Investigation of a novel porcine bacterium by whole genome sequencing and mouse inoculation
A. E. Watt A B , M. S. Marenda A and G. F. Browning AA Asia-Pacific Centre for Animal Health, Melbourne Veterinary School, The University of Melbourne, Parkville, VIC 3010.
B Corresponding author. Email: watta@unimelb.edu.au
Animal Production Science 57(12) 2494-2494 https://doi.org/10.1071/ANv57n12Ab077
Published: 20 November 2017
A bacterial isolate collected from a lung lesion of a pig at slaughter was identified as a member of the family Pasteurellaceae using standard biochemical testing. Antibiotic sensitivity was tested using the CDS method and the isolate was found to be susceptible to ampicillin, cephalexin and enrofloxcain and resistant to tetracycline. Further classification was inconclusive, and the isolate failed to group within any existing genus or species. The lung lesions were typical of those seen after infection with Actinobacillus pleuropneumoniae, but preliminary 16S rRNA gene sequencing indicated that the closest relative was Haemophilus parasuis. The work described here aimed to further investigate this novel bacterium by biochemical testing, full genome sequence analysis and assessment of its pathogenicity in a mouse model of infection.
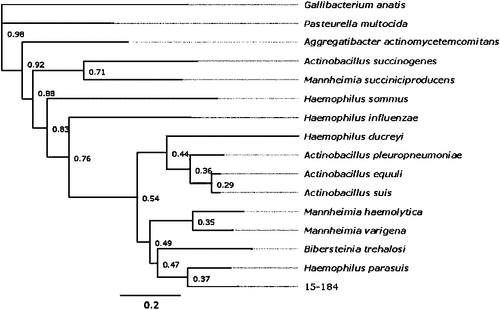
Full genome analysis was completed using Illumina (Illumina Inc., San Diego, CA, USA) and Oxford (Oxford Nanopore Technologies, Oxford, UK) nanopore sequencing, with comparisons of conserved genes suggesting that the novel bacterium was most closely related to H. parasuis. Concatenation and phylogenetic assessment of the housekeeping genes, atpD, infB and rpoB showed that the organism lay in a distinct monophyletic position within the Pasteurellaceae phylogenic tree (Fig. 1). Assessment of the pathogenicity of the organism was performed by inoculation of 6 week old BALB/c mice with the novel bacterium at different concentrations via the intranasal and intraperitoneal routes. The lesions caused after infection were compared to those seen in a positive control group infected with A. pleuropneumoniae. The lungs, liver and spleen were assessed for the severity and type of lesions using histopathological examination. Intranasal inoculation resulted in interstitial pneumonia and bronchitis. The findings suggested that the lesions caused by this yet unclassified member of the Pasteurellaceae differed significantly from those described in previously published studies in which mice were inoculated with G. parasuis (De la Fuente et al. 2007).
|
The results suggested that further research is needed to assess the prevalence of this bacterium in pig populations, as well as to examine its pathogenicity for its natural host, as mouse models only provide an indication of the potential of the organism to cause acute disease. Its potential to cause chronic disease will need to be assessed to fully understand the type and severity of impact this bacterium may have on commercial pig farms.
References
De la Fuente A, Martin C, Martinez S, Frandoloso R, Ferri E (2007) The Open Veterinary Science Journal 1, 11–13.| Crossref | GoogleScholarGoogle Scholar |
Supported by Pork CRC Limited Australia.


