide alignment achieved 99% of the sequence identity to P. semisulcatus, whereas the highest value only reaches 93% in comparing with sequences of P. japonicus obtained from other studies. Therefore the presence of one more cryptic species (other than Forms I and II) of P. japonicus proposed by Tan et al. (2009) is most likely due to misidentification and cannot be substantiated.
Taxonomy
As the type-locality of P. japonicus is Japan, this name should be applied to Form I, i. e. the East China Sea Clade. For Form II, the name P. pulchricaudatus Stebbing, 1914, with type-locality from the Great Fish Point off the eastern coast of South Africa, is available. However, P. pulchricaudatus was known only from the juvenile holotype of ~45 mm body length (Stebbing 1914). As the holotype is a very small juvenile, its sex and exact identity are undetermined. Stebbing (1914) stated that P. pulchricaudatus is nearest to P. japonicus and indeed it resembles the latter in having only one ventral rostral tooth and with the telson laterally spinose. However, in the SW Indian Ocean, Penaeus latisulcatus Kishinouye, 1896 (or P. latisulcatus hathor Burkenroad, 1959) also has one ventral rostral tooth and a laterally spinose telson, and juveniles of P. japonicus and P. latisulcatus cannot be satisfactorily separated without the information of colouration and genetic data (see key provided in Chan 1998). Moreover, the original description and figures provided by Stebbing (1914) indicated that there are eight pairs of lateral spines (four pairs larger) on the telson, a character not known in any other Penaeus s. l. species (i. e. having only 0–3 pairs of lateral telson spines). We examined two small juvenile Form II specimens from Madagascar (7.2, 8.4 mm carapace length, corresponding to ~25, 30 mm body length, NTOU M017512) and they are generally similar to Stebbing’s P. pulchricaudatus but with only three pairs of lateral spines on the telson. Unfortunately, the holotype of P. pulchricaudatus (Iziko South African Museum, A 1056) is now lost (L. Hoenson, W. K. Florence, person. comm.) and its exact identity may never be affirmed. In order to fix the name for Form II, a neotype of a South African female specimen used in the present genetic analysis and with coloured photograph (Fig. 2B) is now selected for P. pulchricaudatus. Therefore, while Form I should be treated as the true P. japonicus, Form II now bears the name P. pulchricaudatus. Below is a taxonomic account for the neotype of P. pulchricaudatus.
Taxonomy
Penaeus pulchricaudatus Stebbing, 1914
Material examined
Neotype. South Africa, Richards Bay, 28°72ʹS, 32°26ʹE, 30 m, 20 March 2010, female, carapace length 44.2 mm, NTOU M01779 (deposited at the National Taiwan Ocean University, Keelung), COI GenBank accession number KJ765796.
Description
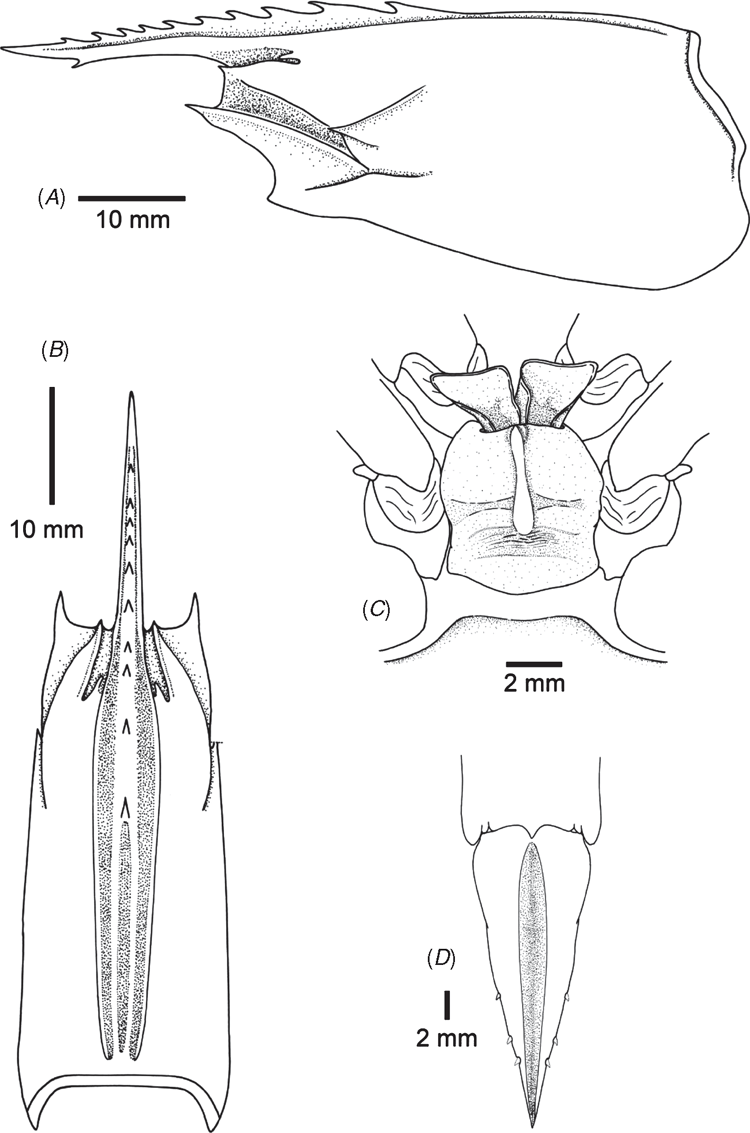
Integument glabrous. Carapace with ridges and grooves well developed but lacking longitudinal and transverse sutures (Fig. 6A, B). Rostrum, without distinct accessory ridge, extending to distal end of antennular peduncle and armed with 9 dorsal teeth (including 3 teeth on carapace) and 1 ventral tooth. Postrostral carina almost reaching posterior carapace and with deep median groove throughout entire length. Adrostral groove as wide as postrostral carina and extending near to posterior carapace. Orbital angle spiniform and antennal spine very pronounced. Postocular sulcus absent. Gastrofrontal groove distinct and with posterior end divided into two. Gastro-orbital carina long, almost reaching anterior margin of carapace. Orbito-antennal groove well defined. Cervical carina sharp, accompanying groove well marked. Hepatic spine very pronounced. Hepatic carina well marked, curved and with anterior part ventrally inclined. Pterygostomian spine and branchiocardiac carina absent. First pereiopod with ischial spine indistinct or absent. Abdominal somite VI bearing 3 cicatrices, lacking dorsolateral groove. Thelycum (Fig. 6C) pouch-like with double tubes and opened anteriorly. Spermatophore deposited on thelycum as a large subtriangular wing-like process with anteriorly disposed broad base and posteriorly long curved dorsal rod inserted into the tube of thelycum. Telson armed with 3 pairs of movable lateral spines (Fig. 6D).
Colouration (Fig. 2B)
Body pale yellowish and crossed with dark brown transverse bands, those on carapace extending from top to about mid-carapace, posterior-most band on abdominal somite VI interrupted. Eyes black-brown. Scaphocerite somewhat greenish with white tips, antennal flagella reddish-brown to yellowish-brown. Pereiopods whitish to yellowish. Pleopods yellowish to reddish, with brown and white spots at bases. Distal part of uropods with patch of bright yellow, followed by narrower patch of bright blue and with margins reddish.
Remarks
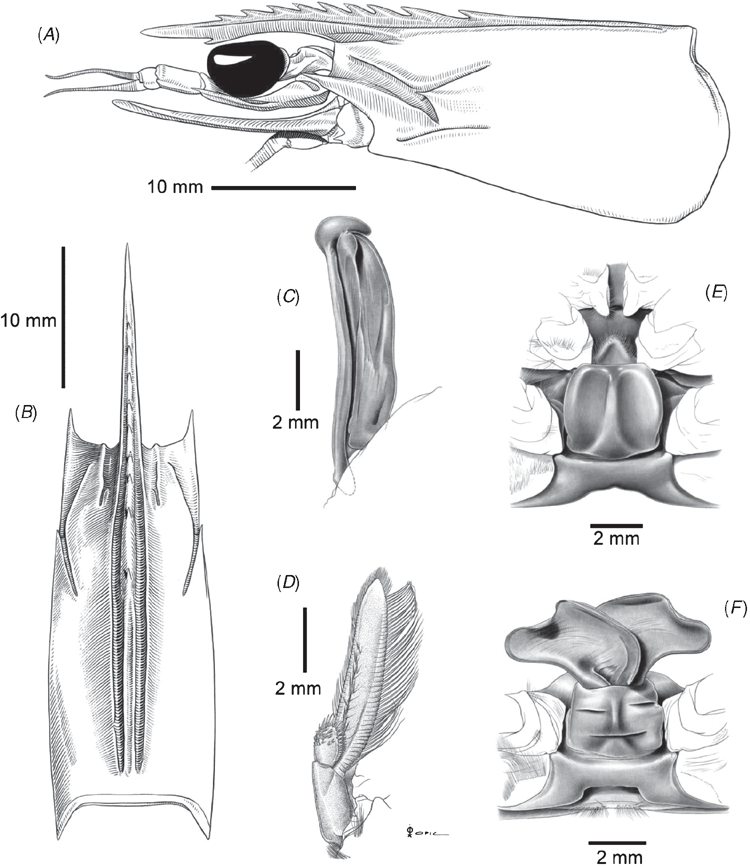
Since materials from Madagascar and South Africa belong to the same genetic group (Figs 4, 5), additional illustrations for P. pulchricaudatus based on Madagascan specimens are provided in Fig. 7 (specimens deposited at the Muséum national d’Histoire naturelle, Paris but could not be located during the present study). Although no male from the topotypic locality eastern coast of South Africa was examined, males from the adjacent locality Madagascar shows that the petasma of this species is symmetrical, semiclosed, and with long distomedian projection overhanging the distal margin of costae (Fig. 7C). The appendix masculina is subelliptical, armed with strong marginal spines and a broad patch of short strong spines (Fig. 7D). As discussed in the Introduction and Tsoi et al. (2005), P. pulchricaudatus is almost identical to P. japonicus and only different in the colouration of the ventrolateral carapace (Figs 1, 2), which has the dark brown transverse bands on the dorsal carapace terminating at mid-carapace and not extending further downwards.
Phylogenetic analysis and population structure
Geographical distribution
Penaeus japonicus is not widely distributed. The previous description ‘P. japonicus is extensively spread across the Indo-West Pacific’ (e. g. Dall et al. 1990; Pérez Farfante and Kensley 1997; Chan 1998) is no longer applicable. Penaeus japonicus represented by Form I is confined to the East China Sea and the northern South China Sea. The species is genetically homogeneous without any significant population structuring. Genetic analyses of specimens along the coasts of mainland China and Taiwan also support the lack of genetic structure of P. japonicus, based on both mtDNA (Shih et al. 2011) and nuclear markers (Chu et al. 2011). Insignificant population differentiation was also observed in Japanese waters and attributed to high levels of gene flow among the localities (Sugaya et al. 2002; Taniguchi and Han 1989).
South-east Asian origin
Penaeus pulchricaudatus is geographically distributed in the South China Sea, Australia and the western Indian Ocean. Phylogenetic analysis shows a strong population structuring. Yet all these clades exhibit a close phylogenetic relationship with the South-east (SE) Asian clade (Fig. 5). Our data reveal a recent population expansion of the species complex. Population clusters of both P. pulchricaudatus and P. japonicus arrange in a star-like pattern radiating from the centrally located South-east Asian cluster in the CR evolution network. The network reveals that the Australian lineage originated from a haplotype 23 (from Nha Trang, Vietnam in SE Asian clade) which is linked to the haplotype 33 (from Singapore in SE Asian clade). Furthermore the observation that some SE Asian individuals (haplotypes 21, 22, 30 and 31) nested within the NW Indian Ocean clade in the BI cladogram (Fig. 4) indicates that the clade likely had SE Asian origin. It is interesting to note that the haplotype 21 exhibits the closest relationship with P. japonicus. Based on the current information, we believe the haplotype 33 and its closely related haplotypes are relatively primitive and associated with the species delineation and population differentiation of this species complex. Our findings on CR evolution network and the highest nucleotide diversity of the SE Asian cluster provide evidence to support that P. japonicus and the lineages of P. pulchricaudatus were derived from the SE Asian region. The mud crab Scylla serrata (Forsskål, 1755) also exhibit the comparable phylogeographic structure and genetically-differentiated geographic clusters in the Indo-West Pacific which were believed to be associated with the seawater level fluctuation and paleo-oceanographic conditions in the Pleistocene (Gopurenko et al. 1999; He et al. 2011). Geographical barriers emanating from land bridges, historic glacial patterns, and oceanic currents would probably have affected the phylogeographic distribution of P. pulchricaudatus and P. japonicus (see Tsoi et al. 2007). The westward dispersal of the ancestral shrimp might be facilitated by the South Equatorial Current across long distance. Yet the evolutionary origins of these two decapods were found to be different, since ancestral S. serrata was originated from the North-west Australia (He et al. 2011) whereas the P. japonicus complex was likely diversified from SE Asia.
Lessepsian migration
‘Penaeus japonicus’ is considered as an invasive alien species (e.g. Galil and Zenetos 2002; Galil 2006) and regarded as one of the 660 Lessepsian migrants (Galil 2012). Also the shrimp is in the list of the ‘top 100 worst invasive alien species’ in the Mediterranean because of its serious impacts on biodiversity and socioeconomic aspects (Streftaris and Zenetos 2006). Yet not a single individual of P. japonicus was found in either the Red Sea or the Mediterranean populations as all our specimens are P. pulchricaudatus. Available photographs and illustrations of ‘P. japonicus’ in the Mediterranean actually represent P. pulchricaudatus (e. g. Otero et al. 2013; p. 81). Thus, the invasive alien species is P. pulchricaudatus, not P. japonicus.
As all our phylogenetic trees demonstrate the close genetic relatedness between the Red Sea and Mediterranean populations, the Mediterranean stocks are believed to have originated from the Red Sea. The founder stock is thus expected to be genetically homogeneous to its source. But in fact, all haplotypes are found to be private in both populations. Moreover, the significant and high pairwise FST values reveal no recent gene flow between these two populations, which are separated by at least 22 mutational steps. Such a differentiation level is equivalent to the divergence between the SE Asian and the Red Sea clades. All the evidence reveals that these two stocks are genetically distinct and the vicariance did not appear to be a very recent event. Referring to the calibrated divergence rate at 19%/Myr for the CR marker (McMillen-Jackson and Bert 2003), the Mediterranean and Red Sea stocks of P. pulchricaudatus are estimated to be isolated from each other at 150 kya, which is much longer than the time elapsed after the establishment of Mediterranean dispersal via the Suez Canal (<150 years). However, such a phenomenon is not unusual in the region. The mussel Brachidontes pharaonis (P. Fischer, 1870) also exhibits a similar lineage divergence in the Mediterranean and Red Sea (Shefer et al. 2004). Sirna Terranova et al. (2006) further suggested the occurrence of ancient polymorphism of this mussel in the region to explain the high genetic divergence. A question is thus raised on these findings: are the data incompatible with the Lessepsian migration hypothesis for explaining the occurrence of P. pulchricaudatus in the Mediterranean?
In fact, private alleles or haplotypes are not uncommon in the Mediterranean pool in various marine fauna e. g. the mussel B. pharaonis mentioned above (Shefer et al. 2004) and the goatfish Upeneus pori Ben-Tuvia and Golani, 1989 (Golani and Ritte 1999). A low level of genetic divergence in the goatfish populations between the Red Sea and the Mediterranean shows good support for the Lessepsian migration hypothesis, yet a rare and single allele undetected in the Red Sea was unexpectedly found in the Mediterranean population (Golani and Ritte 1999). The situation of P. pulchricaudatus may be similar in that these private Mediterranean haplotypes may occur in the Red Sea but are absent in our studied samples. Our Red Sea specimens were only collected from the southern part of the Red Sea (Jizan) which is distant from the Suez Canal. It is thus possible that some Mediterranean haplotypes were missed in a single sampling exercise. In fact, genetic distinctiveness of the populations between the northern and southern populations of the Red Sea has been observed in some organisms, e. g. the Red Sea fishes Saurida undosquamis (Richardson, 1848) (Yağlıoğlu and Turan 2012) and Larabicus quadrilineatus (Rüppell, 1835) (Froukh and Kochzius 2007). Our data thus only provide the genetic information of a southern Red Sea population but not from the entire region. Thus, the migration hypothesis could not be rejected.
Aquaculture implications
Since the Japanese seed stock (i. e. P. japonicus) was introduced to Italy in 1979, the shrimp culture technique has been developed continuously in the Mediterranean (Lumare and Palmegiano 1980; Lumare 1984, 1987, 1998; Türkmen 2007). The kuruma shrimp aquaculture is widespread and even extended to the European coast recently (Türkmen 2007). However most hatcheries still depend upon the breeders collected from nearby wild stocks (Treece 1999; Scordella and Zecca 2002). We believe the farmed stocks might possibly contain P. pulchricaudatus. It is noteworthy that an exceptionally high degree of heterozygosity was detected in stocks of ‘P. japonicus’ that had been ranched and recollected from the lagoon of Lesina (De Matthaeis et al. 1983). Sbordoni et al. (1986) also detected peculiar alleles and an unexpectedly high FST in the introduced hatchery samples. The study mentioned that ‘The genetic structure of the new Japanese stock was consistently different from that of the F1 introduced into Italy’ (Sbordoni et al. 1986: 243), and thus the introduced shrimps were suspected as originating from different geographic localities. Furthermore, a photograph of shrimps from an Italian shrimp farm (http://www.apulashrimp.it/Sito/English/Penaeus_japonicus_eng.htm) shows that it is P. pulchricaudatus. All these scenarios raised uncertainty as to which species, P. japonicus or P. pulchricaudatus is cultured in Italian or other Mediterranean kuruma shrimp farms. A comprehensive survey on the shrimp cultured in different farms of the region is necessary to resolve this issue.
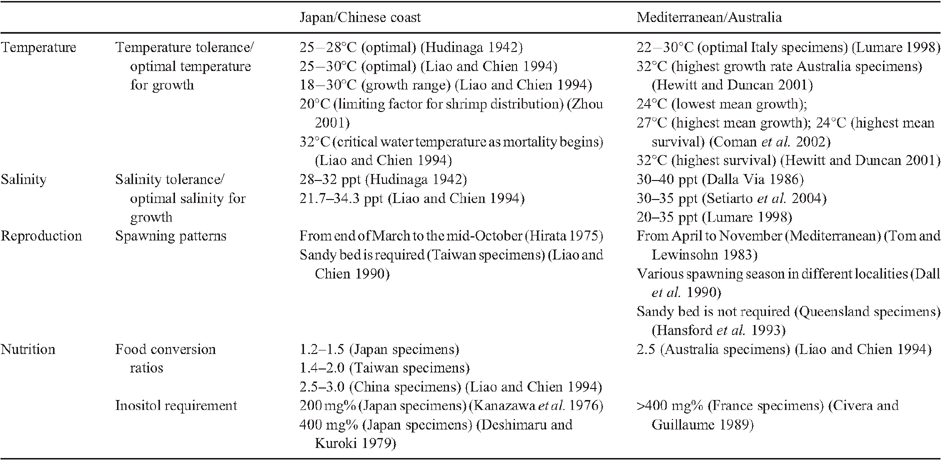
It appears that previous studies on the biology of the kuruma shrimp were undertaken on different species within the species complex. Incoherent descriptions of the environmental conditions required by the shrimp collected from various regions are noted in relevant literature reviews (Table 6). For instance, different optimal temperatures for growth and survival of the shrimp were recorded: 25−28°C from Japan (Hudinaga 1942), 25−30°C (Liao and Chien 1994) from Taiwan; 22−30°C (Lumare 1998) from Italy and 32°C from Australia (Hewitt and Duncan 2001). According to the geographic distribution of the two species, we presume that the first two studies were on P. japonicus or a mixture of the two species, whereas the last two were on P. pulchricaudatus. This vague identity of the species studied may inevitably induce confusion and uncertainty in aquaculture practices. For example, Savini et al. (2008: 61) described the European alien kuruma shrimp as very resistant as ‘cold temperatures are not such a problem for its farming (minimum 5°C)’, yet Zhou (2001) stated that the kuruma shrimp never occurs in regions with sea surface temperature below 20°C in the South China Sea and the water temperature is regarded as the limiting factor affecting its distribution. Furthermore, Quinitio et al. (1991) observed the kuruma shrimp in the Philippines exhibits a faster moulting rate than the Japanese shrimp (Shigueno 1975). Also a sandy bed is suggested as an important factor for ovarian maturation and reproductive success for female shrimp in Taiwan (Liao and Chien 1990), yet the substratum nature is considered not important for females from Australia (Hansford et al. 1993). All these findings suggest that P. japonicus and P. pulchricaudatus may have different environmental requirements and reproductive traits. Further elucidation of the biology of these two species is essential for improving the species-specific culturing techniques in the shrimp farming industry.
|
Conclusion
The kuruma shrimp is a species complex consisting of two sister species: Penaeus japonicus and P. pulchricaudatus. Penaeus japonicus is confined to the East China Sea and the northern South China Sea, whereas P. pulchricaudatus is widely distributed in the South China Sea, Australia, the western Indian Ocean and the Red Sea. Penaeus japonicus is genetically homogeneous, whereas P. pulchricaudatus exhibits a strong phylogeographical structure, and probably originated and differentiated from South-east Asia. Our study shows that the invasive Mediterranean population is P. pulchricaudatus, which entered the Mediterranean through Lessepsian migration from the Red Sea. Further studies on the biological differences of these two cryptic species are necessary for providing information on their aquaculture.
Acknowledgments
Sincere thanks are given to P. O. Ang Jr. C. C. Cheang, Y. H. Yung (The Chinese University of Hong Kong), S. A. Lehnert (CSIRO Division of Livestock Industries, Australia), P. K. L. Ng (National University of Singapore), J. Primavera (SEAFDEC Aquaculture Department, the Philippines), K. Tanaka (Kyorin University, Japan), K. T. Tai (University of Fishery, Vietnam), Z. Y. Wang (Jimei University, China), J. Wong and P. H. Wong (Princess Margaret Hospital, Hong Kong) for collecting specimens. The material from Madagascar for genetic and morphology analysis were collected during the ATIMO VATAE expedition founded by the Total, Prince Albert II of Monaco, and Stavros Niarchos foundations, and conducted by the Muséum national d’Histoire naturelle, Paris (MNHN) and Pro-Natura International. Grateful acknowledgements are extended to these institutions, foundations and the expedition leader P. Bouchet for making the present study possible. We sincerely thank A. Crosnier (ex-MNHN), for allowing us to use his line-drawings prepared by M. Opic (ex-ORSTOM) on the Madagascan specimens; C. C. Lin of the National Taiwan Ocean University for preparing the line-drawings of the neotype. We are indebted to C. P. Li (The Chinese University of Hong Kong), and S. Y. K. Chan (The Hong Kong Institute of Education) for providing technical support. We also like to thank P. A. Tanner (The Hong Kong Institute of Education) for language editing, and anonymous reviewers for constructive comments on the manuscript. This work was supported by grants from the Ministry of Science and Technology Taiwan, R. O. C. to T. Y. Chan and the Research Grants Council, Hong Kong Special Administrative Region (HKSAR), China (Project No. CUHK4157/01M) to K. H. Chu. The experiments complied with the current laws of HKSAR.
References
ASEAN, Association of Southeast Asian Nations (1978). ‘Manual on pond culture of penaeid shrimp. (UNDP: FAO). Available at http://www.fao.org/docrep/field/003/ac006e/ac006e00.htm [Verified September 2014]Balss, H. (1927). Berichte uber die Crustacea Decapoda (Natantia und Anomura). Zoological results of the Cambridge expedition to the Suez Canal, 1924, XIV. Transactions of the Zoological Society of London 22, 221–230.
| Berichte uber die Crustacea Decapoda (Natantia und Anomura). Zoological results of the Cambridge expedition to the Suez Canal, 1924, XIV. Crossref | GoogleScholarGoogle Scholar |
Benzie, J. A. H. (1998). Penaeid genetics and biotechnology. Aquaculture 164, 23–47.
| Penaeid genetics and biotechnology. Crossref | GoogleScholarGoogle Scholar |
Chan, T. Y. (1998). Shrimps and prawns. In ‘The living marine resources of the western central Pacific, Vol. 2. FAO Species identification guide for fishery purposes. (Eds K. E. Carpenter and V. H. Niem) (FAO: Rome).
Chu, K. H., Li, C. P., Tam, Y. K., and Lavery, S. (2003). Application of mitochondrial control region in population genetic studies of the shrimp Penaeus. Molecular Ecology Notes 3, 120–122.
| Application of mitochondrial control region in population genetic studies of the shrimp Penaeus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXivVyitrk%3D&md5=5b06f713d4d2237c0a9fb2c312fc1273CAS |
Chu, T. J., Huang, H. L., Shih, C. H., Lin, F. J., and Tzeng, T. D. (2011). Population structure and expansion of kuruma shrimp (Penaeus japonicus) in the adjacent waters of Taiwan inferred from intron sequences. African Journal of Biotechnology 10, 16994–17009.
| 1:CAS:528:DC%2BC3MXhs1ensb7K&md5=d65084537d8c86b07c8a228e03ab9ec7CAS |
Chuntapa, B., Powtongsook, S., and Menasveta, P. (2003). Water quality control using Spirulina platensis in shrimp culture tanks. Aquaculture 220, 355–366.
| Water quality control using Spirulina platensis in shrimp culture tanks. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXitFagu7w%3D&md5=de031896a89f343e9cbb57a59eec88ffCAS |
Civera, R., and Guillaume, J. (1989). Effect of sodium phytate on growth and tissue mineralization of Penaeus japonicus and Penaeus vannamei juveniles. Aquaculture 77, 145–156.
| Effect of sodium phytate on growth and tissue mineralization of Penaeus japonicus and Penaeus vannamei juveniles. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXkt1anurY%3D&md5=76988789f8c4f4f51f681a50f291aa7fCAS |
Clark, P. F. (1990). Asian prawns go wild in the Channel. New Scientist 125, 30.
Coman, G., Crocos, P., Preston, N., and Fielder, D. (2002). Effect of genotype-environment interaction on the survival and growth of the kuruma shrimp Penaeus japonicus. Aquaculture 204, 197–198.
Dall, W. Hill, J. Rothlisberg, P. C., and Staples, D. J. (1990). The biology of Penaeidae. In ‘Advances in marine biology, Vol. 27’. (Eds J. H. S. Blaxter and A. J. Southward.) (Academic Press: New York.)
Dalla Via, G. J. (1986). Salinity responses of the juvenile penaeid shrimp Penaeus japonicus: I. Oxygen consumption and estimations of productivity. Aquaculture 55, 297–306.
| Salinity responses of the juvenile penaeid shrimp Penaeus japonicus: I. Oxygen consumption and estimations of productivity. Crossref | GoogleScholarGoogle Scholar |
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest2: more models, new heuristics and parallel computing. Nature Methods 9, 772.
| jModelTest2: more models, new heuristics and parallel computing. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFWmsbfP&md5=3cacc445e0743c5953b436bc2cf6a350CAS | 22847109PubMed |
De Francisco, A. K., and Galetti, P. M. J. (2005). Genetic distance between broodstocks of the marine shrimp Litopenaeus vannamei (Decapoda, Penaeidae) by mtDNA analyses. Genetics and Molecular Biology 28, 258–261.
| Genetic distance between broodstocks of the marine shrimp Litopenaeus vannamei (Decapoda, Penaeidae) by mtDNA analyses. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpslGgtLo%3D&md5=500085db182d3f7f8382497064f13eedCAS |
De Matthaeis, M., Allegrucci, G., Caccone, A., Cesaroni, D., Cobolli Sbordoni, M., and Sbordoni, V. (1983). Genetic differentiation between Penaeus kerathurus and P. japonicus (Crustacea, Decapoda). Marine Ecology Progress Series 12, 191–197.
| Genetic differentiation between Penaeus kerathurus and P. japonicus (Crustacea, Decapoda). Crossref | GoogleScholarGoogle Scholar |
Delmendo, M. N. (1989). ‘Some advances attained in shrimp farming research and management practices: Insights to future prospects for expansion of production. (FAO: UN.)
Deshimaru, O., and Kuroki, K. (1979). Requirement of prawn for dietary thiamine, pyridoxine, and choline chloride. Bulletin of the Japanese Society of Scientific Fisheries 45, 363–367.
| Requirement of prawn for dietary thiamine, pyridoxine, and choline chloride. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE1MXktVChs7s%3D&md5=4978fcd95057a03ca19d8f5a7128f943CAS |
DPI, Department of Primary Industry (2013). ‘Prawns aquaculture prospects. Available at: http://www.dpi.nsw.gov.au/fisheries/aquaculture/publications/species-saltwater/prawns [verified September 2014]
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1. 7. Molecular Biology and Evolution 29, 1969–1973.
| Bayesian phylogenetics with BEAUti and the BEAST 1. 7. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFagu7fO&md5=beec25ccf0d2a98903aa512fd45babe5CAS | 22367748PubMed |
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797.
| MUSCLE: multiple sequence alignment with high accuracy and high throughput. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXisF2ks7w%3D&md5=0f644003e0450eab42e285b06371fbb5CAS | 15034147PubMed |
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin version. 3. 0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1, 47–50.
| 1:CAS:528:DC%2BD28XjsFSltg%3D%3D&md5=0bca636798b72e718d584cd640b76be0CAS |
FAO, Food and Agriculture Organization of the United Nations (2014). ‘Species fact sheets Penaeus japonicus (Bate, 1888). Available at: http://www.fao.org/fishery/species/2584/en [verified September 2014]
Flegel, T. W. (2007). The right to refuse revision in the genus Penaeus. Aquaculture 264, 2–8.
| The right to refuse revision in the genus Penaeus.Crossref | GoogleScholarGoogle Scholar |
Flegel, T. W. (2008). Confirmation of the right to refuse revision in the genus Penaeus. Aquaculture 280, 1–4.
| Confirmation of the right to refuse revision in the genus Penaeus.Crossref | GoogleScholarGoogle Scholar |
Froukh, T., and Kochzius, M. (2007). Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Molecular Ecology 16, 1359–1367.
| Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXlt1Gmur4%3D&md5=ba1f43ffc0ae9c6436e7f6fcd1a5e699CAS | 17391261PubMed |
Fu, Y. X., and Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics 133, 693–709.
| 1:STN:280:DyaK3s3gt1Crsw%3D%3D&md5=77d036e2d8d2f79033fb6256afd4960fCAS | 8454210PubMed |
Galil, B. S. (2006). ‘Marsupenaeus japonicus. Delivering Alien Invasive Species Inventories for Europe (DAISIE). Available at: http://www.europe-aliens.org/pdf/Marsupenaeus_japonicus.pdf.
Galil, B. S. (2012). Truth and consequences: The bioinvasion of the Mediterranean Sea. Integrative Zoology 7, 299–311.
| Truth and consequences: The bioinvasion of the Mediterranean Sea. Crossref | GoogleScholarGoogle Scholar | 22938526PubMed |
Galil, B. S. and Zenetos, A. (2002). A sea change exotics in the eastern Mediterranean. In ‘Invasive Aquatic Species of Europe: Distributions, Impacts and Management’. (Eds E. Leppäkoski, S. Olenin and S. Gollasch.) pp. 325–336. (Kluwer Academic Publishers: Netherlands.)
Galil, B. S. Froglia, C., and Noël, P. (2002). CIESM atlas of exotic species in the Mediterranean. In ‘Crustaceans: Decapods and Stomatopods, Vol. 2’. (Ed F. Briand.) p. 192. (CIESM Publishers: Monaco.)
Gilberto, R., and Héctor, S. (2001). Anthropogenic dispersal of decapod crustaceans in aquatic environments. Interciencia 26, 282–288.
Golani, D., and Ritte, U. (1999). Genetic relationship in goatfishes (Mullidae: Perciformes) of the Red Sea and the Mediterranean, with remarks on Suez Canal migrants. Scientia Marina 63, 129–135.
Gopurenko, D., Hughes, J. M., and Keenan, C. P. (1999). Mitochondrial DNA evidence for rapid colonisation of the Indo-West Pacific by the mudcrab Scylla serrata. Marine Biology 134, 227–233.
| Mitochondrial DNA evidence for rapid colonisation of the Indo-West Pacific by the mudcrab Scylla serrata.Crossref | GoogleScholarGoogle Scholar |
Hansford, S. W., McGuren, J. J., and Marsden, G. E. (1993). The effect of substrate type on the ovarian maturation of Penaeus japonicus Bate. Asian Fisheries Science 6, 283–293.
He, L., Zhang, A., Zhu, C., Weese, D., and Qiao, Z. (2011). Phylogeography of the mud crab (Scylla serrata) in the Indo-West Pacific reappraised from mitochondrial molecular and oceanographic clues: transoceanic dispersal and coastal sequential colonization. Marine Ecology (Berlin) 32, 52–64.
| Phylogeography of the mud crab (Scylla serrata) in the Indo-West Pacific reappraised from mitochondrial molecular and oceanographic clues: transoceanic dispersal and coastal sequential colonization. Crossref | GoogleScholarGoogle Scholar |
Hewitt, D. R., and Duncan, P. F. (2001). Effect of high water temperature on the survival, moulting and food consumption of Penaeus (Marsupenaeus) japonicus (Bate, 1888). Aquaculture and Research 32, 305–313.
| Effect of high water temperature on the survival, moulting and food consumption of Penaeus (Marsupenaeus) japonicus (Bate, 1888). Crossref | GoogleScholarGoogle Scholar |
Hirata, H. (1975). An introduction to the reaching methods of prawn Penaeus japonicus Bate, in Japan. Memoirs of the Faculty of Fisheries, Kagoshima University 24, 7–12.
Holthuis, L. B. (1980). Shrimps and prawns of the world. An annotated catalogue of species of interest to fisheries. Fisheries Synopsis No. 125(1), pp. 1–271. (FAO: Rome.)
Hudinaga, M. (1935). Studies on the development of Penaeus japonicus (Bate). Hayatomo Fishery Institute report 1, 1–51.
Hudinaga, M. (1942). Reproduction, development and rearing of Penaeus japonicus Bate. Japan Journal of Zoology 10, 305–393.
Kanazawa, A., Teshima, S., and Tanaka, N. (1976). Nutritional requirements of prawn-V: Requirements for choline and inositol. Memoirs of the Faculty of Fisheries, Kagoshima University 25, 47–51.
| 1:CAS:528:DyaE2sXktlOkt78%3D&md5=e1766937afe1e95f636ca75ab8a90f98CAS |
Kevrekidis, K., and Kevrekidis, T. (1996). The occurrence of Penaeus japonicus in the Aegean Sea. Crustaceana 69, 925–929.
| The occurrence of Penaeus japonicus in the Aegean Sea. Crossref | GoogleScholarGoogle Scholar |
Lavery, S., Chan, T. Y., Tam, Y. K., and Chu, K. H. (2004). Phylogenetic relationships and evolutionary history of the shrimp genus Penaeus s. l. derived from mitochondrial DNA. Molecular Phylogenetics and Evolution 31, 39–49.
| Phylogenetic relationships and evolutionary history of the shrimp genus Penaeus s. l. derived from mitochondrial DNA. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhvFSksrY%3D&md5=c1cbdb57919d171dd053d2d457626110CAS | 15019607PubMed |
Liao, I. C., and Chien, Y. H. (1990). Evaluation and comparison of culture practices for Penaeus japonicus, P. penicillatus and P. chinensis in Taiwan. In ‘The Culture of Cold-tolerant shrimp: Proceedings of an Asian–USA Workshop on Shrimp Culture’. (Eds K. L. Main and W. Fulks.) pp. 49–63. (The Oceanic Institute: Honolulu, HI. )
Liao, I. C., and Chien, Y. H. (1994). Culture of kuruma prawn (Penaeus japonicus) in Asia. World Aquaculture 25, 18–33.
Lumare, F. (1984). Stocking trials of Penaeus japonicus Bate (Decapoda, Natantia) postlarvae in Lesina Lagoon (Southeast coast of Italy). Management of coastal lagoon fisheries, FAO studies and Reviews No. 61, Vol. 2, pp. 593–606. (FAO: Rome.)
Lumare, F. (1987). Reproduction and larval rearing of penaeids. In ‘Production in Marine Hatcheries. Mediterranean regional Aquaculture Project’. (Ed. B. Loix.) pp. 147–179. (FAO: Rome.)
Lumare, F. (1998). Crostacei peneidi: tecnica e gestione dell’allevamento. In ‘Manuale di Divulgazione. Serie Acquacoltura No. 4. (Ente di Sviluppo Agricolo del Veneto: Legnaro.)
Lumare, F., and Casolino, G. (1986). First record of Penaeus japonicus Bate, 1888 (Decapoda Natantia) along Italian coasts. OEbalia (Taranto) 13, 179–183.
Lumare, F., and Palmegiano, G. B. (1980). Acclimatazione di Penaeus japonicus Bate nella Laguna di Lesina (Italia sud orientale). Rivista Italiana di Piscicultura e Ittiopatologia 15, 53–58.
Ma, K. Y., Chan, T. Y., and Chu, K. H. (2011). Refuting the six-genus classification of Penaeus s. l. (Dendrobranchiata, Penaeidae): a combined analysis of mitochondrial and nuclear genes. Zoologica Scripta 40, 498–508.
| Refuting the six-genus classification of Penaeus s. l. (Dendrobranchiata, Penaeidae): a combined analysis of mitochondrial and nuclear genes. Crossref | GoogleScholarGoogle Scholar |
McLaughlin, P. A., Lemaitre, R., Ferrari, F. D., Felder, D. L., and Bauer, R. T. (2008). A reply to T. W. Flegel. Aquaculture 275, 370–373.
| A reply to T. W. Flegel. Crossref | GoogleScholarGoogle Scholar |
McMillen-Jackson, A. L., and Bert, T. M. (2003). Disparate patterns of population genetic structure and population history in two sympatric penaeid shrimp species (Farfantepenaeus aztecus and Litopenaeus setiferus) in the eastern United States. Molecular Ecology 12, 2895–2905.
| Disparate patterns of population genetic structure and population history in two sympatric penaeid shrimp species (Farfantepenaeus aztecus and Litopenaeus setiferus) in the eastern United States. Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3srltFCitw%3D%3D&md5=1e44dcbfe6b7cbb17634a2eaf0aa7138CAS | 14629371PubMed |
Mock, C. R. (2009). The culture of Penaeus japonicus in Japan. Journal of the World Aquaculture Society 3, 285–286.
Nei, M. (1977). F-statistics and analysis of gene diversity in subdivided populations. Annals of Human Genetics 41, 225–233.
| F-statistics and analysis of gene diversity in subdivided populations. Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaE1c%2FnvFWnsg%3D%3D&md5=1603c0852e701cb0670311b7fadfc455CAS | 596830PubMed |
Okauchi, M. Kobayashi, M., and Mizukami, Y. (1995). Water quality management by unicellular algae in shrimp larviculture ponds. U. S. –Japan Cooperative Program in Natural Resources (UJNR). Technical Report 24, pp. 59–64.
Otero, M. Cebrian, E. Francour, P. Galil, B., and Savini, D. (2013). ‘Monitoring marine invasive species in Mediterranean Marine Protected Areas (MPAs): A strategy and practical guide for managers. (IUCN: Spain).
Pérez Farfante, I., and Kensley, B. (1997). Penaeoid and sergestoid shrimps and prawns of the world. Keys and diagnoses for the families and genera. Mémoires du Museum national d’Histoire naturelle 175, 1–233.
Quero, J. C., and Vayne, J. J. (1998). ‘Les fruites de la mer et plantes marines des pèches Françaises. p. 256. (Delachaux Et Niestle: Lausanne.)
Quigley, D. T. G. (2010). Prawns and shrimps in Irish waters. The Irish Skipper August 2010, pp. 22–23.
Quigley, D. T. G., Herdson, D., and Flannery, K. (2013). Occurrence of the kuruma prawn Marsupenaeus japonicus (Spence Bate, 1888) in the Celtic Sea, English Channel, and North-West France. BioInvasions Records 2, 57–61.
| Occurrence of the kuruma prawn Marsupenaeus japonicus (Spence Bate, 1888) in the Celtic Sea, English Channel, and North-West France. Crossref | GoogleScholarGoogle Scholar |
Quinitio, E. T., Parado-Estepa, F. D., and Coniza, E. (1991). Notes on the completion of the life cycle of Penaeus japonicus in captivity in the Philippines. Philippine Journal of Science 120, 155–158.
Rambaut, A., and Drummond, A. J. (2007). ‘Tracer v1. 4’, Available at: http://beast.bio.ed.ac.uk/Tracer [verified September 2014]
Rohlf, F. J. (1973). Algorithm 76. Hierarchical clustering using the minimum spanning tree. The Computer Journal 16, 93–95.
Savini, D. Occhipinti, A. Miossec, L. and Garcia-Berthou, E. (2008). Alien species fact sheets Marsupenaeus japonicus. In ‘Environmental Impacts of Alien Species in Aquaculture Sustainable Management of Europe’s Natural Resources’. (Eds S. Gollasch, I. G. Cowx and A. D. Nunn.) (University of Hull: UK. )
Sbordoni, V., De Matthaeis, E., Cobolli-Sbordoni, M., La Rosa, G., and Mattoccia, M. (1986). Bottleneck effects and the depression of genetic variability in hatchery stocks of Penaeus japonicus (Crustacea, Decapoda). Aquaculture 57, 239–251.
| Bottleneck effects and the depression of genetic variability in hatchery stocks of Penaeus japonicus (Crustacea, Decapoda). Crossref | GoogleScholarGoogle Scholar |
Scordella, G. and Zecca, P. (2002). Shrimp farming in Italy: Status and perspectives. Global Aquaculture Advocate August 2002, 50–52.
Setiarto, A., Strüssmann, C. A., Takashima, F., Watanabe, S., and Yokota, M. (2004). Short-term responses of adult kuruma shrimp Marsupenaeus japonicus (Bate) to environmental salinity: osmotic regulation, oxygen consumption and ammonia excretion. Aquaculture and Research 35, 669–677.
| Short-term responses of adult kuruma shrimp Marsupenaeus japonicus (Bate) to environmental salinity: osmotic regulation, oxygen consumption and ammonia excretion. Crossref | GoogleScholarGoogle Scholar |
Shefer, S., Abelson, A., Mokady, O., and Geffen, E. (2004). Red to Mediterranean Sea bioinvasion: Natural drift through the Suez Canal, or anthropogenic transport? Molecular Ecology 13, 2333–2343.
| Red to Mediterranean Sea bioinvasion: Natural drift through the Suez Canal, or anthropogenic transport?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmsl2rurY%3D&md5=fdefa30a1d7ef105840827d5b6864ef0CAS | 15245405PubMed |
Shigueno, K. (1975). ‘Shrimp culture in Japan. p. 153. (Tokyo: Association for International Technical Promotion)
Shigueno, K. (2001). Farming kuruma shrimp in Japan. Advocate (Boston, Mass.) February, 45–46.
Shih, C. H., Haung, H. L., Chu, T. J., Lee, Y. C., Wang, C. M., and Tzeng, T. D. (2011). Genetic diversity and historical demography of kuruma shrimp (Penaeus japonicus) species complex off China based on mitochondrial DNA analysis. African Journal of Biotechnology 10, 1065–1072.
| 1:CAS:528:DC%2BC3MXisl2gtLk%3D&md5=3ea82cc7ca8cabf811af5bfc8e8470b0CAS |
Sirna Terranova, M., Lo Brutto, S., Arculeo, M., and Mitton, J. B. (2006). Population structure of Brachidontes pharaonis (P. Fischer, 1870) (Bivalvia, Mytilidae) in the Mediterranean Sea, and evolution of a novel mtDNA polymorphism. Marine Biology 150, 89–101.
| Population structure of Brachidontes pharaonis (P. Fischer, 1870) (Bivalvia, Mytilidae) in the Mediterranean Sea, and evolution of a novel mtDNA polymorphism. Crossref | GoogleScholarGoogle Scholar |
Stebbing, T. R. R. (1914). South African Crustacea. Annals of the South African Museum 15, 1–55.
Streftaris, N., and Zenetos, A. (2006). Alien marine species in the Mediterranean the 100 ‘worst invasives’ and their impact. Mediterranean Marine Science 7, 87–118.
| Alien marine species in the Mediterranean the 100 ‘worst invasives’ and their impact. Crossref | GoogleScholarGoogle Scholar |
Sugaya, T., Ikeda, M., Mori, H., and Taniguchi, N. (2002). Inheritance mode of microsatellite DNA markers and their use for kinship estimation in kuruma prawn Penaeus japonicus. Fisheries Science 68, 299–305.
| Inheritance mode of microsatellite DNA markers and their use for kinship estimation in kuruma prawn Penaeus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XjvV2qtLk%3D&md5=27a52c0264f3b06e42d314e8247222bdCAS |
Tajima, F. (1989). Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
| 1:CAS:528:DyaK3cXhslentA%3D%3D&md5=dd07b833dd833e71b0d9429c0036e936CAS | 2513255PubMed |
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739.
| MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1eiu73K&md5=5ee5a71f721d24a7782516dc0ff5fcfcCAS | 21546353PubMed |
Tan, S. H., Wang, G. Z., and Li, S. J. (2009). Sequence variability of mitochondrial 16S rRNA gene and genetic divergence of Marsupenaeus japonicus in the coastal waters of China. Acta Ecologica Sinica 29, 6805–6810.
| 1:CAS:528:DC%2BC3cXhtVWnt7k%3D&md5=f72a93061307e7c20805df1a2f34fed2CAS |
Taniguchi, N., and Han, H. (1989). Population genetic analyzes of fish and shellfish by isozymes. ‘Japanese fisheries resources conservation association report. (Japanese Fisheries Resources Conservation Association: Tokyo)
Tom, M., and Lewinsohn, C. (1983). Aspects of the benthic life cycle of Penaeus (Melicertus) japonicus Bate (Crustacea Decapoda) along the south-eastern coast of the Mediterranean. Fisheries Research 2, 89–101.
| Aspects of the benthic life cycle of Penaeus (Melicertus) japonicus Bate (Crustacea Decapoda) along the south-eastern coast of the Mediterranean. Crossref | GoogleScholarGoogle Scholar |
Treece, D. G. (1999). Shrimp maturation and spawning. The United States–Japan Cooperative Program in Natural Resources (UJNR). Technical Report 28, pp. 123–134.
Tsoi, K. H., Wang, Z. Y., and Chu, K. H. (2005). Genetic divergence between two morphologically similar varieties of the kuruma shrimp Penaeus japonicus. Marine Biology 147, 367–379.
| Genetic divergence between two morphologically similar varieties of the kuruma shrimp Penaeus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkvVClu7o%3D&md5=5d04d56931ff6523305390dc4e7ca12eCAS |
Tsoi, K. H., Chan, T. Y., and Chu, K. H. (2007). Molecular population structure of the kuruma shrimp Penaeus japonicus species complex in western Pacific. Marine Biology 150, 1345–1364.
| Molecular population structure of the kuruma shrimp Penaeus japonicus species complex in western Pacific. Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXitVGqurk%3D&md5=e9ba5d53e8a1da07156708204a4f6dccCAS |
Türkmen, G. (2007). Pond culture of Penaeus semisulcatus and Marsupenaeus japonicus (Decapoda, Penaeidae) on the West coast of Turkey. Turkish Journal of Fisheries and Aquatic Sciences 7, 7–11.
Tzeng, T. D., Yeh, S. Y., and Hui, C. F. (2004). Population genetic structure of the kuruma prawn (Penaeus japonicus) in East Asia inferred from mitochondrial DNA sequences. Journal of Marine Science 61, 913–920.
| 1:CAS:528:DC%2BD2cXotFGnur0%3D&md5=a651667b46e223004c10214ff31b2d53CAS |
Wang, K. X. (1997). ‘Crustacean Biology and Multiplication. (China Agriculture Press: Beijing.) [In Chinese.]
Yağlıoğlu, D., and Turan, C. (2012). Colonization and genetic changes of Indo-Pacific immigrant Saurida undosquamis (Richardson, 1948) (lizardfish) in the Mediterranean Sea. Journal of the Black Sea Mediterranean Environment 18, 329–340.
Yu, H. P., and Chan, T. Y. (1986). ‘The Illustrated Penaeoid Prawns of Taiwan. (Southern Materials Center: Taiwan.)
Zhou, H. (2001). Growth of intensive farmed kuruma shrimp (Penaeus japonicus Bate) in earthen ponds. Journal of Guangxi Academy of Sciences 17, 91–95.
| 1:CAS:528:DC%2BD3MXhvVShsrY%3D&md5=ec1d610f01fbc25642a59482e81ac5dcCAS |