Assessment of genetic structure among Australian east coast populations of snapper Chrysophrys auratus (Sparidae)
Jess A. T. Morgan A F , Wayne D. Sumpton A , Andrew T. Jones B , Alexander B. Campbell A , John Stewart C , Paul Hamer D and Jennifer R. Ovenden
A F , Wayne D. Sumpton A , Andrew T. Jones B , Alexander B. Campbell A , John Stewart C , Paul Hamer D and Jennifer R. Ovenden  E
E
A Animal Science, Department of Agriculture and Fisheries, EcoSciences Precinct, Boggo Road, Dutton Park, Qld 4102, Australia.
B Department of Mathematics, The University of Queensland, Saint Lucia, Qld 4072, Australia.
C New South Wales Department of Primary Industries, Sydney Institute of Marine Science, Chowder Bay Road, Mosman, NSW 2088, Australia.
D Victorian Fisheries Authority, Department of Economic Development Jobs Transport and Resources, Bellarine Highway, Queenscliff, Vic. 3225, Australia.
E Molecular Fisheries Laboratory, School of Biomedical Sciences, The University of Queensland, Saint Lucia, Qld 4072, Australia.
F Corresponding author. Email: jessica.morgan@daf.qld.gov.au
Marine and Freshwater Research 70(7) 964-976 https://doi.org/10.1071/MF18146
Submitted: 4 April 2018 Accepted: 1 November 2018 Published: 19 December 2018
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Snapper Chrysophrys auratus is a high-value food fish in Australia targeted by both commercial and recreational fisheries. Along the east coast of Australia, fisheries are managed under four state jurisdictions (Queensland, Qld; New South Wales, NSW; Victoria, Vic.; and Tasmania, Tas.), each applying different regulations, although it is thought that the fisheries target the same biological stock. An allozyme-based study in the mid-1990s identified a weak genetic disjunction north of Sydney (NSW) questioning the single-stock hypothesis. This study, focused on east-coast C. auratus, used nine microsatellite markers to assess the validity of the allozyme break and investigated whether genetic structure exists further south. Nine locations were sampled spanning four states and over 2000 km, including sites north and south of the proposed allozyme disjunction. Analyses confirmed the presence of two distinct biological stocks along the east coast, with a region of genetic overlap around Eden in southern NSW, ~400 km south of the allozyme disjunction. The findings indicate that C. auratus off Vic. and Tas. are distinct from those in Qld and NSW. For the purpose of stock assessment and management, the results indicate that Qld and NSW fisheries are targeting a single biological stock.A
Additional keywords: effective population size, fisheries management, microsatellite genotyping, stock structure.
Introduction
Chrysophrys auratus (Forster, 1801) is a highly prized fish found in coastal waters off Australia and New Zealand. Throughout Australia the species is a key target in both commercial and recreational fisheries (Kailola et al. 1993). The Australian east coast fishery has components in New South Wales (NSW), Queensland (Qld), Victoria (Vic.) and Tasmania (Tas.) that are currently assessed and managed independently within each jurisdiction (Fowler et al. 2016). Stock assessments, based on a wide array of biological and fishery data, are regularly conducted to provide fisheries managers with the information used to inform the regulation of harvest impacts. Knowledge relating to stock structure is important for stock assessment and management because it informs the spatial units (fishery regions) over which data should be integrated and management should operate. Further, snapper along the east coast of Australia spawn in coastal waters, where their larval dispersal is influenced by the East Australian Current (EAC; Curley et al. 2013). Predicting longer-term evolutionary changes, such as those related to ocean warming or changes to currents (Pecl et al. 2014), depends on knowledge of genetic structure. If populations are in decline, knowledge of genetic connectivity can assist in identifying source or sink populations (Hauser and Carvalho 2008).
The need for clarification of genetic structure of snapper along the Australian east coast was emphasised by recent stock status assessments that indicated that, despite the considerable amount of fishery and biological information available, the stock status of east coast snapper was ‘uncertain’ (Fowler et al. 2016). This uncertainty was largely because of state-based assessments of populations in Qld suggesting they were ‘recruitment overfished’, whereas in NSW they were suggested as possibly ‘growth overfished’ but not ‘recruitment overfished’ because commercial catch rates were increasing and the sizes and ages in landings remained stable (Finn et al. 2015). The status of stocks in Tas. and eastern Vic. was also undefined because of insufficient data (Kemp et al. 2012; Fowler et al. 2016). Although the major east coast C. auratus fisheries in NSW and Qld are managed independently, confirmation of whether or not both fisheries exploit the same biological stock, and thus should be combined for fisheries assessment, is critical in order to reconcile the uncertainty in exploitation status of east coast snapper (Ferrell and Sumpton 1997; Sumpton et al. 2008; Fowler et al. 2016). Further, should the limited information available for fisheries in eastern Vic. and Tas. continue to influence the assessment of the main fisheries further north?
Rapidly evolving molecular technologies have increased in power, and reduced in cost, such that they now play an important role in defining genetic stocks of exploited fish species (Carvalho and Pitcher 2012). Stocks that are genetically different from each other are described as reproductively isolated units or regions within a species distribution (Ovenden 1990). For genetic stocks to be identified, it is necessary that interbreeding among different stocks has been negligible over very long time periods. This definition may or may not relate to ‘fisheries’ or ‘harvest’ stocks, which are described as regions within a species distribution where fishing pressure in one region does not affect the abundance of fish in other regions, implying independence of the processes of replenishment, growth and mortality, but not necessarily different genetics (Gauldie 1988). Identification of genetic stocks provides highly definitive broad spatial regions on which to focus fisheries assessment and identification of management units.
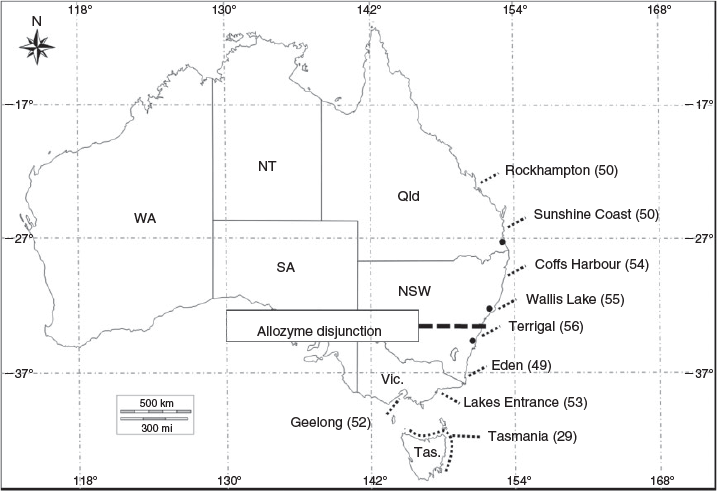
The genetic structure of east coast populations of C. auratus was first investigated in 1980 using allozyme data as part of a study examining the Australian distribution of the species (MacDonald 1980). That study failed to detect differences among the three east coast locations sampled. Sumpton et al. (2008) also used allozyme loci, but sampled the east coast more intensively at seven areas spanning a finer spatial scale between Sydney in NSW and Rockhampton in Qld (Fig. 1). The authors of that study identified a weak genetic disjunction (based largely on a difference at one locus) on the central coast of NSW, north of Sydney but south of Forster (Fig. 1). A signal of isolation by distance (IBD) was found north of the weak genetic disjunction, to the Swains Reef off Rockhampton (Sumpton et al. 2008).
|
Chrysophrys auratus is a long-lived species (up to 40 years; Norriss and Crisafulli 2010) that can grow up to 16 kg and 120 cm in length (MacDonald 1982; Kailola et al. 1993). The species undergoes juvenile sex inversion, with small juveniles having female reproductive tissue and a proportion of individuals becoming male near the age of maturity at (3+ years and at a reasonably large size of 25–30 cm; Francis and Pankhurst 1988; Stewart et al. 2010). These biological characteristics, combined with an intermediate fecundity (Saunders et al. 2012), mean that C. auratus is more susceptible to overfishing than faster-developing species such as sardine and anchovy, particularly if exploitation is not managed at the appropriate spatial scales relevant to population replenishment. Adult reproduction is by broadcast spawning in breeding aggregations, which, depending on the geographic area, can occur in both sheltered inshore bays and coastal waters (Kailola et al. 1993). Tag–recapture and otolith chemistry research suggest that migration of juvenile and adult snapper is highly variable among the geographic regions where studies have been conducted, ranging up to at least hundreds of kilometres (Sanders 1974; Gillanders 2002; Coutin et al. 2003; Moran et al. 2003; Parsons et al. 2003; Sumpton et al. 2003; Hamer et al. 2011; Fowler et al. 2017). Along the east coast of Australia, even if adults showed restricted movements, dispersal of the 20- to 30-day larval stage in prevailing ocean currents, particularly the dominant EAC, provides considerable scope for broad-scale north to south genetic mixing (Ferrell and Sumpton 1997). Despite having a high larval dispersal potential, C. auratus could use local coastal circulation to recruit locally, or could migrate back to its natal origins before breeding, which could create complex genetic structure. Other studies investigating the genetic structure of species with similar pelagic larval periods, namely Pristipomoides multidens (Ovenden et al. 2004), Lutjanus carponotatus (Harrison et al. 2012) and Argyrosomus japonicus (Barnes et al. 2016) have detected genetic structure suggesting low-level mixing despite potential for wide larval dispersal.
Microsatellite markers offer a high level of resolving power because of their potential for high polymorphism. They have been successfully used in New Zealand stocks of C. auratus to investigate population structure (Ashton 2013) and larval recruitment into marine reserves (Le Port et al. 2017). Ashton (2013) found that despite measuring a high level of polymorphism in the 17 microsatellite loci screened, there was little genetic differentiation to distinguish among stocks of C. auratus collected over a 900-km range in New Zealand. Although the genetic results reported by Ashton (2013) supported long distance dispersal, he also reported limited genetic mixing, over a 40-year period, between two locations situated 50 km apart. He proposed site-specific differences in mobility of New Zealand C. auratus to explain the conflicting results (Ashton 2013).
Le Port et al. (2017) used the highly polymorphic microsatellite loci to screen for relatedness among samples to assess the contribution of marine reserves to larval recruitment, finding predominantly small-scale larval dispersal distances of less than 40 km. Microsatellite markers have also been used to investigate the genetic structure of C. auratus in and around Shark Bay, Western Australia (Gardner et al. 2017). As with the New Zealand work, Gardner et al. (2017) also recorded highly polymorphic loci (only 1 of the 12 microsatellites overlapped with the New Zealand studies) and failed to detect significant genetic structure over a 350-km range, despite earlier allozyme studies suggesting that complex genetic structure may exist (MacDonald 1980). Overall, to date individual-based genetic analyses suggest that C. auratus exhibit limited larval dispersal, but lack clear genetic differences, probably due to some level of interbreeding between neighbouring populations over long time periods (an IBD structure).
Genetic markers offer an independent tool that can complement and improve traditional demographic estimators of effective population size (Ne; Luikart et al. 2010). Estimating Ne is challenging in marine species that are highly fecund and have large populations with moderate gene flow (Hare et al. 2011). Despite recent methodological developments (Waples et al. 2011), the relationship between census population size (N) and Ne is still poorly understood (Ovenden et al. 2007; Waples 2016), but is discussed in detail by Ovenden et al. (2016). Hauser et al. (2002) estimated Ne in C. auratus populations from New Zealand and found that Ne was five orders of magnitude below the adult census size (N). Several suggestions have been made regarding the factors that may contribute to very low estimates of Ne, such as high fecundity and biased reproductive success (Hauser et al. 2002; Hauser and Carvalho 2008; Coscia et al. 2016; Waples 2016). Using simulated data, Waples (2016) explored, in turn, the potential effects of increased longevity, fecundity and variance in reproductive success, as well as increased egg quality with age, and found that ‘very tiny’ Ne required some version of Hedgecock’s (1994) ‘sweepstakes’ hypothesis, whereby only a few families reproduce successfully. Waples (2016) also noted that when Ne is large (>10 000), the frequency distribution of estimates of Ne will be bimodal with either infinitely large estimates or otherwise downwardly biased values ranging in the low hundreds to low thousands. Estimating unbiased Ne for empirical data with large N requires good power in terms of sample numbers and number of loci. Although correlating Ne with N may require more biological information than is currently available for C. auratus, relative comparisons of Ne among sample areas is possible and may provide some insight into the effect of local fishing pressure on a population.
The overall objective of this study was to characterise the genetic stock structure of C. auratus along the east coast of Australia using microsatellite markers and to test the specific findings of previously investigated allozyme markers (MacDonald 1980; Sumpton et al. 2008). In addition, more comprehensive sampling than previous allozyme studies has been used to examine the findings of these previous studies, namely the IBD signature, a genetic disjunction north of Sydney and the extent of panmixia among Qld and NSW sites. The genetic stock structure of east coast populations of C. auratus determined in this study will be used to inform stock assessment models currently being developed to better inform cross-jurisdictional assessment and management of east coast C. auratus. Preliminary estimates of Ne are also made to ascertain whether existing methodology is capable of making bounded predictions for C. auratus stocks and provide a potential future proxy for changes in population size.
Materials and methods
Sample collection
Fresh fin clip tissue samples of C. auratus were collected from recreational and commercial fishers who donated samples from fish that were killed during harvest between 2012 and 2016. Samples were stored in individual tubes of 100% molecular-grade ethanol. Sampling was conducted along the east Australian coastline from nine areas, spanning four states and ranging over 2000 km (Fig. 1). The sampling strategy was designed to span a wide geographic range of commercial fisheries including sites from both sides of the genetic disjunction in central NSW identified in the allozyme study of Sumpton et al. (2008; marked on Fig. 1). Most Tasmanian samples (n = 21) were sourced from waters near Devonport in the central north, but because of a paucity of samples, four additional fish from waters off Stanley in the north-west and four from waters closer to Hobart in the south-east were included. All tissue samples were transferred to the Department of Agriculture and Fisheries at the Eco-Sciences Precinct in Brisbane (Qld, Australia) for molecular analysis.
DNA extraction and microsatellite screening
Approximately 3 mm of fin clip tissue was washed in 1 mL of MilliQ water to remove the ethanol preservative before DNA extraction. DNA was extracted using a DNeasy Tissue Kit (Qiagen, Melbourne, Vic., Australia) according to the manufacturer’s instructions into a final elution volume of 50 µL. DNA concentration was quantified using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Melbourne, Vic., Australia).
This project targeted nine microsatellite loci from the larger panel that Le Port et al. (2017) used for their New Zealand C. auratus study. The 17-loci panel of A. Le Port, S. Lavery, N. Kaur, and J. C. Montgomery (unpubl. data) was reduced to exclude loci that were difficult to amplify and those Le Port et al. (2017) had identified as out of Hardy–Weinberg equilibrium (HWE). Primer sequences, annealing temperatures multiplex combinations plus the original source of each locus are detailed in Table S1, available as Supplementary material to this paper.
Rather than individually labelling each primer with a fluorescent probe, the forward primer at every locus was modified with one of four M13 sequences (Table S1). For each assay, the reaction tube contained the primer pair plus a fluorescently labelled M13 primer (with FAM, NED, VIC or PET fluorophores; Oetting et al. 1995; Schuelke 2000; Missiaggia and Grattapaglia 2006; Kirchoff et al. 2008). Thus, the nine loci were polymerase chain reaction (PCR) amplified in three multiplexed PCR reaction tubes (M1–M3) and three single locus reaction tubes (S1–S3; Table S1). A Multiplex PCR Kit (Qiagen) was used to amplify the DNA in a final volume of 6 µL. The PCR samples contained 3 µL of 2× Master Mix, 0.6 µL of 5× Q solution, varying primer concentrations (detailed in Table S1, with the labelled M13 primer concentration the same as the reverse primer) and ~20 ng of Genomic DNA template. Microsatellite PCR amplifications were performed in a Bio-Rad (Sydney, NSW, Australia) thermal cycler (DNA Engine Peltier). The DNA template and enzyme were denatured at 95°C for 15 min, followed by 37 cycles of 94°C for 30 s, 52–62°C (specific annealing temperatures are given in Table S1) for 45 s and 72°C for 90 s. To ensure consistent allele calling during genotyping, a final extension at 72°C for 45 min was used to ensure complete extension of the PCR products. Allele sizing was determined using GeneScan 500 LIZ dye size standard (Thermo Fisher Scientific). Products were separated by electrophoresis on an ABI3130xl sequencer (Applied Biosystems, Thermo Fisher Scientific). According to the manufacturer’s recommendation, LIZ peaks at 35 and 250 were excluded before fragment analysis because of their temperature sensitivity; then, microsatellite peaks were scored using Geneious (ver. 8.1.9, see http://www.geneious.com, accessed 16 May 2016; Kearse et al. 2012). A repeat positive control sample was run on every 96-sample plate to ensure scoring consistency was maintained between electrophoresis runs. Samples returning low (<200 fluorescence units) or no signal strength for a subset of loci were initially subjected to another PCR run with increased starting DNA. If the signal continued to be weak, the multiplex was broken into single-locus reactions.
Microsatellite analysis
To summarise genetic diversity, several different metrics were determined for the loci. A relatedness screen (in GenAlEx, ver. 6.5, see http://biology-assets.anu.edu.au/GenAlEx/Welcome.html, accessed 17 November 2016; Peakall and Smouse 2006) was used to identify duplicate samples, which were removed from subsequent screening. To estimate the level of genetic differentiation between sampling locations the standardised measure of genetic differentiation, Jost’s estimate of differentiation (DEST; Jost 2008) for each location pair was determined (using GenAlEx, ver. 6.5, see http://biology-assets.anu.edu.au/GenAlEx/Welcome.html; Peakall and Smouse 2006). DEST provides a more accurate measure than the fixation index (FST), which tends to be downwardly biased by high allelic diversity at microsatellite loci, small numbers of sampling locations and low sample numbers. The number of alleles was determined to estimate the polymorphism information content (PIC) of the loci, as well as to calculate observed and expected heterozygosity values (using Cervus, ver. 3.0.7, see http://www.fieldgenetics.com/pages/aboutCervus_Overview.jsp, accessed 11 October 2016 Kalinowski et al. 2007).
To determine deviations from HWE, exact tests were used to test each locus, in each sampling location (using Genepop-on-the-Web, ver. 4.2; http://genepop.curtin.edu.au/, accessed 28 November 2017; Raymond and Rousset 1995; Rousset 2008). Although each locus by sampling location test was independent, the same null hypothesis was tested multiple times; thus, a subjective decision was made (following Cabin and Mitchell 2000) to apply an intermediate Bonferroni-type correction (set at 9) for multiple tests (Rice 1989). For loci out of HWE, the direction of bias was determined and an assessment was made to determine whether the result could be attributed to scoring errors, allele dropout or null alleles (using Microchecker, ver. 2.2.3, see http://www.nrp.ac.uk/nrp-strategic-alliances/elsa/software/microchecker/, accessed 5 October 2016; Van Oosterhout et al. 2004). Linkage disequilibrium (LD) was tested using log-likelihood ratio statistics (G-tests) to assess each pair of loci within each sampling location (using Genepop-on-the-Web, ver. 4.2, see http://genepop.curtin.edu.au/; Raymond and Rousset 1995; Rousset 2008) with Bonferroni correction. Two assumptions made by F-statistics and structure analyses are that populations are in HWE and loci are not linked. Loci failing to comply with these assumptions were removed before analysing the data.
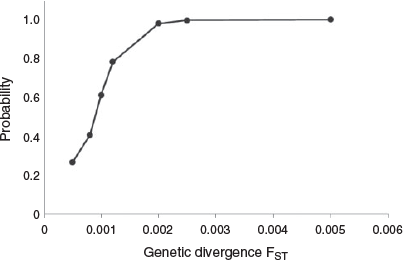
A power analysis to assess the resolving power of the microsatellites to detect genetic differentiation was conducted in POWSIM (ver. 4.1, see http://internt.zoologi.su.se/~ryman/, accessed 5 February 2018; Ryman and Palm 2006; Gardner et al. 2015). As part of the POWSIM evaluation, Ne was set to 10 000 following Gardner et al. (2017) e this value is not based on the linkage disequilibrium based Ne (LDNe) empirical estimates below). Time since divergence (t) was varied to obtain seven divergence levels (FST) between 0.0002 and 0.005. After drift, the base population was subdivided into nine populations for simulations, with the size of each population following the sample data in Fig. 1. The mean of 500 replicates was used to estimate the proportion of samples for which the FST values were significantly different from zero using Fisher’s exact tests.
The population genetic structure of species across the nine sampled locations in eastern Australia was investigated using four approaches. First, Bayesian inference was used to assign individuals to expected stocks using Structure (ver. 2.3.4, see https://web.stanford.edu/group/pritchardlab/structure.html, accessed 6 December 2013; Pritchard et al. 2000). The most likely number of genetic clusters was determined following Evanno et al. (2005), as described below. Second, population pairwise DEST (GenAlEx, ver. 6.5, see http://biology-assets.anu.edu.au/GenAlEx/Welcome.html; Peakall and Smouse 2006) values were estimated for all pairs of sampling locations. Neighbouring locations with non-significant fixation values were then pooled and pairwise fixation values recalculated in an iterative approach to identify possible spatial boundaries to genetic stocks (Broderick et al. 2011). Third, a discriminant analysis of principal components (DAPC; Jombart et al. 2010) available in the Adegenet package (ver. 2.1.1, see https://cran.r-project.org/web/packages/adegenet/index.html, accessed 5 February 2018; Jombart 2008), run through RStudio (ver. 0.99.903, RStudio, Boston, MA, USA, see http://www.RStudio.com/ide, accessed 5 February 2018) was used to distinguish genetic clusters. Finally, an analysis of molecular variance (AMOVA; Arlequin, ver. 3.5.1.2, see http://cmpg.unibe.ch/software/arlequin35/, accessed 18 December 2013; Excoffier and Lischer 2010) was used to determine the percentage genetic variance explained by the groupings deduced from the structure and DEST analyses.
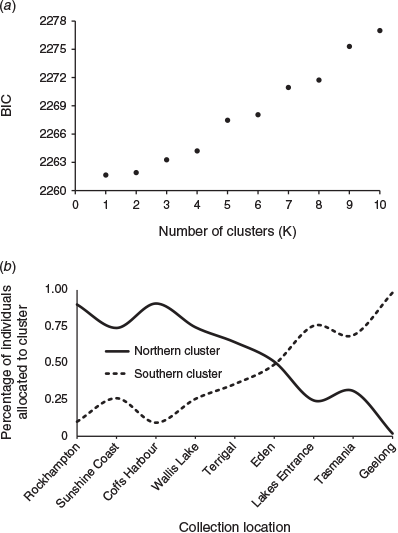
For the structure analysis, a series of Markov chain Monte Carlo (MCMC) simulations was run using models of both admixture and no admixture, and using locations as priors correlated with allele frequencies (Falush et al. 2003). Simulations were run for a range of stock sizes (K = 1–10) to determine the optimal number of clusters following Evanno et al. (2005). Ten repetitions were run for each stock size, burn-in was set to 104 and 106 repetitions were run after burn-in. Using the ΔK estimator approach of Evanno et al. (2005), the rate of change in the log probability of the data between successive K values was calculated (ΔK) and plotted against K to determine the most likely number of genetic stocks.
Multivariate DAPC was conducted using the complete dataset because discriminate analyses are robust to loci out of HWE or in LD. Evidence of genetic clusters was examined in DAPC by running successive K-means clustering in the find.clusters function with scaling activated during the principal component analysis (PCA) to give higher influence in the clustering to loci with more alleles. The optimal number of clusters was determined as the K with the lowest Bayesian information criterion (BIC; Jombart et al. 2010). DAPC was also run for a priori stock number K = 2 based on the outcomes of the structure analysis and population pairwise DEST analysis.
Spatial patterns of genetic divergence were investigated using a genetic model of IBD correlating genetic distance (DEST/(1 – DEST)) to geographic coast distance (km) in Genepop-on-the-Web (ver. 4.2, see http://genepop.curtin.edu.au/; Raymond and Rousset 1995; Rousset 2008). Shoreline distances (km) between sampling locations were estimated manually using Google Maps (API, ver. 3.30, see https://www.google.com.au/maps/ accessed 18 November 2017, Google, Mountain View, CA, USA), factoring in land barriers. The resulting correlation for all sampling locations was plotted in Microsoft Excel 2013 (ver. 15.0.5075.1000, Microsoft, Bellevue, WA, USA) and was assessed using Mantel (1967) tests and distance-based redundancy analysis (dbRDA; Legendre and Anderson 1999) following the recommendation of Kierepka and Latch (2015) to combine statistical tests to assess IBD. Mantel tests were assessed using 5000 permutations in Arlequin (ver. 3.5.1.2, see http://cmpg.unibe.ch/software/arlequin35/, accessed 18 December 2013; Excoffier and Lischer 2010). Distance-based (db)RDA used the Fstat matrix against the sampling locations run through the R package vegan (ver. 2.4–2, J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, R. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, and H. Wagner, see https://cran.r-project.org/web/packages/vegan/index.html, accessed 7 February 2018). IBD analyses were conducted on the complete dataset and on the sampling locations from Terrigal north (based on the K = 2 outcome from the structure and pairwise DEST analyses).
A population genetic self-assignment test using a Rannala and Mountain Bayesian method with threshold 0.05 (Rannala and Mountain 1997) was conducted in GENECLASS2 (Piry et al. 2004) to determine the probability of correctly assigning an individual to a stock in the K = 2 stock model. The test was run both including and excluding the mixed Eden sampling location from the analysis.
Estimating Ne
The linkage disequilibrium method for estimating Ne (LDNe; Waples and Do 2008) was applied using NeEstimator 2.01 (Do et al. 2014) to the samples from each collection area. For each set of samples, all loci and individuals were used and low frequency alleles were discarded if their observed frequency was below Pcrit, the minimum allowed allele frequency. Pcrit was chosen based on the sample size for the particular area according to standard methodology (Waples and Do 2010). Owing to the similar sample sizes from each area, Pcrit = 0.02 for all the LDNe estimates produced. For each estimate, a 95% confidence interval (CI) was also calculated according to the revised jack-knife method of Jones et al. (2016).
Results
In all, 449 C. auratus were collected and genotyped as part of this study. Samples were obtained from nine collection areas over a 2278-km range from Rockhampton in the north to Tas. and Geelong in the south (Fig. 1). Sampled fish ranged in size from 150 to 810 mm fork length (FL), with an approximately equal sex ratio (sex recorded in ~50% of samples). Genotypes were obtained for all animals, across nine loci, with missing values per locus ranging from 0.2 to 9.4% (Table 1). Amplification difficulties due to poor-quality DNA samples were improved by re-extracting and eluting samples into a smaller volume (50 instead of 100 µL), then amplifying the difficult loci as singletons using undiluted DNA instead of multiplexed reactions. A relatedness screen of the samples identified a duplicate animal in the dataset. One of the duplicates was removed from subsequent analyses (taking the total number of C. auratus screened to 448; Table 1).
High allelic diversity was observed in the data, with 18 or more alleles found in two-thirds of the loci (allele number per locus ranged from 8 to 27; Table 1). Observed heterozygosity ranged between 0.342 and 0.879 and was lower than expected for all loci. With Bonferroni correction, only one locus by sampling location comparison was out of HWE (Sunshine Coast with Sal10; Table 2). The inbreeding coefficient (FIS) value for this comparison was positive, indicating a heterozygote deficiency, most likely due to scoring errors. Although P-values were low for some of the other exact tests, there was no strong pattern of deviation from HWE by locus or by location; however, we note that deviations at one location (Sunshine Coast) and one locus (Pma68–23) may not be due to chance. Running the code (x <- replicate(1000000, sum(runif(9) < 0.05)) mean(x > = 3) mean(x > = 2)) in R determined the chance of obtaining three P-values <0.05 from a uniform distribution (0–1) if nine values were selected randomly a million times. The result was extremely unlikely due to chance, suggesting something biological is possibly happening at this site. To evaluate the possible effect of these deviations on population genetic structure, the structure analysis was repeated without Sunshine Coast and locus Pma68–23 (see Fig. S1). The results did not change the overall outcome, or interpretation, suggesting the genetic signal in the data was robust to the deviations. As a result, all loci and collection sites were retained for subsequent analyses.
Screening for LD using exact G-tests (log-likelihood ratio tests) to assess each pair of loci within each collection location identified three significant pairs (Sunshine Coast: Pma1 with CM003195; Eden: GA2A with Pma68–23; Eden: Pma22–9 with Pma68–23). The apparent genetic association of these pairs was not supported in other sampling locations and is likely an artefact of reduced sample numbers at these sites due to missing genotypes (Gordon et al. 1999; Akey et al. 2001). For this reason, at the conclusion of the exploratory analyses, all nine loci were determined to be suitable (largely meeting the assumptions of HWE and no LD) for downstream analyses. Based on the power analysis, the microsatellite data contain sufficient power to detect FST at or above 0.002 with 98% confidence (Fig. 2).
|
Analysing the data for genetic structure using Bayesian modelling (structure; Pritchard et al. 2000) identified a two-cluster model as the most likely fit for all models, with a genetic transition occurring around Eden on the NSW south coast (Fig. 3a). The optimal two-cluster model was determined by the highest average likelihood score (Fig. 3b) and the highest change in mean likelihood score (ΔK; Fig. 3c). The southern stock extends from Geelong, past Tas., then north to Lakes Entrance. The sampling locations flanking Eden (Terrigal, Eden, Lakes Entrance and Tas.) exhibit an intermediate level of mixing between the two stocks.

|
Using pairwise DEST statistics to pool undifferentiated adjacent collection locations, the spatial boundaries of potential genetic stocks were further investigated (Table 3a). As a mixed transition zone between the northern and southern stocks, Eden was excluded from pooling. The first round of pooling grouped Rockhampton with Sunshine Coast, Coffs Harbour with Wallis Lake, and Lakes Entrance with Tas. (note, Lakes Entrance is a linear neighbour to both Tas. and Geelong). The second round of pooling combined Coffs Harbour + Wallis Lake with Terrigal in the north and Lakes Entrance + Tas. with Geelong in the south. The third round of pooling combined Rockhampton + Sunshine Coast with Coffs Harbour + Wallis Lake + Terrigal (diagram summarising pooling order below Fig. 3a Table 3a). The end result of three rounds of pooling was two significantly different stocks (Table 3b), the first extending Rockhampton to Terrigal and the second extending south from Lakes Entrance to Geelong, including Tas. with a significantly different DEST = 0.048 (P = 0.001).

|
The optimum number of clusters (K with the lowest BIC score) determined using the DAPC find.clusters function was K = 1, although the BIC score for K = 2 was only a little higher (Fig. 4a). Results of the analyses run for a priori stock number K = 2, based on the outcomes from the structure analysis and population pairwise DEST analysis, clearly showed a distinct northern and southern stock with a region of overlap at Eden (Fig. 4b).
|
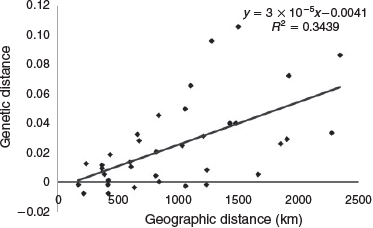
Genetic distance was found to be correlated with linear geographic distance using an IBD genetic model on the complete dataset (Fig. 5). The geographic distance between collection areas explained 34% of their genetic distance (R2 = 0.3439). The IBD signal for the complete dataset was significant using both a Mantel test (5000 permutations; P = 0.0014) and the dbRDA analysis (P = 0.001). Focusing on the five sampling locations from Terrigal north, the northern stock in the K = 2 cluster model, IBD was no longer detected using either test (Mantel, P = 0.27; dbRDA, P = 0.217).
|
Loci Pma1, GA2A and CM000278 were excluded from the AMOVA because they were above the missing data threshold of 5%. Based on the structure and DEST results, Eden was excluded from the analysis because of its mixed nature, and a K = 2 stock model was tested. Analysis of the remaining eight sampling locations and six loci using AMOVA found significant differentiation to support the two-stock model, although stock accounted for only 1.22% of the genetic variability (among group covariance FCT = 0.01219, P < 0.0001). Most of the genetic variation in C. auratus (98.73%) was explained by within-sampling location differences.
Population genetic assignment tests correctly assigned 76.3% (including Eden) and 81.7% (excluding Eden) of samples to the correct stock in a K = 2 model. There was no obvious bias in the direction of incorrect assignments, with 18% of the northern stock incorrectly assigned to the south and 19% of animals from the southern stock incorrectly assigned to the north (K = 2 model excluding Eden).
Using the linkage disequilibrium method, estimates of LDNe were calculated from six of the nine collection areas (Table 4). It was not possible to resolve LDNe for Rockhampton, Wallis Lake or Geelong, possibly due to the fairly low power of the estimator and small sample sizes analysed. The upper 95% CI was infinite (i.e. could not be estimated) for any of the sites. Terrigal returned an LDNe estimate considerably higher than any other site (7- to 40-fold larger). The wide range of sampling in Tas. may also be lowering the LDNe estimate from this region.
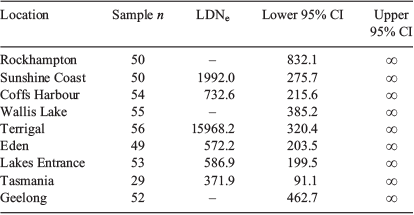
|
Discussion
Testing the null hypothesis of one continuous genetic stock of snapper along Australia’s eastern coast is important to ensure that the most appropriate spatial structure is applied to assessment and management. This study, using microsatellite markers, has confirmed a genetic subdivision of C. auratus populations along the east coast that was previously suggested based on a weak allozyme signal driven by a single locus (Sumpton et al. 2008). Temporal replication of genetic sampling spanning more than a decade has also offered a unique opportunity to investigate the geographic stability of this genetic disjunction.
The present study has revealed a consistent genetic signal, across all methods of statistical analysis (Bayesian modelling, DAPC and FST/DEST statistics) for two distinct genetic stocks. The two genetic stocks transition around Eden in southern NSW. The northern stock spans most of NSW and Qld, and incorporates the region where most of the east coast catches are derived. The southern stock is most distinct in Geelong, but continues through Bass Strait, past Tas., extending up the east coast to Eden, with a small amount of encroachment north to Terrigal. The earlier study by Sumpton et al. (2008) also identified an east coast genetic disjunction but, in that study, it was identified over 400 km further north than in the present study, between Sydney and Forster on the central coast of NSW (Fig. 1). If the genetic disjunction is a consistent feature of this species on the east coast, then the genetic disjunction of Sumpton et al. (2008), based on samples collected in the mid-1990s, is likely the same genetic transition zone identified here, with the shift reflecting a southward movement of the ranges of the two genetic stocks.
The shifting genetic transition zone of the C. auratus stocks may be linked to ocean currents, temperature and possibly salinity. Long-term ocean temperature monitoring shows that the southward penetration of the EAC has increased over the past 60 years, resulting in a poleward advance of warmer and saltier water (Ridgway 2007). Water temperature has been shown to be linked to spawning periods and spawning success in C. auratus (Francis 1993; Lenanton et al. 2009). The southward-shifting EAC has been associated with long-term shifts in the abundance and distribution of other temperate fish species (Last et al. 2011).
The mechanism responsible for creating distinct northern and southern stocks may also be linked to water temperature. Fish living in warmer waters at the northern end of the range spawn during winter, whereas fish living in more temperate waters spawn ~3 months later in spring–summer (Ferrell and Sumpton 1997). As a result, northern fish have an extended growing season and they mature earlier than their southern counterparts (Ferrell and Sumpton 1997; Stewart et al. 2010). Biological parameters for the genetic stock occurring south of Eden are not well characterised; however, there are known spawning aggregations on inshore reefs off eastern Vic. in November, and recruitment of postlarvae and small juveniles occurs in eastern Victorian estuaries in late summer and autumn (Hamer and Jenkins 2004). The structure plot (Fig. 3a) shows that the two stocks overlap and transition around Eden. It is unclear whether the animals in this transition zone are interbreeding to form admixed animals (individual fish exhibit both northern and southern genotypes) or whether they co-occur in this region without interbreeding. There is a suggestion of admixture off Eden, as well as in neighbouring sampling locations, but this may be an artefact of the low number of microsatellite loci and level of missing data (this is the most likely explanation for the apparently admixed animals off the Sunshine Coast). Future studies should focus on screening additional genetic markers and sampling locations at a finer scale between Lakes Entrance and Terrigal to help to resolve this issue. Additional genetic markers may also reveal further genetic signals to differentiate Geelong from Lakes Entrance and Tas.
Standardised fixation measures may be less comparable between studies if mutation rates differ and heterozygosity is high (Leng and Zhang 2011). The total standardised fixation index DEST (0.0232) estimate for the east coast samples was low but significant (P = 0.001). The estimate was almost an order of magnitude higher than the value recorded among Western Australian C. auratus populations from Shark Bay DEST (0.002; Gardner et al. 2017), with the caveat being that only locus Pma1 overlapped between the two studies.
By pooling genetically undifferentiated adjacent collection areas, the spatial boundaries within the two east coast genetic stocks were further investigated using comparisons of pairwise DEST. Following three rounds of pooling, two genetically distinct stocks were identified, the first extending from Rockhampton to Terrigal and the second spanning from Lakes Entrance to Geelong. Prior to pooling, Eden was the only sampling location with no significant DEST values, a result consistent with it having a mixture of both the northern and southern stocks.
A priori expectations based on tagging data were that Geelong would separate from east coast C. auratus stocks (Sanders 1974; Hamer et al. 2011). The microsatellite data do not support this separation. This result is consistent with allozyme results that also failed to differentiate between Victorian sampling locations spanning from Portland in the west to Lake Tyers in the east (Meggs et al. 2003). Although not significant, population pairwise DEST values within the southern stock were greater than those measured between sampling locations in the northern stock. Tasmanian samples were somewhat intermediate genetically between Geelong and Lakes Entrance. Low catch numbers unfortunately resulted in the Tasmanian samples being represented by fish collected from both north-west and eastern waters (see Fig. 1). The Tasmanian sample may have captured more than one genetic stock. With increased sampling, and additional genetic markers, it is possible that genetic structure in southern Australian C. auratus will become apparent.
High levels of connectivity were found among collection locations with a weak IBD signature detected using only the complete dataset. With increasing geographic distance, a linear increase in genetic differences was observed. The slope of the IBD correlation, 7 × 10–6, falls within the interquartile range reported for fish stocks based on a meta-analysis of marine fish (Cooke et al. 2016). The slightly positive slope value indicates that fish are probably not actively recruiting back to natal sites (Cooke et al. 2016), but neither are they moving enormous distances; they are sharing their DNA with neighbouring locations, either by mixed spawning aggregations or larval dispersal. The IBD signature could not be detected when the microsatellite dataset was reduced to the northern stock alone (five sites from Terrigal north) although a weak signal was found by Sumpton et al. (2008) using allozyme analysis. Biological knowledge of the fish would suggest that an IBD pattern likely does exist in the northern stock but has not been detected with the current genetic markers. A similar result was obtained using a microsatellite analyses of New Zealand C. auratus over a 900-km range (Ashton 2013), with stocks in that study found to be largely panmictic with no IBD signal and with low-level genetic differentiation between sites.
This study has demonstrated that suitable genetic markers exist to characterise the population genetic structure of C. auratus stocks in Australian waters. Further research is needed to characterise stocks in southern and western Australian waters. A broad-scale nationwide study is recommended, with a consistent set of variable genetic markers, before fine-scale spatial variation is assessed to assist in the management of local stocks. It would also be valuable to compare Australian C. auratus stocks to populations from New Zealand, which were found to be genetically distinct using allozymes (Meggs et al. 2003). Although the microsatellite loci screened in this study were the same as those used by Le Port et al. (2017), unfortunately different size-scaling ladders were used by the two laboratories for scoring alleles, and consequently the results require cross-validation before they can be combined.
It would be premature to infer population numbers (N) from the LDNe estimates calculated without a better understanding of how the two numbers correlate in C. auratus, but some interesting observations can be made from the results. It is promising that LDNe values were estimated using a dataset limited by low sample numbers, few loci and with a high occurrence of rare alleles (which are currently excluded by the Pcrit). Having infinite upper bounds, the estimates are currently fairly meaningless; however, with more intensive sampling and the addition of more loci, the LDNe estimates could offer a comparative means to assess trends in the abundance of local stocks.
Conclusion
Microsatellite analysis supports a two-stock genetic model for C. auratus along Australia’s east coast. The northern stock extends from Rockhampton to Eden; the northern and southern stocks overlap around Eden and the southern stock then extends south from Eden, past Lakes Entrance and Tas., extending westward to Geelong. The southern stock will likely be found to extend further west. Other studies have suggested that mixing of snapper between stocks to the east and west of Wilsons Promontory in Vic. is limited (Coutin et al. 2003; Hamer et al. 2011). The present study has not detected a significant barrier to gene flow to separate Geelong (Port Phillip Bay) samples from eastern Victorian or Tasmanian samples. The consequence of this finding, in terms of stock assessment and fisheries management along the east coast, is that Qld and NSW fisheries should be assessed together. Preliminary genetic analyses suggest that, with increased sampling, the size of local C. auratus stocks may be able to be independently estimated using DNA tissue samples. This finding could prove valuable for the local management of populations.
Conflicts of interest
Jennifer Ovenden is an Associate Editor for Marine and Freshwater Research. Despite this relationship, she did not at any stage have Associate Editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this Journal. Marine and Freshwater Research encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The remaining authors have no conflicts of interest to declare.
Declaration of funding
This project was supported by the Australian Fisheries Research and Development Corporation (Project number 2015/216).
Acknowledgements
The authors thank Ross Winstanley and Curtis Champion for collecting the fin clips in Victoria and Tasmania. Assistance with the principal component genetic analysis was provided by Marco Kienzle, Valeria Paccapelo and Susan Fletcher. Leanne Clarke (Australian Equine Genetics Research Centre, University of Queensland) provided invaluable capillary electrophoresis support. Thanks also go to Raewyn Street and Agnès Le Port for their preliminary testing of the microsatellite loci and to Robin Waples for his suggestions on interpreting the LDNe estimates. The authors also thank the anonymous reviewers who provided valuable feedback for improving the manuscript; in particular, the authors acknowledge the reviewer who provided the R script for calculating the chance of obtaining low P-values associated with the HWE comparisons.
References
Adcock, G. J., Bernal Ramirez, J. H., Hauser, L., Smith, P. J., and Carvalho, G. R. (2000). Screening of DNA polymorphisms in samples of archived scales from New Zealand snapper. Journal of Fish Biology 56, 1283–1287.| Screening of DNA polymorphisms in samples of archived scales from New Zealand snapper.Crossref | GoogleScholarGoogle Scholar |
Akey, J. M., Zhang, K., Xiong, M., Doris, P., and Jin, L. (2001). The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures. American Journal of Human Genetics 68, 1447–1456.
| The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures.Crossref | GoogleScholarGoogle Scholar |
Ashton, D. T. (2013). Population genetics of New Zealand Pagrus auratus and genetic variation of an aquaculture broodstock. M.Sc. Dissertation, Victoria University of Wellington, New Zealand.
Barnes, T. C., Junge, C., Myers, S. A., Taylor, M. D., Rogers, P. J., Ferguson, G. J., Lieschke, J. A., Donnellan, S. C., and Gillanders, B. M. (2016). Population structure in a wide-ranging coastal teleost (Argyrosomus japonicus, Sciaenidae) reflects marine biogeography across southern Australia. Marine and Freshwater Research 67, 1103–1113.
| Population structure in a wide-ranging coastal teleost (Argyrosomus japonicus, Sciaenidae) reflects marine biogeography across southern Australia.Crossref | GoogleScholarGoogle Scholar |
Broderick, D., Ovenden, J. R., Buckworth, R., Newman, S. J., Lester, R. J. G., and Welch, D. (2011). Genetic population structure of grey mackerel (Scomberomorus semifasciatus) in northern Australia. Journal of Fish Biology 79, 633–661.
| Genetic population structure of grey mackerel (Scomberomorus semifasciatus) in northern Australia.Crossref | GoogleScholarGoogle Scholar |
Brown, R. C., Tsalavouta, M., Terzoglou, V., Magoulas, A., and McAndrew, B. (2005). Additional microsatellites for Sparus aurata and cross‐species amplification within the Sparidae family. Molecular Ecology Notes 5, 605–607.
| Additional microsatellites for Sparus aurata and cross‐species amplification within the Sparidae family.Crossref | GoogleScholarGoogle Scholar |
Cabin, R. J., and Mitchell, R. J. (2000). To Bonferroni or not to Bonferroni: when and how are the questions. Bulletin of the Ecological Society of America 81, 246–248.
Carvalho, G. R., and Pitcher, T. J. (2012). ‘Molecular Genetics in Fisheries.’ (Springer: Dordrecht, Netherlands.)
Chen, S., Liu, Y., Xu, M., and Li, J. (2005). Isolation and characterization of polymorphic microsatellite loci from an EST‐library of red sea bream (Chrysophrys major) and cross‐species amplification. Molecular Ecology Notes 5, 215–217.
| Isolation and characterization of polymorphic microsatellite loci from an EST‐library of red sea bream (Chrysophrys major) and cross‐species amplification.Crossref | GoogleScholarGoogle Scholar |
Cooke, G. M., Schlub, T. E., Sherwin, W. B., and Ord, T. J. (2016). Understanding the spatial scale of genetic connectivity at sea: unique insights from a land fish and a meta-analysis. PLoS One 11, e0150991.
| Understanding the spatial scale of genetic connectivity at sea: unique insights from a land fish and a meta-analysis.Crossref | GoogleScholarGoogle Scholar |
Coscia, I., Chopelet, J., Waples, R. S., Mann, B. Q., and Mariani, S. (2016). Sex change and effective population size: implications for population genetic studies in marine fish. Heredity 117, 251–258.
| Sex change and effective population size: implications for population genetic studies in marine fish.Crossref | GoogleScholarGoogle Scholar |
Coutin, P. C., Cashmore, S., and Sivakumaran, K. P. (2003). Assessment of the snapper fishery in Victoria. Final report to FRDC, Project number 97/128, Marine and Freshwater Resources Institute, Queenscliff, Vic., Australia.
Curley, B. G., Jordan, A. R., Figueira, W. F., and Valenzuela, V. C. (2013). A review of the biology and ecology of key fishes targeted by coastal fisheries in south-east Australia: identifying critical knowledge gaps required to improve spatial management. Reviews in Fish Biology and Fisheries 23, 435–458.
| A review of the biology and ecology of key fishes targeted by coastal fisheries in south-east Australia: identifying critical knowledge gaps required to improve spatial management.Crossref | GoogleScholarGoogle Scholar |
Do, C., Waples, R. S., Peel, D., Macbeth, G., Tillett, B. J., and Ovenden, J. R. (2014). NeEstimator v2: re‐implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14, 209–214.
| NeEstimator v2: re‐implementation of software for the estimation of contemporary effective population size (Ne) from genetic data.Crossref | GoogleScholarGoogle Scholar |
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using the software structure: a simulation study.Crossref | GoogleScholarGoogle Scholar |
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567.
| Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows.Crossref | GoogleScholarGoogle Scholar |
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Ferrell, D., and Sumpton, W. (1997). Assessment of the fishery for snapper (Pagrus auratus) in Queensland and New South Wales. FRDC project report 93/074, Fisheries Research and Development Corporation, Canberra, ACT, Australia.
Finn, M., Flood, M., Stobutzki, I., Maloney, L., Ward, P., Andrews, J., Begg, G., Fletcher, W., Gardner, C., Roelofs, A., Sainsbury, K., Saunders, T., Stewart, J., and Smith, T. (Eds) (2015). Status of key Australian fish stocks 2014. Project number 2014/030, Australian Bureau of Agricultural and Resource Economics and Sciences, Canberra, ACT, Australia.
Fowler, A., Garland, A., Jackson, G., Stewart, J., and Hamer, P. (2016). Snapper, Chrysophrys auratus. In ‘Status of Australian Fish Stocks Reports 2016’. (Eds C. Stewardson, J. Andrews, C. Ashby, M. Haddon, K. Hartmann, P. Hone, P. Horvat, S. Mayfield, A. Roelofs, K. Sainsbury, T. Saunders, J. Stewart, I. Stobutzki, and B. Wise.) (Fisheries Research and Development Corporation: Canberra, ACT, Australia.) Available at http://www.fish.gov.au/report/60-Snapper-2016 [Verified 16 November 2018].
Fowler, A. J., Huveneers, C., and Lloyd, M. T. (2017). Insights into movement behaviour of snapper (Chrysophrys auratus, Sparidae) from a large acoustic array. Marine and Freshwater Research 68, 1438–1453.
| Insights into movement behaviour of snapper (Chrysophrys auratus, Sparidae) from a large acoustic array.Crossref | GoogleScholarGoogle Scholar |
Francis, M. P. (1993). Does water temperature determine year class strength in New Zealand snapper (Pagrus auratus, Sparidae)? Fisheries Oceanography 2, 65–72.
| Does water temperature determine year class strength in New Zealand snapper (Pagrus auratus, Sparidae)?Crossref | GoogleScholarGoogle Scholar |
Francis, M., and Pankhurst, N. (1988). Juvenile sex inversion in the New Zealand snapper Chrysophrys auratus (Bloch and Schneider, 1801) (Sparidae). Marine and Freshwater Research 39, 625–631.
| Juvenile sex inversion in the New Zealand snapper Chrysophrys auratus (Bloch and Schneider, 1801) (Sparidae).Crossref | GoogleScholarGoogle Scholar |
Gardner, M. J., Chaplin, J. A., Potter, I. C., and Fairclough, D. V. (2015). Pelagic early life stages promote connectivity in the demersal labrid Choerodon rubescens. Journal of Experimental Marine Biology and Ecology 472, 142–150.
| Pelagic early life stages promote connectivity in the demersal labrid Choerodon rubescens.Crossref | GoogleScholarGoogle Scholar |
Gardner, M. J., Chaplin, J. A., Potter, I., Fairclough, D. V., and Jackson, G. (2017). The genetic structure of a marine teleost, Chrysophrys auratus, in a large, heterogeneous marine embayment. Environmental Biology of Fishes 100, 1411–1425.
| The genetic structure of a marine teleost, Chrysophrys auratus, in a large, heterogeneous marine embayment.Crossref | GoogleScholarGoogle Scholar |
Gauldie, R. W. (1988). Tagging and genetically isolated stocks of fish: a test of one stock hypothesis and the development of another. Journal of Applied Ichthyology 4, 168–173.
| Tagging and genetically isolated stocks of fish: a test of one stock hypothesis and the development of another.Crossref | GoogleScholarGoogle Scholar |
Gillanders, B. M. (2002). Connectivity between juvenile and adult fish populations: do adults remain near their recruitment estuaries? Marine Ecology Progress Series 240, 215–223.
| Connectivity between juvenile and adult fish populations: do adults remain near their recruitment estuaries?Crossref | GoogleScholarGoogle Scholar |
Gordon, D., Matise, T. C., Heath, S. C., and Ott, J. (1999). Power loss for multiallelic transmission/disequilibrium test when errors introduced: GAW11 simulated data. Genetic Epidemiology 17, S587–S592.
| Power loss for multiallelic transmission/disequilibrium test when errors introduced: GAW11 simulated data.Crossref | GoogleScholarGoogle Scholar |
Hamer, P. A., and Jenkins, G. P. (2004). High levels of spatial and temporal recruitment variation in the temperate sparid Pagrus auratus. Marine and Freshwater Research 55, 663–673.
| High levels of spatial and temporal recruitment variation in the temperate sparid Pagrus auratus.Crossref | GoogleScholarGoogle Scholar |
Hamer, P. A., Acevedo, S., Jenkins, G. P., and Newman, A. (2011). Connectivity of a large embayment and coastal fishery: spawning aggregations in one bay source local and broad-scale fishery replenishment. Journal of Fish Biology 78, 1090–1109.
| Connectivity of a large embayment and coastal fishery: spawning aggregations in one bay source local and broad-scale fishery replenishment.Crossref | GoogleScholarGoogle Scholar |
Hare, M. P., Nunney, L., Schwartz, M. K., Ruzzante, D. E., Burford, M., Waples, R. S., Ruegg, K., and Palstra, F. (2011). Understanding and estimating effective population size for practical application in marine species management. Conservation Biology 25, 438–449.
| Understanding and estimating effective population size for practical application in marine species management.Crossref | GoogleScholarGoogle Scholar |
Harrison, H. B., Williamson, D. H., Evans, R. D., Almany, G. R., Thorrold, S. R., Russ, G. R., Feldheim, K. A., Van Herwerden, L., Planes, S., and Srinivasan, M. (2012). Larval export from marine reserves and the recruitment benefit for fish and fisheries. Current Biology 22, 1023–1028.
| Larval export from marine reserves and the recruitment benefit for fish and fisheries.Crossref | GoogleScholarGoogle Scholar |
Hatanaka, A., Yamada, S. I., Sakamoto, T., and Mitsuboshi, T. (2006). Isolation and application of microsatellite DNA markers for pedigree tracing of seedlings of red sea bream (Pagrus major). Journal of the World Aquaculture Society 37, 139–143.
| Isolation and application of microsatellite DNA markers for pedigree tracing of seedlings of red sea bream (Pagrus major).Crossref | GoogleScholarGoogle Scholar |
Hauser, L., and Carvalho, G. R. (2008). Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish and Fisheries 9, 333–362.
| Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts.Crossref | GoogleScholarGoogle Scholar |
Hauser, L., Adcock, G. J., Smith, P. J., Bernal Ramírez, J. H., and Carvalho, G. R. (2002). Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proceedings of the National Academy of Sciences of the United States of America 99, 11742–11747.
| Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus).Crossref | GoogleScholarGoogle Scholar |
Hedgecock, D. (1994). Does variance in reproductive success limit effective population sizes of marine organisms. In ‘Genetics and Evolution of Aquatic Organisms’. (Ed. A. Beaumont.) pp. 122–134. (Chapman & Hall: London, UK.)
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405.
| adegenet: a R package for the multivariate analysis of genetic markers.Crossref | GoogleScholarGoogle Scholar |
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11, 94.
| Discriminant analysis of principal components: a new method for the analysis of genetically structured populations.Crossref | GoogleScholarGoogle Scholar |
Jones, A., Ovenden, J., and Wang, Y. (2016). Improved confidence intervals for the linkage disequilibrium method for estimating effective population size. Heredity 117, 217–223.
| Improved confidence intervals for the linkage disequilibrium method for estimating effective population size.Crossref | GoogleScholarGoogle Scholar |
Jost, L. (2008). GST and its relatives do not measure differentiation. Molecular Ecology 17, 4015–4026.
| GST and its relatives do not measure differentiation.Crossref | GoogleScholarGoogle Scholar |
Kailola, P. J., Williams, M. J., Stewart, P. C., Reichelt, R. E., McNee, A., and Grieve, C. (1993). ‘Australian Fisheries Resources.’ (Bureau of Resources Sciences and Fisheries Research and Development Corporation: Canberra, ACT. Australia.)
Kalinowski, S., Taper, M., and Marshall, T. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16, 1099–1106.
| Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment.Crossref | GoogleScholarGoogle Scholar |
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., and Drummond, A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Kemp, J., Conron, S., Hamer, P., Bruce, T., Bridge, N., and Brown, L. (2012) Victorian snapper stock assessment 2011. Fisheries Victoria Assessment Report Series 64. The State of Victoria, Department of Primary Industries, Queenscliff, Vic., Australia.
Kierepka, E., and Latch, E. (2015). Performance of partial statistics in individual‐based landscape genetics. Molecular Ecology Resources 15, 512–525.
| Performance of partial statistics in individual‐based landscape genetics.Crossref | GoogleScholarGoogle Scholar |
Kirchoff, V. S., Peacock, M. M., and Teglas, M. B. (2008). Permanent genetic resources: identification and characterization of 14 polymorphic microsatellite loci in the argasid tick Ornithodoros coriaceus. Molecular Ecology Resources 8, 446–448.
| Permanent genetic resources: identification and characterization of 14 polymorphic microsatellite loci in the argasid tick Ornithodoros coriaceus.Crossref | GoogleScholarGoogle Scholar |
Last, P. R., White, W. T., Gledhill, D. C., Hobday, A. J., Brown, R., Edgar, G. J., and Pecl, G. (2011). Long‐term shifts in abundance and distribution of a temperate fish fauna: a response to climate change and fishing practices. Global Ecology and Biogeography 20, 58–72.
| Long‐term shifts in abundance and distribution of a temperate fish fauna: a response to climate change and fishing practices.Crossref | GoogleScholarGoogle Scholar |
Le Port, A., Montgomery, J., Smith, A., Croucher, A., McLeod, I., and Lavery, S. (2017). Temperate marine protected area provides recruitment subsidies to local fisheries. Proceedings of the Royal Society of London – B. Biological Sciences 284, 20171300.
| Temperate marine protected area provides recruitment subsidies to local fisheries.Crossref | GoogleScholarGoogle Scholar |
Legendre, P., and Anderson, M. J. (1999). Distance‐based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs 69, 1–24.
| Distance‐based redundancy analysis: testing multispecies responses in multifactorial ecological experiments.Crossref | GoogleScholarGoogle Scholar |
Lenanton, R., St John, J., Keay, I., Wakefield, C., Jackson, G., Wise, B., and Gaughan, D. (2009). Spatial scales of exploitation among populations of demersal scalefish: implications for management. Part 2: stock structure and biology of two indicator species, West Australian dhufish (Glaucosoma hebraicum) and pink snapper (Pagrus auratus), in the West Coast Bioregion. Final Fisheries Research and Development Corporation Report, Project number 2003/052, Fisheries Research Report number 174. Department of Fisheries, Perth, WA, Australia.
Leng, L., and Zhang, D. X. (2011). Measuring population differentiation using GST or D? A simulation study with microsatellite DNA markers under a finite island model and nonequilibrium conditions. Molecular Ecology 20, 2494–2509.
| Measuring population differentiation using GST or D? A simulation study with microsatellite DNA markers under a finite island model and nonequilibrium conditions.Crossref | GoogleScholarGoogle Scholar |
Luikart, G., Ryman, N., Tallmon, D. A., Schwartz, M. K., and Allendorf, F. W. (2010). Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conservation Genetics 11, 355–373.
| Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches.Crossref | GoogleScholarGoogle Scholar |
MacDonald, C. M. (1980). Population structure, biochemical adaptation and systematics in temperate marine fishes of the genera Arripis and Chrysophrys (Pisces: Perciformes). Ph.D. thesis, Australian National University, Canberra, ACT, Australia.
MacDonald, C. (1982). Life history characteristics of snapper Chrysophrys auratus (Bloch and Scheider, 1801) in Australian waters. Ministry for Conservation, Fisheries and Wildlife Division, Melbourne, Vic., Australia.
Mantel, N. (1967). The detection of disease clustering and a generalised regression approach. Cancer Research 27, 209–220.
Meggs, L. B., Austin, C. M., and Coutin, P. C. (2003). Low allozyme variation in snapper, Pagrus auratus, in Victoria, Australia. Fisheries Management and Ecology 10, 155–162.
| Low allozyme variation in snapper, Pagrus auratus, in Victoria, Australia.Crossref | GoogleScholarGoogle Scholar |
Missiaggia, A., and Grattapaglia, D. (2006). Plant microsatellite genotyping with 4-color fluorescent detection using multiple-tailed primers. Genetics and Molecular Research 5, 72–78.
Moran, M., Burton, C., and Jenke, J. (2003). Long-term movement patterns of continental shelf and inner gulf snapper (Pagrus auratus, Sparidae) from tagging in the Shark Bay region of Western Australia. Marine and Freshwater Research 54, 913–922.
| Long-term movement patterns of continental shelf and inner gulf snapper (Pagrus auratus, Sparidae) from tagging in the Shark Bay region of Western Australia.Crossref | GoogleScholarGoogle Scholar |
Norriss, J. V., and Crisafulli, B. (2010). Longevity in Australian snapper Pagrus auratus (Sparidae). Journal of the Royal Society of Western Australia 93, 129–132.
Oetting, W. S., Lee, H., Flanders, D., Wiesner, G., Sellers, T., and King, R. (1995). Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30, 450–458.
| Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers.Crossref | GoogleScholarGoogle Scholar |
Ovenden, J. R. (1990). Mitochondrial DNA and marine stock assessment: a review. Australian Journal of Marine and Freshwater Research 41, 835–853.
| Mitochondrial DNA and marine stock assessment: a review.Crossref | GoogleScholarGoogle Scholar |
Ovenden, J. R., Salini, J., O’Connor, S., and Street, R. (2004). Pronounced genetic population structure in a potentially vagile fish species (Pristipomoides multidens, Teleostei; Perciformes; Lutjanidae) from the East Indies triangle. Molecular Ecology 13, 1991–1999.
| Pronounced genetic population structure in a potentially vagile fish species (Pristipomoides multidens, Teleostei; Perciformes; Lutjanidae) from the East Indies triangle.Crossref | GoogleScholarGoogle Scholar |
Ovenden, J. R., Peel, D., Street, R., Courtney, A. J., Hoyle, S. D., Peel, S. L., and Podlich, H. (2007). The genetic effective and adult census size of an Australian population of tiger prawns (Penaeus esculentus). Molecular Ecology 16, 127–138.
| The genetic effective and adult census size of an Australian population of tiger prawns (Penaeus esculentus).Crossref | GoogleScholarGoogle Scholar |
Ovenden, J., Leigh, G., Blower, D., Jones, A., Moore, A., Bustamante, C., Buckworth, R., Bennett, M., and Dudgeon, C. (2016). Can estimates of genetic effective population size contribute to fisheries stock assessments? Journal of Fish Biology 89, 2505–2518.
| Can estimates of genetic effective population size contribute to fisheries stock assessments?Crossref | GoogleScholarGoogle Scholar |
Parsons, D., Babcock, R., Hankin, R., Willis, T. J., Aitken, J., O’Dor, R., and Jackson, G. (2003). Snapper Pagrus auratus (Sparidae) home range dynamics: acoustic tagging studies in a marine reserve. Marine Ecology Progress Series 262, 253–265.
| Snapper Pagrus auratus (Sparidae) home range dynamics: acoustic tagging studies in a marine reserve.Crossref | GoogleScholarGoogle Scholar |
Peakall, R., and Smouse, P. E. (2006). GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6, 288–295.
| GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research.Crossref | GoogleScholarGoogle Scholar |
Pecl, G., Ward, T., Briceno, F., Fowler, A., Frusher, S., Gardner, C., Hamer, P., Hartmann, K., Hartog, J., Hobday, A., Hoshino, E., Jennings, S., Le Bouhellec, B., Linnane, A., Marzloff, M., Mayfield, S., Mundy, C., Ogier, E., Sullivan, A., Tracey, S., Tuck, G., and Wayte, S. (2014). Preparing fisheries for climate change: identifying adaptation options for four key fisheries in south eastern Australia. Final Report to the Fisheries Research and Development Corporation, Project 2011/039, Canberra, ACT, Australia.
Piry, S., Alapetite, A., Cornuet, J. M., Paetkau, D., Baudouin, L., and Estoup, A. (2004). GENECLASS2: a software for genetic assignment and first-generation migrant detection. The Journal of Heredity 95, 536–539.
| GENECLASS2: a software for genetic assignment and first-generation migrant detection.Crossref | GoogleScholarGoogle Scholar |
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rannala, B., and Mountain, J. L. (1997). Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences of the United States of America 94, 9197–9201.
| Detecting immigration by using multilocus genotypes.Crossref | GoogleScholarGoogle Scholar |
Raymond, M., and Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. The Journal of Heredity 86, 248–249.
| GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism.Crossref | GoogleScholarGoogle Scholar |
Rice, W. R. (1989). Analysing tables of statistical tests. Evolution 43, 223–225.
| Analysing tables of statistical tests.Crossref | GoogleScholarGoogle Scholar |
Ridgway, K. (2007). Long‐term trend and decadal variability of the southward penetration of the East Australian Current. Geophysical Research Letters 34, L13613.
| Long‐term trend and decadal variability of the southward penetration of the East Australian Current.Crossref | GoogleScholarGoogle Scholar |
Rousset, F. (2008). Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8, 103–106.
| Genepop’007: a complete re-implementation of the genepop software for Windows and Linux.Crossref | GoogleScholarGoogle Scholar |
Ryman, N., and Palm, S. (2006). POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Molecular Ecology Notes 6, 600–602.
| POWSIM: a computer program for assessing statistical power when testing for genetic differentiation.Crossref | GoogleScholarGoogle Scholar |
Sanders, M. (1974). Tagging indicates at least two stocks of snapper Chrysophrys auratus in south‐east australian waters. New Zealand Journal of Marine and Freshwater Research 8, 371–374.
| Tagging indicates at least two stocks of snapper Chrysophrys auratus in south‐east australian waters.Crossref | GoogleScholarGoogle Scholar |
Saunders, R. J., Fowler, A. J., and Gillanders, B. M. (2012). The spawning dynamics of snapper (Chrysophrys auratus) in northern Spencer Gulf, South Australia. New Zealand Journal of Marine and Freshwater Research 46, 491–510.
| The spawning dynamics of snapper (Chrysophrys auratus) in northern Spencer Gulf, South Australia.Crossref | GoogleScholarGoogle Scholar |
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology 18, 233–234.
| An economic method for the fluorescent labeling of PCR fragments.Crossref | GoogleScholarGoogle Scholar |
Stewart, J., Rowling, K., Hegarty, A.-M., and Nuttall, A. (2010). Size and age at sexual maturity of snapper Pagrus auratus in New South Wales 2008/09. Fisheries Research Report Series 27, Industry & Investment NSW, Sydney, NSW, Australia.
Sumpton, W. D., Sawynok, B., and Carstens, N. (2003). Localised movement of snapper (Pagrus auratus, Sparidae) in a large subtropical marine embayment. Marine and Freshwater Research 54, 923–930.
| Localised movement of snapper (Pagrus auratus, Sparidae) in a large subtropical marine embayment.Crossref | GoogleScholarGoogle Scholar |
Sumpton, W. D., Ovenden, J. R., Keenan, C. P., and Street, R. (2008). Evidence for a stock discontinuity of snapper (Pagrus auratus) on the East coast of Australia. Fisheries Research 94, 92–98.
| Evidence for a stock discontinuity of snapper (Pagrus auratus) on the East coast of Australia.Crossref | GoogleScholarGoogle Scholar |
Takagi, M., Taniguchi, N., Cook, D., and Doyle, R. W. (1997). Isolation and characterization of microsatellite loci from red sea bream Pagrus major and detection in closely related species. Fisheries Science 63, 199–204.
| Isolation and characterization of microsatellite loci from red sea bream Pagrus major and detection in closely related species.Crossref | GoogleScholarGoogle Scholar |
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICROCHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4, 535–538.
| MICROCHECKER: software for identifying and correcting genotyping errors in microsatellite data.Crossref | GoogleScholarGoogle Scholar |
Waples, R. S. (2016). Tiny estimates of the Ne/N ratio in marine fishes: are they real? Journal of Fish Biology 89, 2479–2504.
| Tiny estimates of the Ne/N ratio in marine fishes: are they real?Crossref | GoogleScholarGoogle Scholar |
Waples, R. S., and Do, C. (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Molecular Ecology Resources 8, 753–756.
| LDNE: a program for estimating effective population size from data on linkage disequilibrium.Crossref | GoogleScholarGoogle Scholar |
Waples, R. S., and Do, C. (2010). Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evolutionary Applications 3, 244–262.
| Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution.Crossref | GoogleScholarGoogle Scholar |
Waples, R. S., Do, C., and Chopelet, J. (2011). Calculating Ne and Ne/N in age-structured populations: a hybrid Felsenstein–Hill approach. Ecology 92, 1513–1522.
| Calculating Ne and Ne/N in age-structured populations: a hybrid Felsenstein–Hill approach.Crossref | GoogleScholarGoogle Scholar |
A This paper is copyright of The State of Queensland (through the Department Agriculture and Fisheries).




